Chemical and Pharmacological Potential of Coccoloba cowellii, an Endemic Endangered Plant from Cuba
Abstract
1. Introduction
2. Results
2.1. UHPLC-HRMS Analysis
2.2. Antibacterial and Cytotoxic Activity
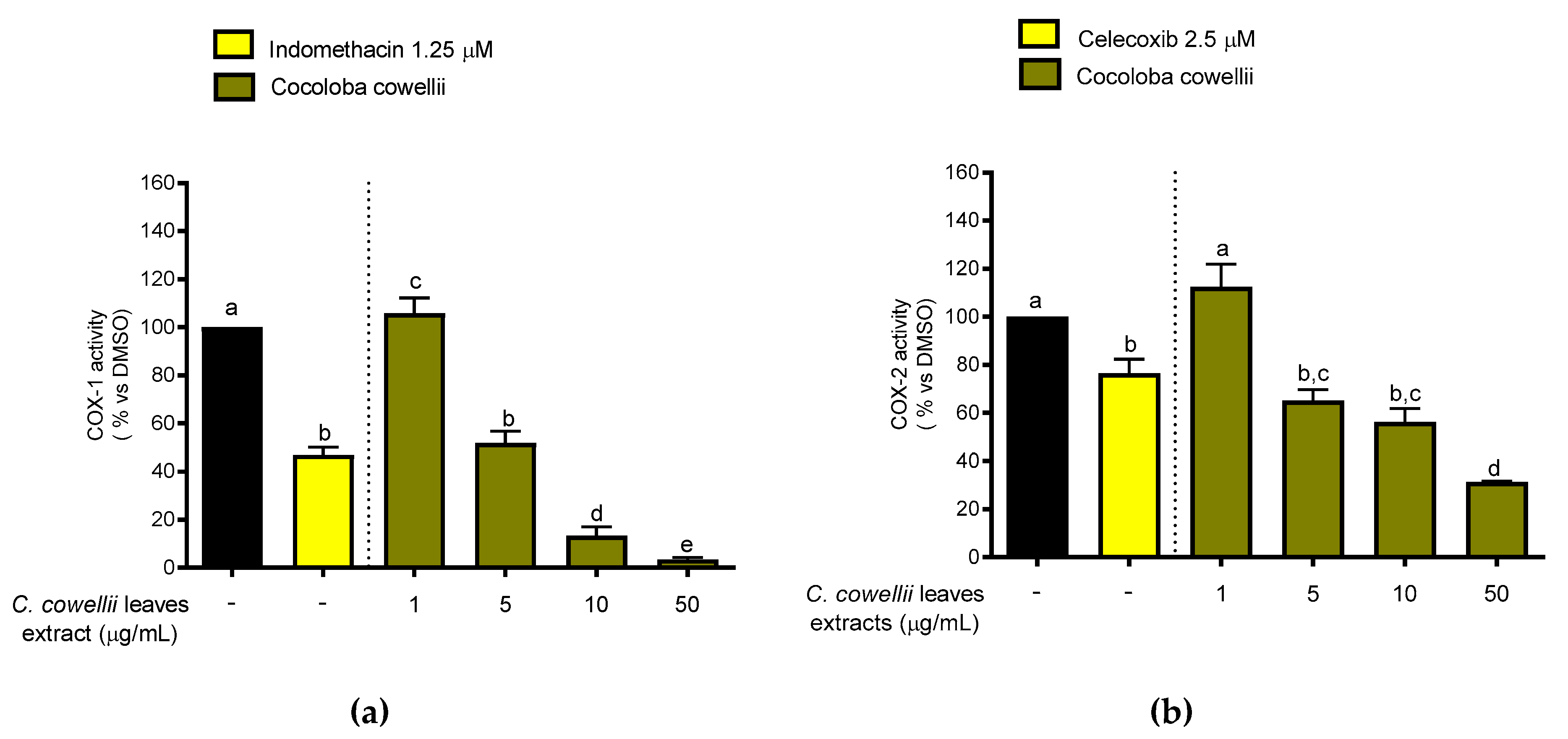
2.3. Inhibition of COX-1 and COX-2 Enzymatic Activity
3. Materials and Methods
3.1. Chemicals and Plant Material
3.2. Leaf Extraction
3.3. UHPLC-HRMS Analysis
Data Processing
3.4. Antimicrobial Assay
3.4.1. Microorganisms and Dilutions
3.4.2. Antibacterial and Antifungal Assay
3.4.3. Antitrypanosomal Assay
3.5. Cytotoxicity Assay
3.6. COX-1 and COX-2 Enzymatic Inhibition Assay
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Publisher’s Note
References
- FAO. The State of the World’s Biodiversity for Food and Agriculture. Available online: http://www.fao.org/3/CA3129EN/CA3129EN.pdf (accessed on 28 October 2020).
- Coelho, N.; Gonçalves, S.; Romano, A. Endemic plant species conservation: Biotechnological approaches. Plants 2020, 9, 345. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.J.; Fernández-Palacios, J.M. Island Biogeography. Ecology, Evolution, and Conservation, 2nd ed.; Oxford University Press: Oxford, UK, 2007; ISBN 9780444641304. [Google Scholar]
- Maury, G.L.; Rodríguez, D.M.; Hendrix, S.; Arranz, J.C.E.; Boix, Y.F.; Pacheco, A.O.; Díaz, J.G.; Morris-Quevedo, H.J.; Dubois, A.F.; Aleman, E.I.; et al. Antioxidants in plants: A valorization potential emphasizing the need for the conservation of plant biodiversity in Cuba. Antioxidants 2020, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Huxley, A.J. The New RHS Dictionary of Gardening; McMillan Press: London, UK, 1999; ISBN 0333770188. [Google Scholar]
- Halberstein, R.A.; Saunders, A.B. Traditional medical practices and medicinal plant usage on a Bahamian island. Cult. Med. Psychiatry 1978, 2, 177–203. [Google Scholar] [CrossRef]
- Coe, F.G. Ethnomedicine of the Rama of southeastern Nicaragua. J. Ethnobiol. 2008, 28, 1–38. [Google Scholar] [CrossRef]
- Mors, W.B.; Rizzini, C.T.; Pereira, N.A.; Defilipps, R.A. Medicinal Plants of Brazil; Reference Publications: Algonac, MI, USA, 2000; ISBN 0917256425. [Google Scholar]
- El-Kawe, B.M.A. A Pharmacognostical Study of Coccoloba peltata Schott Family Polygonaceae. Master’s Thesis, Cairo University, Cairo, Egypt, 2019. [Google Scholar]
- Kawasaki, M.; Kanomata, T.; Yoshitama, K. Flavonoids in the leaves of twenty-eight polygonaceous plants. Bot. Mag. Tokyo 1986, 99, 63–74. [Google Scholar] [CrossRef]
- Li, X.-C.; ElSohIy, H.N.; Nimrod, A.C.; Clark, A.M. Antifungal activity of (−)-epigallocatechin gallate from Coccoloba dugandiana. Planta 1999, 65, 212–217. [Google Scholar] [CrossRef]
- Brandi, M.L. Flavonoids: Biochemical effects and therapeutic applications. Bone Miner. 1992, 19, S3–S14. [Google Scholar] [CrossRef]
- Barros Cota, B.; Braga De Oliveira, A.; Dias De Souza-Filho, J.; Castro Braga, F. Antimicrobial activity and constituents of Coccoloba acrostichoides. Fitoterapia 2003, 74, 729–731. [Google Scholar] [CrossRef]
- Dan, S.; Dan, S.S. Phytochemical study of Adansonia digitata, Coccoloba excoriata, Psychotria adenophylla and Schleichera oleosa. Fitoterapia 1986, 57, 445–446. [Google Scholar]
- Malathi, S.; Masilamani, P.; Balasubramanian, V.; Rao, R.B.; Brindha, P. Constituents of Coccoloba uvifera leaves. Fitoterapia 1995, 66, 277. [Google Scholar]
- de Barros, I.B.; de Souza Daniel, J.F.; Pinto, J.P.; Rezende, M.I.; Filho, R.B.; Ferreira, D.T. Phytochemical and antifungal activity of anthraquinones and root and leaf extracts of coccoloba mollis on phytopathogens. Braz. Arch. Biol. Technol. 2011, 54, 535–541. [Google Scholar] [CrossRef][Green Version]
- Shaw, P.E.; Moshonas, M.G.; Baldwin, E.A. Volatile constituents of Coccolobo uvifera. Phytochemistry 1992, 31, 3495–3497. [Google Scholar] [CrossRef]
- Moreno Morales, S.; Crescente Vallejo, O.; Henríquez Guzmán, W.; Liendo Polanco, G.; Herrera Mata, H. Three constituents with biological activity from Coccoloba uvifera seeds. Ciencia 2008, 16, 84–89. [Google Scholar]
- Corbett, Y.; Herrera, L.; Gonzalez, J.; Cubilla, L.; Capson, T.L.; Coley, P.D.; Kursar, T.A.; Romero, L.I.; Ortega-Barria, E. A novel DNA-based microfluorimetric method to evaluate antimalarial drug activity. Am. J. Trop. Med. Hyg. 2004, 70, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Noa, I.C. Coccoloba howardii (Polygonaceae), a new species from Cuba. Willdenowia 2012, 42, 95–98. [Google Scholar] [CrossRef]
- Roig y Mesa, J.T. Plantas Medicinales, Aromáticas o Venenosas de Cuba (Tomo II), 2nd ed.; Editorial Científico-Técnica: La Habana, Cuba, 2012; ISBN 978-959-05-0814-1. [Google Scholar]
- González-Torres, L.; Palmarola, A.; González-Oliva, L.; Bécquer, E.; Testé, E.; Barrios, D. Lista Roja de la Flora de Cuba. Bissea 2016, 10, 352. [Google Scholar] [CrossRef]
- Méndez Rodríguez, D.; Molina Pérez, E.; Spengler Salabarria, I.; Escalona-Arranz, J.C.; Cos, P. Chemical composition and antioxidant activity of Coccoloba cowellii Britton Lic. Rev. Cuba. Química 2019, 32, 185–198. [Google Scholar]
- De Oliveira, G.G.; Carnevale Neto, F.; Demarque, D.P.; De Sousa Pereira-Junior, J.A.; Sampaio Peixoto Filho, R.C.; De Melo, S.J.; Da Silva Almeida, J.R.G.; Lopes, J.L.C.; Lopes, N.P. Dereplication of flavonoid glycoconjugates from Adenocalymma imperatoris-maximilianii by untargeted tandem mass spectrometry-based molecular networking. Planta Med. 2017, 83, 636–646. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Vukics, V.; Guttman, A. structural characterization of flavonoid glycosides by multi-stage mass spectrometry. Wiley Intersci. 2008, 221–235. [Google Scholar] [CrossRef]
- Ablajan, K.; Abliz, Z.; Shang, X.Y.; He, J.M.; Zhang, R.P.; Shi, J.G. Structural characterization of flavonol 3,7-di-O-glycosides and determination of the glycosylation position by using negative ion electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2006, 41, 352–360. [Google Scholar] [CrossRef]
- Ma, Y.L.; Li, Q.M.; Van Den Heuvel, H.; Claeys, M. Characterization of flavone and flavonol aglycones by collision-induced dissociation tandem mass spectrometry. Rapid Commun. Mass Spectrom. 1997, 11, 1357–1364. [Google Scholar] [CrossRef]
- Callemien, D.; Collin, S. Use of RP-HPLC-ESI(−)-MS/MS to differentiate various proanthocyanidin isomers in lager beer extracts. J. Am. Soc. Brew. Chem. 2008, 66, 109–115. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, A.L.D.; Kaplum, V.; Rossi, D.C.P.; da Silva, L.B.R.; Melhem, M.d.S.C.; Taborda, C.P.; de Mello, J.C.P.; Nakamura, C.V.; Ishida, K. Proanthocyanidin polymeric tannins from Stryphnodendron adstringens are effective against Candida spp. isolates and for vaginal candidiasis treatment. J. Ethnopharmacol. 2018, 216, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Cota, B.B.; De Oliveira, A.B.; Ventura, C.P.; Mendonça, M.P.; Braga, F.C. Antimicrobial activity of plant species from a Brazilian hotspot for conservation priority. Pharm. Biol. 2002, 40, 542–547. [Google Scholar] [CrossRef]
- Facey, P.C.; Pascoe, K.O.; Porter, R.B.; Jones, A.D. Investigation of plants used in Jamaican folk medicine for anti-bacterial activity. J. Pharm. Pharmacol. 1999, 51, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Jacob, M.; Walker, L.; Tekwani, B. Screening North American plant extracts in vitro against Trypanosoma brucei for discovery of new antitrypanosomal drug leads. BMC Complement. Altern. Med. 2016, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Amisigo, C.M.; Antwi, C.A.; Adjimani, J.P.; Gwira, T.M. In vitro anti-trypanosomal effects of selected phenolic acids on Trypanosoma brucei. PLoS ONE 2019, 14, e0216078. [Google Scholar] [CrossRef] [PubMed]
- Sosa, A.M.; Moya Álvarez, A.; Bracamonte, E.; Korenaga, M.; Marco, J.D.; Barroso, P.A. Efficacy of topical treatment with (−)-epigallocatechin gallate, a green tea catechin, in mice with cutaneous leishmaniasis. Molecules 2020, 25, 1741. [Google Scholar] [CrossRef] [PubMed]
- Paveto, C.; Güida, M.C.; Esteva, M.I.; Martino, V.; Coussio, J.; Flawiá, M.M.; Torres, H.N. Anti-Trypanosoma cruzi Activity of Green Tea (Camellia sinensis) Catechins. Antimicrob. Agents Chemother. 2004, 48, 69–74. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Cyclooxygenases: Structural and functional insights. J. Lipid Res. 2009, 50, S29–S34. [Google Scholar] [CrossRef]
- Fiebich, B.L.; Grozdeva, M.; Hess, S.; Hüll, M.; Danesch, U.; Bodensieck, A.; Bauer, R. Petasites hybridus extracts in vitro inhibit COX-2 and PGE2 release by direct interaction with the enzyme and by preventing p42/44 MAP kinase activation in rat primary microglial cells. Planta Med. 2005, 71, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, F.; Sun, Y.; Li, B.; Oi, N.; Chen, H.; Lubet, R.A.; Bode, A.M.; Dong, Z. Select dietary phytochemicals function as inhibitors of COX-1 but not COX-2. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Waffo-Téguo, P.; Hawthorne, M.E.; Cuendet, M.; Mérillon, J.M.; Kinghorn, A.D.; Pezzuto, J.M.; Mehta, R.G. Potential cancer-chemopreventive activities of wine stilbenoids and flavans extracted from grape (Vitis vinifera) cell cultures. Nutr. Cancer 2001, 40, 173–179. [Google Scholar] [CrossRef]
- Wallace, J.L.; Vong, L. NSAID-induced gastrointestinal damage and the design of GI-sparing NSAIDs Selective COX-2 inhibitors: A promise unfulfilled new approaches: Terminal PG synthase inhibitors. Curr. Opin. Investig. Drugs 2008, 9, 1151–1156. [Google Scholar]
- Wallace, J.L.; Ferraz, J.G.P. New pharmacologic therapies in gastrointestinal disease. Gastroenterol. Clin. N. Am. 2010, 39, 709–720. [Google Scholar] [CrossRef]
- Solomon, S.D.; McMurray, J.J.V.; Pfeffer, M.A.; Wittes, J.; Fowler, R.; Finn, P.; Anderson, W.F.; Zauber, A.; Hawk, E.; Bertagnolli, M. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N. Engl. J. Med. 2005, 352, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Bresalier, R.S.; Sandler, R.S.; Quan, H.; Bolognese, J.A.; Oxenius, B.; Horgan, K.; Lines, C.; Riddell, R.; Morton, D.; Lanas, A.; et al. Cardiovascular events associated with rofecoxib in a colorectal adenoma chemoprevention trial. N. Engl. J. Med. 2005, 352, 1092–1102. [Google Scholar] [CrossRef]
- Bijttebier, S.; Van Der Auwera, A.; Voorspoels, S.; Noten, B.; Hermans, N.; Pieters, L. A first step in the quest for the active constituents in Filipendula ulmaria (meadowsweet): Comprehensive phytochemical identification by liquid chromatography coupled to quadrupole-orbitrap mass spectrometry. Planta Med. 2016, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Baldé, M.A.; Tuenter, E.; Matheeussen, A.; Traoré, M.S.; Balde, A.M.; Pieters, L.; Foubert, K. Bioassay-guided isolation of antiplasmodial and antimicrobial constituents from the roots of Terminalia albida. J. Ethnopharmacol. 2020, 113624. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Cos, P.; Vlietinck, A.J.; Berghe, D.V.; Maes, L. Anti-infective potential of natural products: How to develop a stronger in vitro “proof-of-concept”. J. Ethnopharmacol. 2006, 106, 290–302. [Google Scholar] [CrossRef]
- Díaz, J.G.; Tuenter, E.; Cesar, J.; Arranz, E.; Llauradó, G.; Cos, P.; Pieters, L. Antimicrobial activity of leaf extracts and isolated constituents of Croton linearis. J. Ethnopharmacol. 2019. [Google Scholar] [CrossRef]
- Buckner, F.S.; Verlinde, C.L.M.J.; La Flamme, A.C.; Van Voorhis, W.C. Efficient technique for screening drugs for activity against Trypanosoma cruzi using parasites expressing β-galactosidase. Antimicrob. Agents Chemother. 1996, 40, 2592–2597. [Google Scholar] [CrossRef]
- Berenguer-rivas, C.A.; Escalona-arranz, J.C.; Llauradó-maury, G.; Van Der Auwera, A.; Piazza, S.; Méndez-rodríguez, D.; Foubert, K.; Cos, P.; Pieters, L. Anti-inflammatory effect of Adelia ricinella L. aerial parts. J. Pharm. Pharmacol. 2021, 1–7. [Google Scholar] [CrossRef]
- Li, Z.H.; Guo, H.; Xu, W.B.; Ge, J.; Li, X.; Alimu, M.; He, D.J. Rapid identification of flavonoid constituents directly from PTP1B inhibitive extract of raspberry (Rubus idaeus L.) leaves by HPLC-ESI-QTOF-MS-MS. J. Chromatogr. Sci. 2016, 54, 805–810. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
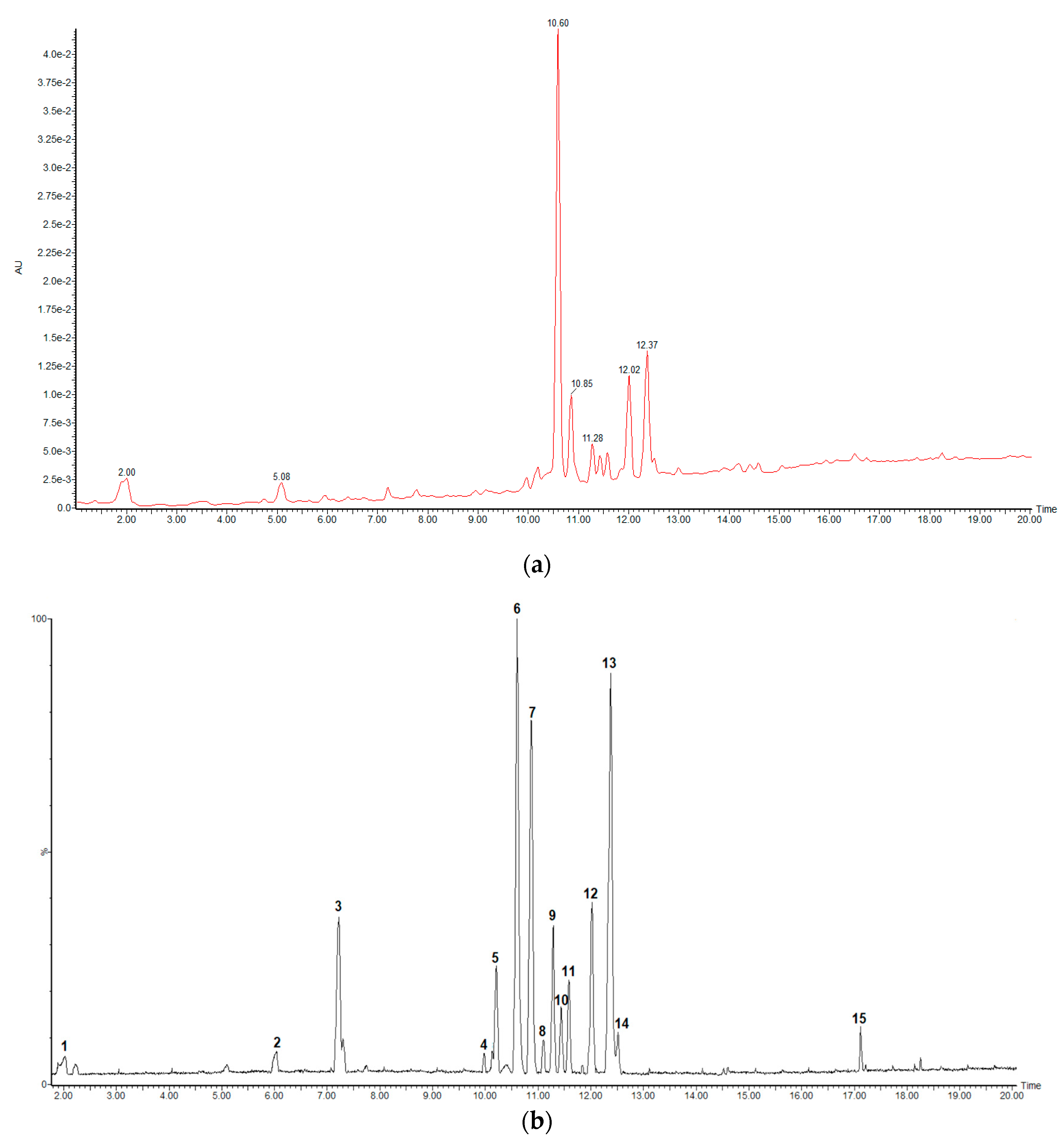
| Peak No. | Rt (min) | Measured Mass (m/z) | Theoretical Mass (m/z) | Accuracy (ppm) | MS/MS Ions | MF | Tentative Identification |
|---|---|---|---|---|---|---|---|
| 1 | 2.03 | 169.0130 | 169.0137 | −4.1 | 125.0268 | C7H5O5 | Gallic acid (std) |
| 2 | 6.04 | 289.0728 | 289.0712 | 5.5 | 125.8721 | C15H13O6 | Catechin (std) |
| 3 | 7.22 | 289.0693 | 289.0712 | −6.6 | 245.0787/137.0222/125.0238 | C15H13O6 | Epicatechin (std) |
| 4 | 9.98 | 479.0845 | 479.0826 | 4.0 | 317.0249/316.0233/287.0161/271.0255 | C21H19O13 | Myricetin-3-O-galactoside |
| 5 | 10.21 | 729.1411 | 729.1456 | −6.2 | 577.1219/451.1033/441.0815/407.0768/289.0728/287.0542 | C37H29O16 | Procyanidin B1 monogallate |
| 6 | 10.60 | 493.0612 | 493.0618 | −1.2 | 317.0285/287.0196/178.9975 | C21H17O14 | Myricetin-O-glucuronide |
| 7 | 10.87 | 441.0815 | 441.0822 | −1.6 | 289.0693/169.0157/125.0238 | C22H17O10 | Epicatechin-3-O-gallate (std) |
| 8 | 11.11 | 567.2066 | 567.2078 | −2.1 | 341.1396/326.11326/160.8430 | C27H35O13 | Unknown |
| 9 | 11.29 | 463.0859 | 463.0877 | −3.9 | 317.0285/316.0233/271.0255 | C21H19O12 | Myricetin-3-O-deoxyhexoside |
| 10 | 11.43 | 463.0859 | 463.0877 | −3.9 | 301.0344/300.0265/271.0221 | C21H19O12 | Quercetin-O-hexoside 1 |
| 11 | 11.58 | 463.0859 | 463.0877 | −3.9 | 301.0344/300.0265/271.0221 | C21H19O12 | Quercetin-O-hexoside 2 |
| 12 | 12.02 | 477.0659 | 477.0669 | −2.1 | 301.0344/299.0204/271.0255 | C21H17O13 | Quercetin-3-O-glucuronide |
| 13 | 12.38 | 433.0745 | 433.0771 | −6.0 | 301.0344/300.0265/271.0255/255.0287 | C20H17O11 | Quercetin-O-pentoside 1 |
| 14 | 12.51 | 433.0745 | 433.0771 | −6.0 | 301.0344/300.0265/271.0221/255.0287 | C20H17O11 | Quercetin-O-pentoside 2 |
| 15 | 17.12 | 331.2498 | 331.2484 | 4.2 | 313.2348/160.8430 | C18H35O5 | Unknown |
| Test Sample | Cytotoxicity (CC50 µg/mL) | Antimicrobial Screening (IC50 µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| MRC-5 | S. aureus | E. coli | C. albicans | A. fumigatus | C. neoformans | T. cruzi | T. brucei | |
| TE | >64.0 | >64.0 | >64.0 | 1.7 ± 0.6 | >64.0 | 2.7 ± 2.0 | 38.3 ± 6.8 | 33.1 ± 0.4 |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez, D.; Escalona-Arranz, J.C.; Foubert, K.; Matheeussen, A.; Van der Auwera, A.; Piazza, S.; Cuypers, A.; Cos, P.; Pieters, L. Chemical and Pharmacological Potential of Coccoloba cowellii, an Endemic Endangered Plant from Cuba. Molecules 2021, 26, 935. https://doi.org/10.3390/molecules26040935
Méndez D, Escalona-Arranz JC, Foubert K, Matheeussen A, Van der Auwera A, Piazza S, Cuypers A, Cos P, Pieters L. Chemical and Pharmacological Potential of Coccoloba cowellii, an Endemic Endangered Plant from Cuba. Molecules. 2021; 26(4):935. https://doi.org/10.3390/molecules26040935
Chicago/Turabian StyleMéndez, Daniel, Julio C. Escalona-Arranz, Kenn Foubert, An Matheeussen, Anastasia Van der Auwera, Stefano Piazza, Ann Cuypers, Paul Cos, and Luc Pieters. 2021. "Chemical and Pharmacological Potential of Coccoloba cowellii, an Endemic Endangered Plant from Cuba" Molecules 26, no. 4: 935. https://doi.org/10.3390/molecules26040935
APA StyleMéndez, D., Escalona-Arranz, J. C., Foubert, K., Matheeussen, A., Van der Auwera, A., Piazza, S., Cuypers, A., Cos, P., & Pieters, L. (2021). Chemical and Pharmacological Potential of Coccoloba cowellii, an Endemic Endangered Plant from Cuba. Molecules, 26(4), 935. https://doi.org/10.3390/molecules26040935







