Abstract
Proviral integration site for Moloney murine leukemia virus (Pim)-1/2 kinase overexpression has been identified in a variety of hematologic (e.g., multiple myeloma or acute myeloid leukemia (AML)) and solid (e.g., colorectal carcinoma) tumors, playing a key role in cancer progression, metastasis, and drug resistance, and is linked to poor prognosis. These kinases are thus considered interesting targets in oncology. We report herein the design, synthesis, structure–activity relationships (SAR) and in vitro evaluations of new quinoxaline derivatives, acting as dual Pim1/2 inhibitors. Two lead compounds (5c and 5e) were then identified, as potent submicromolar Pim-1 and Pim-2 inhibitors. These molecules were also able to inhibit the growth of the two human cell lines, MV4-11 (AML) and HCT-116 (colorectal carcinoma), expressing high endogenous levels of Pim-1/2 kinases.
1. Introduction
Proviral integration site for Moloney murine leukemia virus (Pim) kinases 1, 2 and 3 are homologous constitutively active proto-oncogenic serine/threonine protein kinases, which share a high level of sequence homology, and subsequent functional redundancies [1,2]. Nevertheless, they differ partially in their tissue distribution. Pim-1 and Pim-2 are mainly expressed in hematopoietic cells, while Pim-3 is more abundant in breast, brain, kidneys, and epithelia [3]. These oncoproteins are positive regulators of cell cycle progression, and inhibit apoptosis, acting as oncogenic survival factors [4,5]. They play a critical role in the control of cell proliferation, survival, differentiation, and migration [6,7]. Pim kinases are aberrantly up-regulated in a variety of hematologic (e.g., multiple myeloma, acute and chronic myeloid leukemias) and solid (e.g., prostate, breast, colon) tumors [8,9,10,11,12], contributing to malignant transformation, cancer progression, metastasis, drug resistance and often poor prognosis. The Pim kinases overexpression observed in cancer cells results from the abnormal activation of upstream kinases (e.g., BCR-Abl, Jak2) or receptor tyrosine kinases (RTK) (e.g., fms-like tyrosine kinase 3 (FLT3)-ITD), responsible for the activation of the signal transducer and activator of transcription (STAT) family transcription factors (particularly STAT 3/5) [13].
Interestingly, it has been demonstrated that Pim kinases are involved in the expression, activation, and stabilization of drug efflux transporters, contributing to multidrug resistance. More specifically, Pim-1 phosphorylates the P-glycoprotein (Pgp), resulting in its protection from ubiquitination and proteasomal degradation. Pim-1 also mediates phosphorylation of the breast-cancer-resistant protein (BCRP), promoting its multimerization and stable membrane expression [14,15]. Consequently, Pim kinases, by promoting survival signals, and multiple drug resistance can reduce the efficacy of chemotherapeutic drugs.
All these findings, and the fact that pim genes triple knockout mice demonstrated mild phenotypic changes [16], highlight the therapeutic potential of these kinases as targets in oncology, especially in drug-resistant tumors, in order to restore chemosensitivity to classical chemotherapies. Moreover, because of the redundant functions of the three Pim isoforms, there is currently a great interest for the development of pan-Pim inhibitors.
A number of different chemical classes of Pim inhibitors have been reported to date, including, for example, imidazo [1,2-b]pyridazines, thiazolidine-2,4-diones, benzo[d]imidazoles or pyridines [17,18] (Figure 1). For the most promising candidates, clinical trials were carried out or are currently ongoing [18,19,20,21]. Thus, the pan-PIM inhibitors PIM447 (LGH447, Novartis) demonstrated antitumor activity in monotherapy in patients with relapsed/refractory multiple myeloma with a good tolerance profile [21].
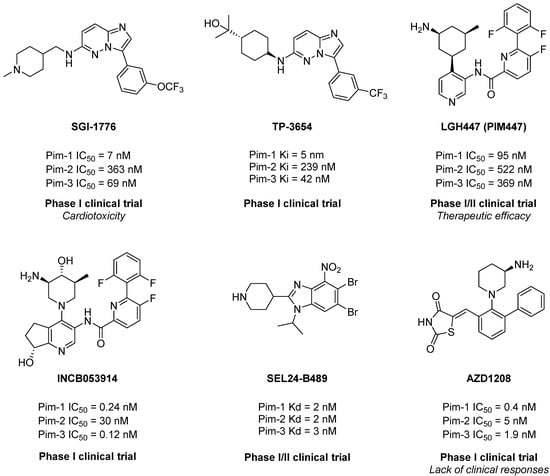
Figure 1.
Selection of the most representative and/or promising proviral integration site for Moloney murine leukemia virus (Pim) inhibitors.
Crystallographic studies of Pim-1 and Pim-2 revealed unique structural features of Pim kinases. Thus, in all three Pim kinases, the hinge region is characterized both by an atypical conformation and by the lack of a hydrogen bond (H-bond) donor in the hinge. Indeed, this region contains two proline residues (Pro123 and Pro125, in Pim-1), with no H-bond donor property. Consequently, Pim kinases bound ATP, the natural substrate, via only one hinge H-bond, involving the ATP adenine amino substituent and the hinge glutamate (Glu121, in Pim-1) backbone carbonyl. Moreover, the lateral chain of the hinge prolines and other hydrophobic amino acid residues in their environment, create a hydrophobic pocket in the hinge region. Such differences between Pim kinases and other kinases provide an opportunity for the design of selective pan-Pim inhibitors [22,23].
As part of our laboratory drug discovery program aimed at identifying new targeted therapies for the treatment of hematologic malignancies, and, regarding the particular potential of Pim-1 and Pim-2 as targets in leukemia [24,25], we decided to develop new dual Pim-1/Pim-2 specific inhibitors.
In a previous work, we identified the quinoxaline-2-carboxylic acid 1 as a new lead compound (Figure 2). This molecule potently inhibits Pim-1 enzymatic activity at submicromolar concentrations (IC50 of 74 nM) [26]. However, recent studies showed that this compound exerted only a modest activity against Pim-2 with an IC50 of 2.10 µM (data not published). In the light of these results, we planned to prepare analogues of compound 1, with an optimized activity on both Pim-1 and Pim-2 isoforms.
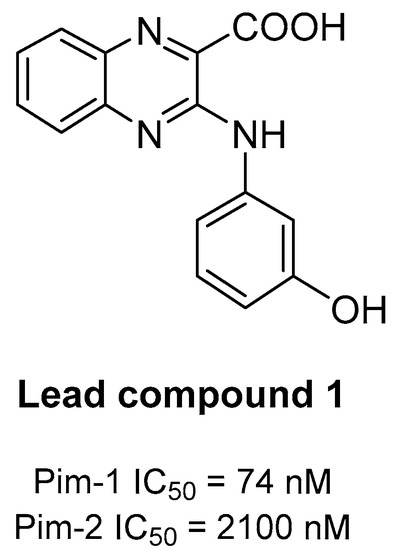
Figure 2.
Chemical structure of lead compound 1 [26].
Pim inhibitors of the literature can be classified into two classes: ATP mimetics, which establish an H-bond with the hinge glutamate residue (Glu121, in Pim-1), and non-ATP mimetics, which bind distant to the hinge or interact with the hinge through weak (mainly hydrophobic) interactions, with multiple residues of the unique hydrophobic pocket in the hinge environment. It has been shown that non-ATP mimetic Pim inhibitors could achieve higher selectivity over other kinases [27].
Consequently, in this work, we decided to probe the unique hydrophobic pocket of the Pim kinases hinge environment, by adding halogenated substituents in positions 6 or 7 of the quinoxaline scaffold, oriented towards the hinge region of the ATP binding site, and able to form weak interactions in this area. Eight new derivatives were then prepared and tested for their Pim-1 and Pim-2 inhibitory activity. The selectivity profiles and anti-proliferative activities of the most active compounds were further evaluated against a panel of mammalian kinases and against four different cell lines, respectively.
2. Results and Discussion
2.1. Synthesis
The preparation of the 6- or 7-substituted-quinoxaline-2-carboxylic acids was performed as described in Scheme 1, from commercial o-phenylenediamines according to literature procedures [28,29].
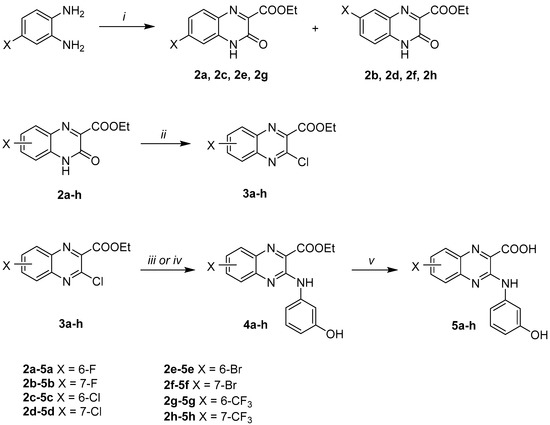
Scheme 1.
Reagents and conditions: (i) diethyl 2-oxomalonate (1 eq), citric acid (15 mol%), EtOH, rt, 2–24 h, 16–54%; (ii) N,N-dimethylformamide (DMF) (cat.), POCl3, 0 °C, and then reflux, 3 h, 51–100%; (iii) 3-aminophenol (1.1–3 eq), EtOH, MW 150 °C, 3 h, 29–99%; (iv) 3-aminophenol (3 eq), p-toluenesulfonic acid (cat.), EtOH, MW 100 °C, 8 h, 70%; (v) K2CO3 (3 eq), MeOH/H2O (4/1), reflux, 4 h, 94–100%.
First, the appropriate 4-substituted-1,2-phenylenediamine was condensed with diethyl 2-oxomalonate in the presence of citric acid (15 mol%) at room temperature in ethanol to give, after separation by column chromatography, ethyl 6-substituted-3-oxo-3,4-dihydroquinoxaline-2-carboxylates 2a, 2c, 2e, and 2g, and their seven-substituted isomers 2b, 2d, 2f, and 2h, respectively. The identification of each isomer was realized by comparison with literature spectroscopic data [30,31].
Subsequent chlorination in position 3 of compounds 2a–h using N,N-dimethylformamide (DMF) as a catalyst in refluxing phosphorous oxychloride afforded intermediates 3a–h, which were used directly for the next step because of a lack of stability.
Access to quinoxaline-2-carboxylic acids was then performed using a two-step synthetic pathway. Intermediates 3a–h undergo initial nucleophilic aromatic substitution with 3-aminophenol in ethanol under microwave irradiation, to yield esters 4a–h. For compound 4g synthesis, a catalytic amount of p-toluenesulfonic acid (p-TSA) was used. Then, hydrolysis of intermediate ethyl esters 4a–h with potassium carbonate in refluxing 80% aqueous methanol was performed, followed by an acidification with a 15% hydrochloric acid aqueous solution to afford acids 5a–h. The 3D structural determination of the substituted-quinoxaline-2-carboxylic acids 5b, 5d and 5e was established by X-ray crystallography (Figure 3) [32] and confirmed the structure in the solid state.
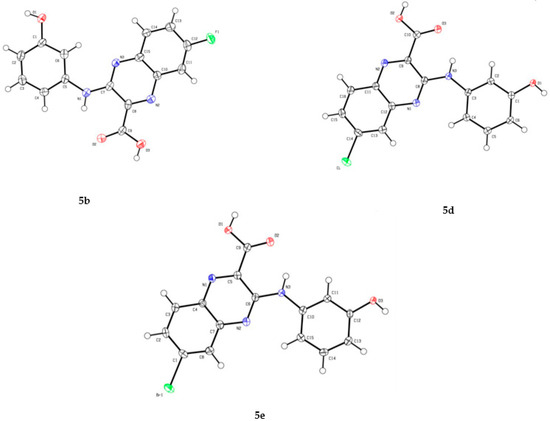
Figure 3.
Oak Ridge Thermal-Ellipsoid Plot Program (ORTEP) drawing of substituted-quinoxaline-2-carboxylic acids 5b, 5d and 5e with thermal ellipsoids at 30% level [32].
2.2. Enzymatic Assays
2.2.1. Pim-1 and Pim-2 Enzymatic Activity Inhibition
Synthesized compounds were first evaluated for their ability to inhibit the in vitro enzymatic activity of Homo sapiens Pim-1 (HsPim-1) and HsPim-2, using a luminescence-based kinase assay [33]. SGI-1776, a small-molecule pan-Pim protein kinase inhibitor [18], commercially available, was used as a control for the in vitro studies. To increase our inhibitor affinity on Pim kinases, we first decided to structurally vary the substitution patterns of the quinoxaline scaffold of 1 in positions 6 and 7, introducing halogenated substituents, trying to establish hydrophobic interactions with the unique hinge hydrophobic pocket (Table 1).

Table 1.
Enzymatic assays on HsPim-1 and HsPim-2 (ADP-Glo method at 10 μM ATP).
In position 7, it appears that the nature of the halogenated group has important repercussions on compounds activity. Thus, introduction, in this position, of a bromine led to the potent derivative 5f. This compound maintained a submicromolar activity on Pim-1 (IC50 of 180 nM) with approximately the same level of activity than lead compound 1 on both Pim isoforms (Table 1, entries 1 and 7). In contrast, the 7-substitution by a fluorine or a chlorine atom or by a trifluoromethyl group was not favorable for the activity, as evidenced by derivatives 5b, 5d and 5h (IC50 > 1 µM on both Pim-1 and Pim-2 isoforms) (Table 1, entries 3, 5 and 9).
In position 6, similar Pim-1 inhibition trends were observed with all halogenated substituents whatever their size (Table 1, entries 2, 4, 6 and 8), with IC50 values in the range of 130 to 200 nM, in every case, and nearly the same level of potency than lead compound 1 (Table 1, entry 1). Interestingly, the inhibition profile on Pim-2 isoform was significantly improved for smaller 5a (6-F) and bulkier 5e (6-Br) derivatives, with IC50 values 3- and 4-fold lower than lead 1, respectively. Finally, the most active compound 5c (6-Cl) was 12-fold more active on Pim-2 isoform than lead 1, and exhibited the same level of activity than the reference drug SGI-1776 on both isoforms (Table 1, entry 10).
2.2.2. Selectivity over a Panel of Mammalian Protein Kinases
To compare the selectivity profile of our inhibitors, most active candidates were further evaluated against a selected panel of mammalian protein kinases (comprising RnDYRK1A, HsCDK5/p25, HsCDK9/CyclinT, HsHaspin, MmCLK1, HsCK1ε and HsGSK3β) using a luminescence-based kinase assay [33].
As it can be observed with lead compound 1, derivatives 5c and 5e maintained an interesting selectivity profile against the five potential off-target kinases HsCDK5/p25, HsCDK9/CyclinT, HsHaspin, MmCLK1 and HsCK1ε with IC50 inhibition values > 10 µM in every case (Table 2, entries 1, 2 and 3).

Table 2.
Kinase selectivity profile of the most active quinoxalines (ADP-Glo method at 10 μM ATP).
The new quinoxalines tested have limited activity against HsGSK3β, displaying micromolar inhibition, in contrast to lead 1 (IC50 > 10 µM) (Table 2, entries 1, 2 and 3). Our best inhibitor 5c exhibited nevertheless an IC50 value at least 21- and 16-fold higher for HsGSK3β than for HsPim-1 and HsPim-2, respectively.
In contrast, while compound 1 potently inhibits RnDYRK1A (IC50 of 0.27 µM), the most potent quinoxalines, 5c and 5e, displayed only micromolar inhibition of RnDYRK1A (Table 2, entries 1, 2 and 3).
Importantly, the general selectivity profile of our best inhibitors (5c and 5e) was significantly improved in comparison to the reference drug SGI-1776, which inhibited eight of the nine mammalian kinases tested, with IC50 values in the submicromolar to low micromolar range (0.05–9.53 µM) (Table 2, entry 4).
2.3. Docking Studies
As already hypothesized for compound 1, docking analysis of compound 5c (6-Cl) into the ATP pocket of Pim-1 (Figure 4a) showed a key salt bridge between the carboxylate function in position 2 of the quinoxaline scaffold and the ammonium side chain of the catalytically essential Lys67. As expected, the 6-chloro substituent is oriented towards the hydrophobic pocket formed by Leu44 (P-loop residue), Ala65, the alkyl side chain of Arg122, Val126 and Leu174 and contributes to enhance van der Waals interactions. This hypothesis can be extrapolated for the other 6-halogenated substituents (5a (6-F), 5e (6-Br)) and for the 6-trifluoromethyl group (5g). Isomerization of the bromine atom from the 6- to the 7-position (5f) does not modify the IC50 values in Pim-1, while it impacts ones with a fluorine atom (5b), chlorine atom (5d) or a trifluoromethyl group (5h). The docking results of compound 5f (Figure 4b) into the ATP pocket of Pim-1 revealed a similar salt bridge interaction with Lys67, even if the 7-bromo substituent can influence both conformation and binding in this sterically restricted area of the hinge region, probably to promote a halogen-bond interaction with the carbonyl group of Glu121. This hypothesis is consistent with the halogen-bonding theory where heavier halogens bound to aryl groups show a partial positive charge opposite the C-X bond (with the exception of fluorine atoms due to their high electronegativity) that allows for interactions with classical H-bond acceptors [34].
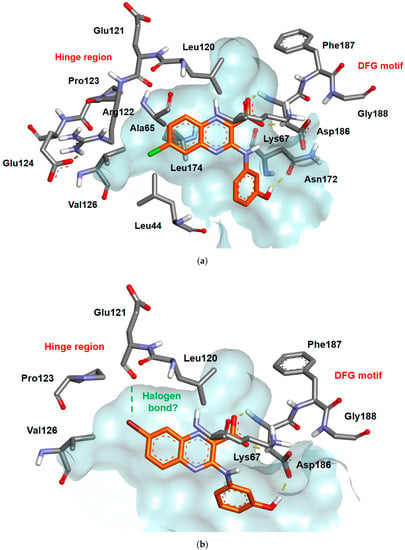
Figure 4.
(a) Illustration of the possible binding mode within the ATP pocket of Pim-1 of compound 5c (b) Illustration of the possible binding mode within the ATP pocket of Pim-1 of compound 5f. Hydrogen bonds are indicated as yellow lines. Pim-1 PDB ID: 3A99.
2.4. In Vitro Cell-Based Assays
Most active Pim inhibitors were then tested in vitro on the human acute myeloid leukemia (AML) cell line MV4-11, harboring the FLT3-ITD mutation, and overexpressing Pim kinases. Indeed, Pim 1 and Pim-2 kinases have been shown to be key downstream effectors of FLT3-induced signaling [35,36]. Moreover, the FLT3-ITD mutation confers poor prognosis in patients with AML [36]. Cytotoxic effects were evaluated using an MTT assay, and living cells were also counted with the trypan blue dye exclusion method (Table 3).

Table 3.
Cell-based assays of the most active quinoxalines.
The most potent Pim1/2 inhibitors, 5c and 5e, exhibited in vitro cytotoxic effects on the MV4-11 cell line with EC50 values in the micromolar range (35.5 ± 1.1 µm and 32.9 ± 9.6 µM, respectively), and a better activity than the lead compound 1 (EC50 of 61.2 ± 3.9 µM) (Table 3, entries1, 2 and 3).
To determine if the growth inhibitory activity of these compounds was restricted to leukemic cells, we analyzed the effects of these quinoxalines on normal bone marrow stromal and mesenchymal stem cells, major cellular components of the leukemic and hematopoietic microenvironment. To this purpose, we used bone marrow mesenchymal stem cells (MSCs), freshly isolated from brain-dead donors, and the human bone marrow stromal cell line HS-27a, sharing similar biological properties with MSCs.
The results show that, as can be observed with lead compound 1, quinoxaline 5c demonstrated a moderate inhibitory activity on these cells’ growth with EC50 values of 63.2 ± 13.1 µM and 86.3 ± 3.3 µM, on MSCs and HS-27a, respectively (Table 3, entries 1 and 2). Interestingly, quinoxaline 5e did not significantly inhibit HS-27a and MSC cell growth (EC50 values > 100 µM) (Table 3, entry 3), suggesting a selective effect of this compound on myeloid leukemia cell growth, sparing normal bone marrow stromal cells of the leukemic niche.
The best candidates were finally evaluated on the human colorectal carcinoma cell line HCT-116, a solid tumor cell line, highly expressing Pim-1 and Pim-2 kinases [12,37], using an MTT assay. Compounds 1, 5c, and 5e exhibited the same level of activity on this cell line with EC50 values ranging from 32.9 ± 10.2 µM to 45.3 ± 1.0 µM (Table 3, entries 1, 2 and 3).
It is interesting to notice that, even if the reference drug SGI-1776 demonstrated higher activity on leukemic and colorectal carcinoma cells’ growth than our derivatives, this compound also potently inhibited MSC and HS-27a cells’ growth (Table 3, entry 4), which is consistent with its lack of selectivity on the selected panel of mammalian protein kinases studied (Table 2).
3. Materials and Methods
3.1. General Remarks
All solvents were anhydrous reagents from commercial sources. Unless otherwise noted, all chemicals and reagents were obtained commercially and used without purification. Microwave heating was carried out with a single-mode Initiator Alstra (Biotage) unit. Melting points (Mp) were determined on a Stuart capillary apparatus and are uncorrected. High-resolution mass spectra (HRMS) were performed in positive mode with an ESI source on a Q-TOF mass spectrometer (Bruker maXis) with an accuracy tolerance of 2 ppm. NMR spectra were recorded at 300 MHz (1H) or 75 MHz (13C) on a Bruker Avance (300 MHz) spectrometer. The chemical shifts are reported in parts per million (ppm, δ) relative to residual deuterated solvent peaks. The abbreviations s = singlet, d = doublet, t = triplet, q = quadruplet, qt = quintuplet, m = multiplet and bs = broad signal were used throughout. In the 1H-NMR spectra of quinoxalines 5a–5h, the carboxylic acid proton was not observed. The presence of the carbonyl peak has been confirmed by 13C-NMR and the compound structure by HRMS. The identification of compounds 2b, 2d, 2f, and 2g was realized by comparison with literature spectroscopic data [30,31]. The reference of the literature-relevant spectroscopic data is given below the characterization of each concerned compound.
3.2. Chemistry
Ethyl 6-fluoro-3-oxo-3,4-dihydroquinoxaline-2-carboxylate (2a) and ethyl 7-fluoro-3-oxo-3,4-dihydroquinoxaline-2-carboxylate (2b), Method A: a mixture of 4-fluoro-1,2-phenylenediamine (366.8 mg, 2.91 mmol), diethyl 2-oxomalonate (506.5 mg, 2.91 mmol) and citric acid (83.8 mg, 0.44 mmol) in ethanol (15 mL) was stirred magnetically at room temperature overnight. Ethanol was then evaporated under reduced pressure, and the resulting residue was purified by silica column chromatography using cyclohexane with ethyl acetate gradient (0–60%) as eluents to give compounds 2a (190.6 mg, 28%) and 2b (372.0 mg, 54%) as yellow powders.
Compound 2a: Mp 186.8 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.96 (bs, 1H, NH), 7.91 (dd, 1H, J = 9.0, 5.7 Hz), 7.24 (ddd, 1H, J = 9.0, 2.7 Hz), 7.07 (dd, 1H, J = 9.0, 2.7 Hz), 4.36 (q, 2H, J = 7.1 Hz, CH2), 1.32 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 164.0 (d, J = 248.9 Hz), 163.9, 152.7, 149.8, 135.0 (d, J = 13.0 Hz), 132.4 (d, J = 11.0 Hz), 128.3 (d, J = 1.5 Hz), 112.7 (d, J = 24.2 Hz), 102.2 (d, J = 26.7 Hz), 62.3, 14.4. 19F NMR (282 MHz, DMSO-d6) δ −106.0. HRMS (ESI) m/z: [M+H]+ calcd for C11H10FN2O3, 237.0670; found: 237.0668.
Compound 2b: Mp 207.6 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.97 (bs, 1H, NH), 7.73 (dd, 1H, J = 9.0, 2.9 Hz), 7.59 (ddd, 1H, J = 9.0, 2.9 Hz), 7.38 (dd, 1H, J = 9.0, 5.1 Hz), 4.38 (q, 2H, J = 7.1 Hz, CH2), 1.32 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.9, 158.4 (d, J = 239.3 Hz), 152.5, 152.3, 131.4 (d, J = 11.6 Hz), 130.1, 120.9 (d, J = 24.6 Hz), 117.8 (d, J = 9.2 Hz), 114.7 (d, J = 22.7 Hz), 62.4, 14.4. 19F NMR (282 MHz, DMSO-d6) δ −118.4. HRMS (ESI) m/z: [M+H]+ calcd for C11H10FN2O3, 237.0670; found: 237.0667.
Literature spectroscopic data reference [30].
Ethyl 6-chloro-3-oxo-3,4-dihydroquinoxaline-2-carboxylate (2c) and ethyl 7-chloro-3-oxo-3,4-dihydroquinoxaline-2-carboxylate (2d): The title compound was synthesized according to the general method A from 4-chloro-1,2-phenylenediamine (285.2 mg, 2.00 mmol), diethyl 2-oxomalonate (348.3 mg, 2.00 mmol) and citric acid (57.7 mg, 0.30 mmol) in ethanol (10 mL). The reaction mixture was stirred magnetically at room temperature for 2 h. The solvent was then removed under reduced pressure, and the resulting residue was purified by silica column chromatography using cyclohexane with ethyl acetate gradient (0–40%) as eluent to give compounds 2c (161.7 mg, 32%) and 2d (274.2 mg, 54%) as yellow powders.
Compound 2c: Mp 236.0 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.94 (bs, 1H, NH), 7.85 (d, 1H, J = 8.7 Hz), 7.40 (dd, 1H, J = 8.7, 2.2 Hz), 7.34 (d, 1H, J = 2.2 Hz), 4.37 (q, 2H, J = 7.1 Hz, CH2), 1.32 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.8, 152.6, 151.1, 136.8, 134.3, 131.4, 130.0, 124.5, 115.6, 62.3, 14.4. HRMS (ESI) m/z: [M+H]+ calcd for C11H10ClN2O3: 253.0374; found: 253.0374.
Compound 2d: Mp 213.2 °C. 1H NMR (300 MHz, DMSO-d6) δ 13.00 (bs, 1H, NH), 7.93 (d, 1H, J = 2.3 Hz), 7.69 (dd, 1H, J = 8.8, 2.3 Hz), 7.36 (d, 1H, J = 8.8 Hz), 4.38 (q, 2H, J = 7.1 Hz, CH2), 1.32 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.8, 152.6, 152.2, 132.6, 132.2, 131.7, 128.6, 128.0, 118.0, 62.4, 14.4. HRMS (ESI) m/z: [M+H]+ calcd for C11H10ClN2O3: 253.0374; found: 253.0373.
Literature spectroscopic data reference [30].
Ethyl 6-bromo-3-oxo-3,4-dihydroquinoxaline-2-carboxylate (2e) and ethyl 7-bromo-3-oxo-3,4-dihydroquinoxaline-2-carboxylate (2f): The title compounds were synthesized according to the general method A from 4-bromo-1,2-phenylenediamine (748.2 mg, 4.00 mmol), diethyl 2-oxomalonate (696.6 mg, 4.00 mmol) and citric acid (115.3 mg, 0.60 mmol) in ethanol (15 mL). The reaction mixture was stirred magnetically at room temperature for 24 h. The solvent was then removed under reduced pressure, and the resulting residue was purified by silica column chromatography using cyclohexane with ethyl acetate gradient (40–70%) as eluent to give compound 2e (393.5 mg, 33%) as a beige powder and compound 2f (304.5 mg, 26%) as an orange powder.
Compound 2e: Mp 234.8°C. 1H NMR (300 MHz, DMSO-d6) δ 12.93 (bs, 1H, NH), 7.77 (d, 1H, J = 8.6 Hz), 7.52 (dd, 1H, J = 8.6, 2.0 Hz), 7.49 (d, 1H, J = 2.0 Hz), 4.36 (q, 2H, J = 7.1 Hz, CH2), 1.31 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.9, 152.6, 151.3, 134.5, 131.5, 130.2, 127.3, 125.6, 118.6, 62.3, 14.4. HRMS (ESI) m/z: [M+H]+ calcd for C11H10BrN2O3: 296.9869; found: 296.9869.
Compound 2f: Mp 234.8°C. 1H NMR (300 MHz, DMSO-d6) δ 13.00 (bs, 1H, NH), 8.06 (d, 1H, J = 2.2 Hz), 7.80 (dd, 1H, J = 8.8, 2.2 Hz), 7.29 (d, 1H, J = 8.8 Hz), 4.37 (q, 2H, J = 7.1 Hz, CH2), 1.32 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.8, 152.6, 152.1, 135.2, 132.6, 132.1, 131.6, 118.2, 115.6, 62.4, 14.4. HRMS (ESI) m/z: [M+H]+ calcd for C11H10BrN2O3: 296.9869; found: 296.9868.
Literature spectroscopic data reference [30].
Ethyl 3-oxo-6-(trifluoromethyl)-3,4-dihydroquinoxaline-2-carboxylate (2g) and ethyl 3-oxo-7-(trifluoromethyl)-3,4-dihydroquinoxaline-2-carboxylate (2h): The title compounds were synthesized according to the general method A from 4-trifluoromethyl-1,2-phenylenediamine (387.5 mg, 2.20 mmol), diethyl 2-oxomalonate (383.1 mg, 2.20 mmol) and citric acid (63.4 mg, 0.33 mmol) in ethanol (15 mL). The reaction mixture was stirred magnetically at room temperature overnight. The solvent was then removed under reduced pressure, and the resulting residue was purified by silica column chromatography using dichloromethane with ethyl acetate gradient (0–20%) as eluent to give the desired compounds 2g (325.8 mg, 52%) and 2h (98.9 mg, 16%) as beige powders.
Compound 2g: Mp 204.1 °C. 1H NMR (300 MHz, DMSO-d6) δ 13.11 (bs, 1H, NH), 8.06 (d, 1H, J = 8.4 Hz), 7.67 (dd, 1H, J = 8.4, 1.8 Hz), 7.63 (d, 1H, J = 1.8 Hz), 4.40 (q, 2H, J = 7.2 Hz, CH2), 1.33 (t, 3H, J = 7.2 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.7, 153.6, 152.6, 133.4, 133.0, 131.7 (q, J = 32.3 Hz), 131.1, 124.0 (q, J = 271.0 Hz), 120.3 (q, J = 3.5 Hz), 113.5 (q, J = 3.8 Hz), 62.5, 14.4. 19F RMN (282 MHz, DMSO-d6) δ −61.4. HRMS (ESI) m/z: [M+H]+ calcd for C12H10F3N2O3: 287.0638; found: 287.0637.
Literature spectroscopic data reference [31].
Compound 2h: Mp 161.2 °C. 1H NMR (300 MHz, DMSO-d6) δ 13.18 (bs, 1H, NH), 8.20 (d, 1H, J = 1.5 Hz), 7.96 (dd, 1H, J = 8.7, 1.5 Hz), 7.51 (d, 1H, J = 8.7 Hz), 4.39 (q, 2H, J = 7.2 Hz, CH2), 1.33 (t, 3H, J = 7.2 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 163.7, 152.9, 152.6, 136.1, 130.4, 128.7 (q, J = 3.3 Hz), 127.0 (q, J = 4.1 Hz), 124.6 (q, J = 32.7 Hz), 124.3 (q, J = 270.2 Hz), 117.7, 62.5, 14.4. 19F RMN (282 MHz, DMSO-d6) δ −60.4. HRMS (ESI) m/z: [M+H]+ calcd for C12H10F3N2O3: 287.0638; found: 287.0635.
Ethyl 3-chloro-6-fluoroquinoxaline-2-carboxylate (3a), Method B: into a dry three-neck round bottom flask was introduced compound 2a (141.3 mg, 0.60 mmol) in phosphorous oxychloride (1.4 mL) at ice bath temperature. N,N-dimethylformamide (140 µL) was then added at 0 °C and the reaction mixture was refluxed for 3 h. After cooling, the resulting mixture was poured over ice, stirred magnetically at room temperature overnight, and extracted with dichloromethane. The combined organic layers were dried over MgSO4, filtered, and evaporated under reduced pressure to obtain derivative 3a (112.9 mg, 74%) as a brown oil. 1H NMR (300 MHz, DMSO-d6) δ 8.32 (dd, 1H, J = 9.2, 5.8 Hz), 8.02–7.90 (m, 2H), 4.49 (q, 2H, J = 7.1 Hz, CH2), 1.38 (t, 3H, J = 7.1 Hz, CH3). 19F RMN (282 MHz, DMSO-d6) δ −103.6.
Ethyl 3-chloro-7-fluoroquinoxaline-2-carboxylate (3b): The title compound was synthesized according to the general method B from compound 2b (236.2mg, 1.00 mmol), phosphorous oxychloride (2.3 mL) and DMF (230 µL). Compound 3b was obtained (254.6 mg, 100%) as a brown oil. 1H NMR (300 MHz, DMSO-d6) δ 8.23 (dd, 1H, J = 9.2, 5.6 Hz, 1H), 8.09 (dd, 1H, J = 9.2, 2.8 Hz), 8.00 (ddd, 1H, J = 9.2, 8.4, 2.8 Hz), 4.50 (t, 2H, J = 7.1 Hz, CH2), 1.39 (t, 3H, J = 7.1 Hz, CH3). 19F RMN (282 MHz, DMSO-d6) δ −105.8.
Ethyl 3,6-dichloroquinoxaline-2-carboxylate (3c): The title compound was synthesized according to the general method B from compound 2c (50.0 mg, 0.20 mmol), phosphorous oxychloride (461 µL) and DMF (46 µL). Compound 3c was obtained (53.7 mg, 100%) as a brown oil. 1H NMR (300 MHz, DMSO-d6) δ 8.29 (d, 1H, J = 2.3 Hz), 8.26 (d, 1H, J = 9.0 Hz), 8.03 (dd, 1H, J = 9.0, 2.3 Hz), 4.50 (q, 2H, J = 7.1 Hz, CH2), 1.39 (t, 3H, J = 7.1 Hz, CH3).
Ethyl 3,7-dichloroquinoxaline-2-carboxylate (3d): The title compound was synthesized according to the general method B from compound 2d (200.0 mg, 0.79 mmol), phosphorous oxychloride (1.9 mL) and DMF (185 µL). Compound 3d was obtained (202.0 mg, 94%) as a brown oil. 1H NMR (300 MHz, DMSO-d6) δ 8.37 (d, 1H, J = 2.2 Hz), 8.17 (d, 1H, J = 9.0 Hz), 8.07 (dd, 1H, J = 9.0, 2.2 Hz), 4.51 (q, 1H, J = 7.1 Hz, CH2), 1.39 (t, 1H, J = 7.1 Hz, CH3).
Ethyl 6-bromo-3-chloroquinoxaline-2-carboxylate (3e): The title compound was synthesized according to the general method B from compound 2e (200.0 mg, 0.67 mmol), phosphorous oxychloride (1.6 mL) and DMF (157 µL). Compound 3e was obtained (212.4 mg, 100%) as a brown oil. 1H NMR (300 MHz, DMSO-d6) δ 8.39 (dd, 1H, J = 1.9, 0.8 Hz), 8.16–8.08 (m, 2H), 4.49 (q, 2H, J = 7.1 Hz, CH2), 1.38 (t, 3H, J = 7.1 Hz, CH3).
Ethyl 7-bromo-3-chloroquinoxaline-2-carboxylate (3f): The title compound was synthesized according to the general method B from compound 2f (200.0 mg, 0.67 mmol), phosphorous oxychloride (1.6 mL) and DMF (157 µL). Compound 3f was obtained (212.2 mg, 100%) as a brown oil. 1H NMR (300 MHz, DMSO-d6) δ 8.52 (dd, 1H¸ J = 2.1, 0.5 Hz), 8.18 (dd, 1H¸ J = 9.0, 2.1 Hz), 8.08 (dd, 1H, J = 9.0, 0.5 Hz), 4.50 (q, 2H, J = 7.1 Hz, CH2), 1.38 (t, 3H, J = 7.1 Hz, CH3).
Ethyl 3-chloro-6-(trifluoromethyl)quinoxaline-2-carboxylate (3g): The title compound was synthesized according to the general method B from compound 2g (278.0 mg, 0.97 mmol), phosphorous oxychloride (2.3 mL) and DMF (230 µL). Compound 3g was obtained (295.8 mg, 100%) as a yellow oil. 1H NMR (300 MHz, DMSO-d6) δ 8.57 (d, 1H, J = 2.0 Hz), 8.44 (d, 1H, J = 8.8 Hz), 8.25 (dd, 1H, J = 8.8, 2.0 Hz), 4.52 (q, 2H, J = 7.1 Hz, CH2), 1.40 (t, 3H, J = 7.1 Hz, CH3). 19F RMN (282 MHz, DMSO-d6) δ −61.3.
Ethyl 3-chloro-7-(trifluoromethyl)quinoxaline-2-carboxylate (3h): The title compound was synthesized according to the general method B from compound 2h (50.0 mg, 0.18 mmol), phosphorous oxychloride (0.4 mL) and DMF (40 µL). Compound 3h was obtained (27.0 mg, 51%) as a yellow oil. 1H NMR (300 MHz, DMSO-d6) δ 8.64 (d, 1H, J = 0.8 Hz), 8.33 (d, 1H, J = 8.9 Hz), 8.28 (dd, 1H, J = 8.9, 0.8 Hz), 4.52 (q, 2H, J = 7.1 Hz, CH2), 1.39 (t, 3H, J = 7.1 Hz, CH3). 19F RMN (282 MHz, DMSO-d6) δ −61.3 (s).
Ethyl 6-fluoro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylate (4a), Method C1: a solution of compound 3a (84.6 mg, 0.33 mmol) and 3-aminophenol (39.8 mg, 0.365 mmol) in absolute ethanol (2 mL) was stirred under microwave irradiation for 3 h at 150 °C. Ethanol was then evaporated under reduced pressure, and the resulting residue was purified by silica column chromatography using cyclohexane with ethyl acetate gradient (0–30%) as eluent to give compound 4a (88.9 mg, 82%) as an orange powder. Mp 199.8 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.15 (bs, 1H), 9.49 (bs, 1H), 8.06 (ddd, 1H, J = 9.1, 6.1, 0.5 Hz), 7.55 (t, 1H, J = 2.0 Hz), 7.53–7.42 (m, 2H), 7.24–7.14 (m, 2H), 6.53 (ddd, 1H, J = 7.7, 2.0, 1.3 Hz), 4.48 (q, 2H, J = 7.1 Hz, CH2), 1.41 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.9, 164.9 (d, J = 247.5 Hz), 158.3, 149.2, 143.7 (d, J = 14.6 Hz), 140.3, 133.3, 132.7 (d, J = 11.0 Hz), 132.2, 130.1, 116.5 (d, J = 25.6 Hz), 111.4, 110.9, 110.4 (d, J = 21.8 Hz), 107.6, 60.2, 14.5. 19F RMN (282 MHz, DMSO-d6) δ −105.3. HRMS (ESI) m/z: [M+H]+ calcd for C17H15FN3O3: 328.1092; found: 328.1094.
Ethyl 7-fluoro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylate (4b): The title compound was synthesized according to the general method C1 from compound 3b (254.6 mg, 1.00 mmol) and 3-aminophenol (120.0 mg, 1.10 mmol) in absolute ethanol (5 mL). Compound 4b was obtained (138.6 mg, 42%) as a red powder. Mp 223.4 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.01 (bs, 1H), 9.47 (bs, 1H), 7.85–7.70 (m, 3H), 7.55 (t, 1H, J = 1.8 Hz), 7.22–7.13 (m, 2H), 6.51 (ddd, 1H, J = 7.3, 1.8 Hz), 4.48 (q, 2H, J = 7.1 Hz, CH2), 1.41 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.8, 160.1 (d, J = 243.1 Hz), 158.3, 148.5, 140.6, 139.6, 136.0 (d, J = 12.8 Hz), 133.6, 130.0, 128.7 (d, J = 9.6 Hz), 123.1 (d, J = 26.7 Hz), 113.5 (d, J = 21.4 Hz), 111.2, 110.6, 107.3, 62.9, 14.5. 19F RMN (282 MHz, DMSO-d6) δ -114.4. HRMS (ESI) m/z: [M+H]+ calcd for C17H15FN3O3: 328.1092; found: 328.1092.
Ethyl 6-chloro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylate (4c): The title compound was synthesized according to the general method C1 from compound 3c (203.1 mg, 0.75 mmol) and 3-aminophenol (245.2 mg, 2.25 mmol) in absolute ethanol (5 mL). After purification by silica column chromatography using cyclohexane with ethyl acetate gradient (0–40%) as eluent, compound 4c was obtained (253.9 mg, 99%) as an orange powder. Mp 226.8 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.14 (bs, 1H), 9.48 (bs, 1H), 8.01 (d, 1H, J = 8.9 Hz), 7.82 (dd, 1H, J = 2.3, 1.0), 7.63 (m, 1H), 7.57 (dd, 1H, J = 8.9, 2.2 Hz), 7.22–7.14 (m, 2H), 6.56–6.50 (m, 1H), 4.48 (q, 2H, J = 7.1 Hz, CH2), 1.40 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.8, 158.3, 149.2, 142.9, 140.3, 137.8, 134.6, 133.3, 131.8, 130.1, 127.0, 125.3, 111.3, 110.9, 107.5, 62.9, 14.5. HRMS (ESI) m/z: [M+H]+ calcd for C17H15ClN3O3: 344.0796; found: 344.0794.
Ethyl 7-chloro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylate (4d): The title compound was synthesized according to the general method C1 from compound 3d (167 mg, 0.62 mmol) and 3-aminophenol (74 mg, 0.68 mmol) in absolute ethanol (5 mL). After purification by silica column chromatography using dichloromethane with ethyl acetate gradient (0–10%) as eluent, compound 4d was obtained (192 mg, 91%) as a red powder. Mp 216.9 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.07 (bs, 1H), 9.49 (bs, 1H), 8.06 (d, 1H, J = 2.2 Hz), 7.82 (dd, 1H, J = 8.8, 2.2 Hz), 7.77 (d, 1H, J = 8.8 Hz), 7.56 (t, 1H, J = 2.1 Hz), 7.22–7.14 (m, 2H), 6.52 (ddd, 1H, J = 7.0, 2.1 Hz), 4.48 (q, 2H, J = 7.1 Hz, CH2), 1.40 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.7, 158.3, 148.8, 141.1, 140.4, 136.1, 133.9, 133.6, 130.4, 130.0, 128.5, 128.3, 111.3, 110.8, 107.5, 62.9, 14.5. HRMS (ESI) m/z: [M+H]+ calcd for C17H15ClN3O3: 344.0796; found: 344.0799.
Ethyl 6-bromo-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylate (4e): The title compound was synthesized according to the general method C1 from compound 3e (203.2 mg, 0.64 mmol) and 3-aminophenol (210.8 mg, 1.93 mmol) in absolute ethanol (5 mL). After cooling, the reaction mixture was filtered, giving compound 4e (72.4 mg, 29%) as a red powder. Mp 230.6 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.14 (bs, 1H), 9.48 (bs, 1H), 7.98 (d, 1H, J = 2.1 Hz), 7.91 (d, 1H, J = 8.8 Hz), 7.70–7.64 (m, 2H), 7.21–7.13 (m, 2H), 6.55–6.49 (m, 1H), 4.47 (q, 2H, J = 7.1 Hz, CH2), 1.40 (t, 2H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.8, 158.3, 149.1, 143.1, 140.3, 134.8, 133.3, 131.8, 130.1, 129.6, 128.6, 126.7, 111.3, 110.8, 107.5, 62.9, 14.5. HRMS (ESI) m/z: [M+H]+ calcd for C17H15BrN3O3: 388.0291; found: 388.0291.
Ethyl 7-bromo-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylate (4f): The title compound was synthesized according to the general method C1 from compound 3f (210.9 mg, 0.67 mmol) and 3-aminophenol (218.7 mg, 2.00 mmol) in absolute ethanol (5 mL). After purification by silica column chromatography using cyclohexane with ethyl acetate gradient (0–50%) as eluent, compound 4f was obtained (88.4 mg, 34%) as a red powder. Mp 244.1 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.08 (bs, 1H), 9.50 (bs, 1H), 8.20 (d, 1H, J = 2.2 Hz), 7.91 (dd, 1H, J = 8.9, 2.2 Hz), 7.70 (d, 1H, J = 8.9 Hz), 7.58–7.56 (m, 1H), 7.22–7.14 (m, 2H), 6.52 (ddd, 1H, J = 7.1, 2.1 Hz), 4.48 (q, 2H, J = 7.1 Hz, CH2), 1.40 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.7, 158.3, 148.9, 141.4, 140.4, 136.6, 136.1, 133.8, 131.7, 130.0, 128.5, 118.5, 111.3, 110.8, 107.5, 62.9, 14.5. HRMS (ESI) m/z: [M+H]+ calcd for C17H15BrN3O3: 388.0291; found: 388.0290.
Ethyl 3-((3-hydroxyphenyl)amino)-6-(trifluoromethyl)quinoxaline-2-carboxylate (4g), Method C2: a solution of compound 3g (256.9 mg, 0.84 mmol), 3-aminophenol (276.0 mg, 2.53 mmol) and p-TSA, as a catalyst, in absolute ethanol (1.5 mL) was stirred under microwave irradiation for 8 h at 100 °C. Ethanol was then evaporated under reduced pressure, and the resulting residue was purified by silica column chromatography using cyclohexane with ethyl acetate gradient (0–30%) as eluent to give the desired compound 4g (222.7 mg, 70%) as a red powder. Mp 184.7 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.16 (bs, 1H), 9.52 (bs, 1H), 8.20 (d, 1H, J = 8.6 Hz), 8.10 (d, 1H, J = 1.8 Hz), 7.79 (dd, 1H, J = 8.6, 1.8 Hz), 7.67 (m, 1H), 7.24–7.16 (m, 2H), 6.54 (ddd, 1H, J = 6.9, 2.4 Hz), 4.50 (q, 2H, J = 7.2 Hz, CH2), 1.42 (t, 3H, J = 7.2 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.5, 158.3, 149.2, 141.7, 140.2, 137.2, 135.6, 132.7 (q, J = 31.9 Hz), 131.6, 130.1, 124.2 (q, J = 271.1 Hz), 124.1 (q, J = 3.5 Hz), 121.6 (q, J = 4.0 Hz), 111.4, 111.0, 107.6, 63.1, 14.4. 19F RMN (282 MHz, DMSO-d6) δ −61.4. HRMS (ESI) m/z: [M+H]+ calcd for C18H15F3N3O3: 378.1060; found: 378.1059.
Ethyl 3-((3-hydroxyphenyl)amino)-7-(trifluoromethyl)quinoxaline-2-carboxylate (4h): The title compound was synthesized according to the general method C1 from compound 3h (86.2 mg, 0.28 mmol) and 3-aminophenol (33.9 mg, 0.31 mmol) in absolute ethanol (2 mL). After purification by silica column chromatography using cyclohexane with ethyl acetate gradient (0–20%) as eluent, compound 4h was obtained (38.8 mg, 36%) as a yellow powder. Mp 178.5 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.22 (bs, 1H), 9.56 (bs, 1H), 8.34 (d, 1H, J = 1.8 Hz), 8.05 (dd, 1H, J = 8.7, 1.8 Hz), 7.93 (d, 1H, J = 8.7 Hz), 7.60 (m, 1H), 7.23–7.17 (m, 2H), 6.56 (ddd, 1H, J = 7.2, 1.8 Hz), 4,50 (q, 2H, J = 7.1 Hz, CH2), 1.42 (t, 3H, J = 7.1 Hz, CH3). 13C NMR (75 MHz, DMSO-d6) δ 165.5, 158.3, 149.7, 144.2, 140.1, 134.8, 134.7, 130.1, 128.4 (q, J = 3.1 Hz), 128.1, 127.7 (q, J = 4.4 Hz), 126.3 (q, J = 32.3 Hz), 124.4 (q, J = 270.0 Hz), 111.6, 111.2, 107.8, 63.0, 14.4. 19F RMN (282 MHz, DMSO-d6) δ −60.6 (t, J = 2.9 Hz). HRMS (ESI) m/z: [M+H]+ calcd for C18H15F3N3O3: 378.1060; found: 378.1059.
6-Fluoro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylic acid (5a), Method D: to ester 4a (43.2 mg, 0.13 mmol) in aqueous methanol (80%, 15 mL), potassium carbonate (54.7 mg, 0.40 mmol) was added and the reaction mixture was refluxed for 4 h. After cooling, the solvent was removed under reduced pressure. Then, the residue was acidified with a 15% hydrochloric acid aqueous solution, and extracted with dichloromethane. The combined organic layers were washed with brine, dried over MgSO4, filtered, and evaporated under reduced pressure to yield the acid 5a (39.5 mg, 100%) as a dark red powder. Mp 200.4 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.59 (bs, 1H), 9.47 (bs, 1H), 8.04 (m, 1H), 7.56 (t, 1H J = 2.0 Hz), 7.52–7.40 (m, 2H), 7.24 (ddd, 1H, J = 7.9, 2.0, 1.0 Hz), 7.17 (t, 1H, J = 7.9 Hz), 6.52 (ddd, 1H, J = 7.9, 2.0, 1.0 Hz). 13C NMR (75 MHz, DMSO-d6) δ 168.0, 164.8 (d, J = 249.7 Hz), 158.3, 149.7, 144.0, 143.8, 140.4, 133.2, 132.5 (d, J = 11.6 Hz), 130.1, 116.2 (d, J = 25.9 Hz), 111.2, 110.8, 110.3 (d, J = 21.8 Hz), 107.4. 19F RMN (282 MHz, DMSO-d6) δ −105.6. HRMS (ESI) m/z: [M+H]+ calcd for C15H11FN3O3: 300.0779; found: 300.0779.
7-Fluoro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylic acid (5b): The title compound was synthesized according to the general method D from compound 4b (107.3 mg, 0.33 mmol) and potassium carbonate (135.9 mg, 0.98 mmol) in aqueous methanol (80%, 15 mL). Compound 5b was obtained (97.1 mg, 99%) as a red powder. Mp 180.1 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.44 (bs, 1H), 9.47 (bs, 1H), 7.85–7.69 (m, 3H), 7.56 (t, 1H, J = 2.0 Hz), 7.22 (ddd, 1H, J = 7.7, 2.0, 1.2 Hz), 7.16 (t, 1H, J = 7.7 Hz), 6.50 (ddd, 1H, J = 7.7, 2.0, 1.2 Hz). 13C NMR (75 MHz, DMSO-d6) δ 167.9, 160.0 (d, J = 243.1 Hz), 158.3, 149.0, 140.7, 139.7, 136.0 (d, J = 12.2 Hz), 133.9, 130.0, 128.6 (d, J = 9.5 Hz), 122.9 (d, J = 25.7 Hz), 113.4 (d, J = 21.2 Hz), 111.0, 110.5, 107.2. 19F RMN (282 MHz, DMSO-d6) δ −114.7. HRMS (ESI) m/z: [M+H]+ calcd for C15H11FN3O3: 300.0779; found: 300.0779.
6-Chloro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylic acid (5c): The title compound was synthesized according to the general method D from compound 4c (29.7 mg, 0.09 mmol) and potassium carbonate (35.7 mg, 0.26 mmol) in aqueous methanol (80%, 5 mL). Compound 5c was obtained (27.2 mg, 100%) as a dark red powder. Mp 194.6 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.60 (bs, 1H), 9.48 (bs, 1H), 7.98 (d, 1H, J = 8.8 Hz), 7.82 (d, 1H, J = 2.3 Hz), 7.64 (t, 1H, J = 1.9 Hz), 7.57 (dd, 1H, J = 8.8, 2.3 Hz), 7.23–7.13 (m, 2H), 6.51 (ddd, 1H, J = 7.2, 1.9 Hz). 13C NMR (75 MHz, DMSO-d6) δ 167.9, 158.3, 149.6, 143.1, 140.4, 137.6, 134.6, 133.5, 131.7, 130.1, 126.9, 125.3, 111.2, 110.7, 107.4. HRMS (ESI) m/z: [M+H]+ calcd for C15H11ClN3O3: 316.0483; found: 316.0484.
7-Chloro-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylic acid (5d): The title compound was synthesized according to the general method D from compound 4d (128 mg, 0.37 mmol) and potassium carbonate (154 mg, 1.12 mmol) in aqueous methanol (80%, 15 mL). Compound 5d was obtained (110 mg, 94%) as a dark red powder. Mp 173–174 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.48 (bs, 1H), 9.49 (bs, 1H), 8.03 (d, 1H, J = 2.2 Hz), 7.84–7.76 (m, 2H), 7.57 (t, 1H, J = 1.9 Hz), 7.23–7.14 (m, 2H), 6.53–6.49 (m, 1H). 13C NMR (75 MHz, DMSO-d6) δ 167.3, 157.8, 148.8, 140.8, 140.0, 135.7, 135.5, 133.0, 129.7, 129.6, 128.0, 127.8, 110.7, 110.2, 106.9. HRMS (ESI) m/z: [M+H]+ calcd for C15H11ClN3O3: 316.04855; found: 316.04778.
6-Bromo-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylic acid (5e): The title compound was synthesized according to the general method D from compound 4e (39.0 mg, 0.10 mmol) and potassium carbonate (41.5 mg, 0.30 mmol) in aqueous methanol (80%, 5 mL). Compound 5e was obtained (36.0 mg, 100%) as a red powder. Mp 199.0 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.54 (bs, 1H), 9.47 (bs, 1H), 7.98 (d, 1H, J = 2.2 Hz), 7.90 (d, 1H, J = 8.8 Hz), 7.70–7.64 (m, 2H), 7.22–7.13 (m, 2H), 6.51 (ddd, 1H, J = 6.6, 2.4 Hz). 13C NMR (75 MHz, DMSO-d6) δ 167.9, 158.3, 149.5, 143.3, 140.4, 134.7, 133.4, 131.7, 130.1, 129.5, 128.6, 126.6, 111.2, 110.7, 107.4. HRMS (ESI) m/z: [M+H]+ calcd for C15H11BrN3O3: 359.9978; found: 359.9977.
7-Bromo-3-((3-hydroxyphenyl)amino)quinoxaline-2-carboxylic acid (5f): The title compound was synthesized according to the general method D from compound 4f (40.7 mg, 0.11 mmol) and potassium carbonate (43.5 mg, 0.32 mmol) in aqueous methanol (80%, 5 mL). Compound 5f was obtained (37.8 mg, 100%) as an orange powder. Mp 193.1 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.48 (bs, 1H), 9.53 (bs, 1H), 8.16 (d, 1H, J = 2.3 Hz), 7.91 (dd, 1H, J = 8.9, 2.3 Hz), 7.71 (d, 1H, J = 8.9 Hz), 7.58 (t, 1H, J = 1.9 Hz), 7.23–7.13 (m, 2H), 6.52 (dd, 1H, J = 7.5, 1.9 Hz). 13C NMR (75 MHz, DMSO-d6) δ 167.8, 158.3, 149.3, 141.5, 140.4, 136.6, 136.0, 133.9, 131.7, 130.1, 128.5, 118.3, 111.1, 110.7, 107.4. HRMS (ESI) m/z: [M+H]+ calcd for C15H11BrN3O3: 359.9978; found: 359.9979.
3-((3-Hydroxyphenyl)amino)-6-(trifluoromethyl)quinoxaline-2-carboxylic acid (5g): The title compound was synthesized according to the general method D from ester 4g (30.8 mg, 0.08 mmol) and potassium carbonate (33.6 mg, 0.24 mmol) in aqueous methanol (80%, 15 mL). After cooling, the solvent was removed under reduced pressure. Then, the residue was acidified with a 15% hydrochloric acid aqueous solution, and extracted with diethyl ether. The combined organic layers were washed with brine, dried over MgSO4, filtered and evaporated under reduced pressure to yield the acid 5g (28.6 mg, 100%) as a dark red powder. Mp 196.2 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.56 (bs, 1H), 9.51 (bs, 1H), 8.17 (d, 1H, J = 8.4 Hz), 8.09 (d, 1H, J = 1.8 Hz), 7.79 (dd, 1H, J = 8.4, 1.8 Hz), 7.68 (m, 1H), 7.24–7.15 (m, 2H), 6.54 (ddd, 1H, J = 7.2, 1.8 Hz). 13C NMR (75 MHz, DMSO-d6) δ 167.7, 158.3, 149.7, 141.8, 140.3, 137.2, 135.7, 132.4 (q, J = 37.5 Hz), 131.6, 130.1, 124.3 (q, J = 270.0 Hz), 124.1 (q, J = 3.0 Hz), 121.4 (q, J = 2.3 Hz), 111.3, 110.9, 107.4. 19F RMN (282 MHz, DMSO-d6) δ −61.4. HRMS (ESI) m/z: [M+H]+ calcd for C16H11F3N3O3: 350.0747; found: 350.0749.
3-((3-Hydroxyphenyl)amino)-7-(trifluoromethyl)quinoxaline-2-carboxylic acid (5h): The title compound was synthesized according to the general method D from ester 4h (30.8 mg, 0.08 mmol) and potassium carbonate (33.6 mg, 0.25 mmol) in aqueous methanol (80%, 15 mL). After cooling, the solvent was removed under reduced pressure. Then, the residue was acidified with a 15% hydrochloric acid aqueous solution, and extracted with diethyl ether. The combined organic layers were washed with brine, dried over MgSO4, filtered and evaporated under reduced pressure to yield the acid 5h (33.2 mg, 100%) as a red ocher powder. Mp 178.7 °C. 1H NMR (300 MHz, DMSO-d6) δ 10.90 (bs, 1H), 9.52 (bs, 1H), 8.29 (s, 1H), 8.03 (dd, 1H, J = 8.8, 1.7 Hz), 7.92 (d, 1H, J = 8.8 Hz), 7.61 (t, 1H, J = 2.0 Hz), 7.26–7.16 (m, 2H), 6.54 (ddd, 1H, J = 7.7, 1.3 Hz). 13C NMR (75 MHz, DMSO-d6) δ 167.4, 158.3, 150.4, 144.5, 140.3, 136.0, 134.6, 130.1, 128.0 (2 × C), 127.4, 125.9 (q, J = 30 Hz), 124.7 (q, J = 270 Hz), 111.3, 111.0, 107.5. 19F RMN (282 MHz, DMSO-d6) δ—60.5. HRMS (ESI) m/z: [M+H]+ calcd for C16H11F3N3O3: 350.0747; found: 350.0747.
3.3. Docking Studies
Molecular modeling studies were performed using SYBYL-X 1.3 software [38] running on a Dell precision T3400 workstation. The three-dimensional structure of compounds 5c and 5f (under their carboxylate forms to imitate physiological conditions) were built from a standard fragments library and optimized using the Tripos force field [39] including the electrostatic term calculated from Gasteiger and Hückel atomic charges. Powell’s method available in Maximin2 procedure was used for energy minimization until the gradient value was smaller than 0.001 kcal/(mol*Å). The crystal structure of Pim-1 in complex with 5′-adenylyl-β,γ-imidodiphosphate (AMP-PNP) at 1.6 Å resolution (PDB ID 3A99) [40] was used as template for docking. Water molecules were removed from the coordinates set since no information about conserved water molecules is known for this chemical series in Pim-1. Flexible docking of compounds 5c and 5f into the ATP-binding site was performed using GOLD software [41]. The most stable docking models were selected according to the best scored conformation predicted by the Chemscore scoring function implemented in GOLD. Finally, the complexes were energy-minimized using Powell’s method available in Maximin2 procedure with the Tripos force field and a dielectric constant of 4.0, until the gradient value reached 0.1 kcal/mol.Å. Biovia Discovery Studio Visualizer [42] was used for graphical display.
3.4. Biology
3.4.1. Mammalian Protein Kinase Assays
Kinase enzymatic activities were assayed with 10 µM ATP in 384-well plates using the luminescent ADP-GloTM assay (Promega, Madison, WI, USA) according to the recommendations of the manufacturer (see [32] for details on this method). The transmitted signal was measured using the Envision (PerkinElmer, Waltham, MA, USA) microplate luminometer and expressed in Relative Light Unit (RLU). In order to determine the half maximal inhibitory concentration (IC50), the assays were performed in duplicate in the absence or presence of increasing doses of the tested compounds. GraphPad Prism6 software (GraphPad Software, San Diego, CA, USA) was used to fit dose–response curves and to determine the IC50 values. Kinase activities are expressed in % of maximal activity, i.e., measured in the absence of inhibitor. Peptide substrates were obtained from Proteogenix (Schiltigheim, France).
The following kinases were analyzed during this study: HsPim-1 and HsPim-2 (human proto-oncogene, recombinant, expressed in bacteria) were assayed with 0.20 µg/µL of consensus peptide substrate: ARKRRRHPSGPPTA; RnDYRK1A-kd (Rattus norvegicus, amino acids 1 to 499 including the kinase domain, recombinant, expressed in bacteria, DNA vector kindly provided by Dr. W. Becker, Aachen, Germany) was assayed with 0.033 μg/µL of the following peptide: KKISGRLSPIMTEQ as substrate; HsCDK2/CyclinA (human cyclin-dependent kinase-2, kindly provided by Dr. A. Echalier-Glazer, Leicester, UK) was assayed with 0.8 µg/µL of histone H1 as substrate; HsCDK9/CyclinT (human, recombinant, expressed by baculovirus in Sf9 insect cells) was assayed with 0.20 µg/µL of the following peptide: YSPTSPSYSPTSPSYSPTSPSKKKK, as substrate; HsHaspin-kd (human, kinase domain, amino acids 470 to 798, recombinant, expressed in bacteria) was assayed with 0.007 µg/µL of Histone H3 (1–21) peptide (ARTKQTARKSTGGKAPRKQLA) as substrate; MmCLK1 (from Mus musculus, recombinant, expressed in bacteria) was assayed with 0.027 μg/µL of the following peptide: GRSRSRSRSRSR as substrate; HsCK1ε (human casein kinase 1ε, recombinant, expressed by baculovirus in Sf9 insect cells) was assayed with 0.02 µg/µL of the following peptide: RRKHAAIGSpAYSITA (“Sp” stands for phosphorylated serine) as CK1-specific substrate; HsGSK-3β (human glycogen synthase kinase-3) was assayed with 0.010 µg/µL of GS-1 peptide, a GSK-3-selective substrate (YRRAAVPPSPSLSRHSSPHQSpEDEEE). For kinases expressed in bacteria, Escherichia coli BL21 DE3 pLysS strain (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) was transformed with pGEX-2T-1 (Sigma-Aldrich, St. Louis, MO, USA) containing the coding region of the corresponding kinase. For kinases expressed in Sf9 insect cells, the coding region of the corresponding kinase was cloned in pFastBacTM vector. Purification of the recombinant kinases were performed following the protocol “Bac-to-Bac® Baculovirus Expression System” provided by the manufacturer (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA).
To validate the kinase assay, model inhibitors were used for each tested enzyme: Staurosporine from Streptomyces sp. (#S5921, purity ≥ 95%, Sigma-Aldrich) for HsCK1ε; Indirubin-3′-oxime (#I0404, purity ≥ 98%, Sigma-Aldrich) for HsGSK-3β, HsPim-1 and 2, human cyclin-dependent kinases, RnDYRK1A and MmCLK1; CHR-6494 (#SML0648, purity ≥ 98%, Sigma-Aldrich) for Haspin.
3.4.2. Cell Cultures and Reagents
MV4-11 cell lines were obtained from the Deutshe Sammlung von Mikroorganismens und Zellkulturen (DSMZ). HS-27a cell lines were obtained from the American Type Culture Collection (ATCC). All cell lines were cultured in Roswell Park Memorial Institute medium (RPMI), with 10% fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin at 37 °C and 5% CO2. Bone marrow (BM) MSCs from brain-dead donors were isolated and cultured as described [43]. Informed consent was obtained before BM samples were taken. The HCT-116 cell line (colorectal carcinoma) was obtained from ATCC and cultured in McCoy’s medium with 10% fetal bovine serum without antibiotics at 37 °C and 5% CO2.
3.4.3. In Vitro Cell-Based Assays
Cell viability was studied using an MTT cell proliferation assay. To determine the concentration effect of the molecules, 0.2 × 105 MV4-11 leukemic cells or 0.1 × 105 HS-27a cells were incubated in 100 μL of RPMI red phenol-free medium (ThermoFisher Scientific), 0.035 × 105 MSCs were incubated in 100 µL of alphaMEM medium (Gibco) and 0.05 × 105 HCT-116 cells were incubated in 100 µL of McCoy’s medium, in 96-well plates for 48 h and then treated with quinoxalines (stock solution at 50 mM in DMSO) or SGI-1776 (SAB Signalway antibody, stock solution at 50 mM in DMSO), as reference, with concentrations ranging from 100 nM to 100 μM for 48 h.
Cells were incubated with 10 μL of MTT working solution (5 g/L of methylthiazolyldiphenyl-tetrazolium bromide from Sigma Aldrich, Lyon, France) during 4 h. Cells were then lysed overnight at 37 °C with 100 μL of 10% sodium dodecyl sulfate (SDS) and 0.003% HCl. Optical density (OD) at 570 nm was measured using a spectrophotometer CLARIOstar® (BMG Labtech, Offenburg, Germany) or Mithras Multimode Microplate Reader LB940 (Berthold, Versailles, France). Living cells were also counted with the trypan blue dye exclusion method. When a dose-dependent activity was observed, EC50 values were calculated using Graphpad PRISM 7 software (n = 3 in triplicate). Data were collected from at least three independent experiments and the values reported are means ± standard errors of the mean (SEM).
4. Conclusions
In summary, using a structure-based design approach, we have developed a new promising quinoxaline-2-carboxylic acid series of dual Pim-1/2 inhibitors. Starting from the lead compound 1, moderately active on Pim-2, we significantly improved the inhibition profile on Pim-2 isoform by adding halogenated substituents in position 6. Docking studies demonstrated that this 6-halogenated group was oriented towards the unique hydrophobic pocket of the Pim kinases hinge environment, contributing to increase van der Waals interactions in this area. Two lead compounds, 5c (6-Cl) and 5e (6-Br), were then identified, exhibiting submicromolar potency on both Pim-1 and Pim-2 isoforms, with an interesting selectivity profile against the panel of mammalian kinases studied. In vitro cell-based assays on human hematologic (AML) and solid tumor (colorectal carcinoma) cell lines overexpressing Pim-1/2 kinases were then realized, showing growth inhibitory activities at micromolar concentrations. These encouraging results make them promising new leads for further pharmacomodulation studies.
Supplementary Materials
The following are available online, spectroscopic data for final compounds, and Figure S1: Inhibition curves of compound 5c on HsPim1 (A), HsPim2 (B), RnDYRK1A (C), and HsGSK3β (D), Figure S2: Inhibition curves of compound 5e on HsPim1 (A), HsPim2 (B), RnDYRK1A (C), and HsGSK3β (D).
Author Contributions
Conceptualization, C.D.-S. and C.L.; methodology, C.D.-S. and M.-C.V.-M.; investigation, B.O., M.B.-B., C.L., C.D.-S., F.G., S.I., W.R., C.C., J.G., N.P., T.R. and S.B.; writing—original draft preparation, C.D.-S. and C.L.; writing—review and editing, C.D.-S. and C.L.; supervision, C.D.-S. and M.-C.V.-M.; project administration, C.D.-S. and M.-C.V.-M.; funding acquisition, M.-C.V.-M., P.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the « Association pour le Développement de la Recherche et de l’Innovation dans le NORD PAS DE CALAIS » (ADRINORD) (to B.O.). The authors also thank the Cancéropôle Grand Ouest (3MC network—Marine Molecules, Metabolism and Cancer), GIS IBiSA (Infrastructures en Biologie Santé et Agronomie) and Biogenouest (Western France life science and environment core facility network) for supporting the KISSf screening facility. The authors wish to thank the Région Centre Val de Loire and LabEx SynOrg (ANR-11-LABX-0029) for financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors thank the « Plateforme Scientifique et Technique Analyses des Systèmes Biologiques » (PST-ASB), Tours (France), for NMR spectrometry and the « Fédération de Recherche » ICOA/CBM (FR2708) platform for HRMS analyses.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Theo Cuypers, H.; Selten, G.; Quint, W.; Zijlstra, M.; Maandag, E.R.; Boelens, W.; van Wezenbeek, P.; Melief, C.; Berns, A. Murine Leukemia Virus-Induced T-Cell Lymphomagenesis: Integration of Proviruses in a Distinct Chromosomal Region. Cell 1984, 37, 141–150. [Google Scholar] [CrossRef]
- Bachmann, M.; Kosan, C.; Xing, P.X.; Montenarh, M.; Hoffmann, I.; Möröy, T. The Oncogenic Serine/Threonine Kinase Pim-1 Directly Phosphorylates and Activates the G2/M Specific Phosphatase Cdc25C. Int. J. Biochem. Cell Biol. 2006, 38, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Narlik-Grassow, M.; Blanco-Aparicio, C.; Carnero, A. The PIM Family of Serine/Threonine Kinases in Cancer. Med. Res. Rev. 2014, 34, 136–159. [Google Scholar] [CrossRef] [PubMed]
- Nawijn, M.C.; Alendar, A.; Berns, A. For Better or for Worse: The Role of Pim Oncogenes in Tumorigenesis. Nat. Rev. Cancer 2010, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Tursynbay, Y.; Zhang, J.; Li, Z.; Tokay, T.; Zhumadilov, Z.; Wu, D.; Xie, Y. Pim-1 Kinase as Cancer Drug Target: An Update. Biomed. Rep. 2016, 4, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Kitanaka, C.; Noguchi, K.; Muramatsu, T.; Asai, A.; Kuchino, Y. Physical and Functional Interactions between Pim-1 Kinase and Cdc25A Phosphatase: Implications for the pim-1-mediated activation of the c-myc signaling pathway. J. Biol. Chem. 1999, 274, 18659–18666. [Google Scholar] [CrossRef]
- Leung, C.O.; Wong, C.C.; Fan, D.N.; Kai, A.K.; Tung, E.K.; Xu, I.M.; Ng, I.O.; Lo, R.C. PIM1 Regulates Glycolysis and Promotes Tumor Progression in Hepatocellular Carcinoma. Oncotarget 2015, 6, 10880–10892. [Google Scholar] [CrossRef]
- Keane, N.A.; Reidy, M.; Natoni, A.; Raab, M.S.; O’Dwyer, M. Targeting the Pim Kinases in Multiple Myeloma. Blood Cancer J. 2015, 5, e325. [Google Scholar] [CrossRef]
- Amson, R.; Sigaux, F.; Przedborski, S.; Flandrin, G.; Givol, D.; Telerman, A. The Human Protooncogene Product P33pim Is Expressed during Fetal Hematopoiesis and in Diverse Leukemias. Proc. Natl. Acad. Sci. USA 1989, 86, 8857–8861. [Google Scholar] [CrossRef]
- Dhanasekaran, S.M.; Barrette, T.R.; Ghosh, D.; Shah, R.; Varambally, S.; Kurachi, K.; Pienta, K.J.; Rubin, M.A.; Chinnaiyan, A.M. Delineation of Prognostic Biomarkers in Prostate Cancer. Nature 2001, 412, 822–826. [Google Scholar] [CrossRef]
- Brasó-Maristany, F.; Filosto, S.; Catchpole, S.; Marlow, R.; Quist, J.; Francesch-Domenech, E.; Plumb, D.A.; Zakka, L.; Gazinska, P.; Liccardi, G.; et al. PIM1 Kinase Regulates Cell Death, Tumor Growth and Chemotherapy Response in Triple-Negative Breast Cancer. Nat. Med. 2016, 22, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, U.; Beckmann, N.; Thomas, M.; Grünweller, A.; Huber, K.; Bracher, F.; Hartmann, R.K.; Aigner, A. Functional Role and Therapeutic Potential of the Pim-1 Kinase in Colon Carcinoma. Neoplasia 2013, 15, 783-IN28. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Li, Y.-Y. Use of Regulators and Inhibitors of Pim-1, a Serine/Threonine Kinase, for Tumour Therapy (Review). Mol. Med. Rep. 2014, 9, 2051–2060. [Google Scholar] [CrossRef] [PubMed]
- Isaac, M.; Siu, A.; Jongstra, J. The Oncogenic PIM Kinase Family Regulates Drug Resistance through Multiple Mechanisms. Drug Resist. Updat. 2011, 14, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Darby, R.A.J.; Unsworth, A.; Knapp, S.; Kerr, I.D.; Callaghan, R. Overcoming ABCG2-Mediated Drug Resistance with Imidazo-[1,2-b]-Pyridazine-Based Pim1 Kinase Inhibitors. Cancer Chemother. Pharmacol. 2015, 76, 853–864. [Google Scholar] [CrossRef]
- Mikkers, H.; Nawijn, M.; Allen, J.; Brouwers, C.; Verhoeven, E.; Jonkers, J.; Berns, A. Mice Deficient for All PIM Kinases Display Reduced Body Size and Impaired Responses to Hematopoietic Growth Factors. Mol. Cell. Biol. 2004, 24, 6104–6115. [Google Scholar] [CrossRef]
- Asati, V.; Mahapatra, D.K.; Bharti, S.K. PIM Kinase Inhibitors: Structural and Pharmacological Perspectives. Eur. J. Med. Chem. 2019, 172, 95–108. [Google Scholar] [CrossRef]
- Blanco-Aparicio, C.; Carnero, A. Pim Kinases in Cancer: Diagnostic, Prognostic and Treatment Opportunities. Biochem. Pharmacol. 2013, 85, 629–643. [Google Scholar] [CrossRef]
- Foulks, J.M.; Carpenter, K.J.; Luo, B.; Xu, Y.; Senina, A.; Nix, R.; Chan, A.; Clifford, A.; Wilkes, M.; Vollmer, D.; et al. A Small-Molecule Inhibitor of PIM Kinases as a Potential Treatment for Urothelial Carcinomas. Neoplasia 2014, 16, 403–412. [Google Scholar] [CrossRef]
- Cortes, J.; Tamura, K.; DeAngelo, D.J.; de Bono, J.; Lorente, D.; Minden, M.; Uy, G.L.; Kantarjian, H.; Chen, L.S.; Gandhi, V.; et al. Phase I Studies of AZD1208, a Proviral Integration Moloney Virus Kinase Inhibitor in Solid and Haematological Cancers. Br. J. Cancer 2018, 118, 1425–1433. [Google Scholar] [CrossRef]
- Raab, M.S.; Thomas, S.K.; Ocio, E.M.; Guenther, A.; Goh, Y.-T.; Talpaz, M.; Hohmann, N.; Zhao, S.; Xiang, F.; Simon, C.; et al. The First-in-Human Study of the Pan-PIM Kinase Inhibitor PIM447 in Patients with Relapsed and/or Refractory Multiple Myeloma. Leukemia 2019, 33, 2924–2933. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mandiyan, V.; Suzuki, Y.; Zhang, C.; Rice, J.; Tsai, J.; Artis, D.R.; Ibrahim, P.; Bremer, R. Crystal Structures of Proto-Oncogene Kinase Pim1: A Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma. J. Mol. Biol. 2005, 348, 183–193. [Google Scholar] [CrossRef]
- Bullock, A.N.; Russo, S.; Amos, A.; Pagano, N.; Bregman, H.; Debreczeni, J.É.; Lee, W.H.; von Delft, F.; Meggers, E.; Knapp, S. Crystal Structure of the PIM2 Kinase in Complex with an Organoruthenium Inhibitor. PLoS ONE 2009, 4, e7112. [Google Scholar] [CrossRef] [PubMed]
- Koblish, H.; Li, Y.; Shin, N.; Hall, L.; Wang, Q.; Wang, K.; Covington, M.; Marando, C.; Bowman, K.; Boer, J.; et al. Preclinical Characterization of INCB053914, a Novel Pan-PIM Kinase Inhibitor, Alone and in Combination with Anticancer Agents, in Models of Hematologic Malignancies. PLoS ONE 2018, 13, e0199108. [Google Scholar] [CrossRef] [PubMed]
- Czardybon, W.; Windak, R.; Gołas, A.; Gałęzowski, M.; Sabiniarz, A.; Dolata, I.; Salwińska, M.; Guzik, P.; Zawadzka, M.; Gabor-Worwa, E.; et al. A Novel, Dual Pan-PIM/FLT3 Inhibitor SEL24 Exhibits Broad Therapeutic Potential in Acute Myeloid Leukemia. Oncotarget 2018, 9. [Google Scholar] [CrossRef]
- Oyallon, B.; Brachet-Botineau, M.; Logé, C.; Bonnet, P.; Souab, M.; Robert, T.; Ruchaud, S.; Bach, S.; Berthelot, P.; Gouilleux, F.; et al. Structure-Based Design of Novel Quinoxaline-2-Carboxylic Acids and Analogues as Pim-1 Inhibitors. Eur. J. Med. Chem. 2018, 154, 101–109. [Google Scholar] [CrossRef]
- Le, B.T.; Kumarasiri, M.; Adams, J.R.; Yu, M.; Milne, R.; Sykes, M.J.; Wang, S. Targeting Pim Kinases for Cancer Treatment: Opportunities and Challenges. Future Med. Chem. 2015, 7, 35–53. [Google Scholar] [CrossRef]
- Mahesh, R.; Dhar, A.K.; Sasank, T.V.N.V.T.; Thirunavukkarasu, S.; Devadoss, T. Citric Acid: An Efficient and Green Catalyst for Rapid One Pot Synthesis of Quinoxaline Derivatives at Room Temperature. Chin. Chem. Lett. 2011, 22, 389–392. [Google Scholar] [CrossRef]
- Mahesh, R.; Devadoss, T.; Dhar, A.K.; Venkatesh, S.M.; Mundra, S.; Pandey, D.K.; Bhatt, S.; Jindal, A.K. Ligand-Based Design, Synthesis, and Pharmacological Evaluation of 3-Methoxyquinoxalin-2-Carboxamides as Structurally Novel Serotonin Type-3 Receptor Antagonists. Arch. Pharm. (Weinheim) 2012, 345, 687–694. [Google Scholar] [CrossRef]
- Takano, Y.; Shiga, F.; Asano, J.; Ando, N.; Uchiki, H.; Fukuchi, K.; Anraku, T. Design, Synthesis, and AMPA Receptor Antagonistic Activity of a Novel 6-Nitro-3-Oxoquinoxaline-2-Carboxylic Acid with a Substituted Phenyl Group at the 7 Position. Bioorg. Med. Chem. 2005, 13, 5841–5863. [Google Scholar] [CrossRef]
- Takano, Y.; Shiga, F.; Asano, J.; Hori, W.; Fukuchi, K.; Anraku, T.; Uno, T. Design and Synthesis of Novel 7-Heterocycle-6-Trifluoromethyl-3-Oxoquinoxaline-2-Carboxylic Acids Bearing a Substituted Phenyl Group as Superior AMPA Receptor Antagonists with Good Physicochemical Properties. Bioorg. Med. Chem. 2006, 14, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Cambridge Crystallographic Data Centre. Supplementary X-ray Crystallographic Data (CCDC-2051184, CCDC-2051185 and CCDC-2051186); Cambridge Crystallographic Data Centre: Cambridge, UK, 2020; Available online: https://www.ccdc.cam.ac.uk/ (accessed on 18 December 2020).
- Zegzouti, H.; Zdanovskaia, M.; Hsiao, K.; Goueli, S.A. ADP-Glo: A Bioluminescent and Homogeneous ADP Monitoring Assay for Kinases. ASSAY Drug Dev. Technol. 2009, 7, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Bissantz, C.; Kuhn, B.; Stahl, M. A Medicinal Chemist’s Guide to Molecular Interactions. J. Med. Chem. 2010, 53, 5061–5084. [Google Scholar] [CrossRef]
- Mizuki, M.; Schwäble, J.; Steur, C.; Choudhary, C.; Agrawal, S.; Sargin, B.; Steffen, B.; Matsumura, I.; Kanakura, Y.; Böhmer, F.D.; et al. Suppression of Myeloid Transcription Factors and Induction of STAT Response Genes by AML-Specific Flt3 Mutations. Blood 2003, 101, 3164–3173. [Google Scholar] [CrossRef]
- Kim, K.-T.; Baird, K.; Ahn, J.-Y.; Meltzer, P.; Lilly, M.; Levis, M.; Small, D. Pim-1 Is up-Regulated by Constitutively Activated FLT3 and Plays a Role in FLT3-Mediated Cell Survival. Blood 2005, 105, 1759–1767. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, H.; Wang, F.; Han, Y.; Yang, W. Pim-2 Modulates Aerobic Glycolysis and Energy Production during the Development of Colorectal Tumors. Int. J. Med. Sci. 2015, 12, 487–493. [Google Scholar] [CrossRef]
- Tripos Associates, Inc. SYBYL-X 1.3; Tripos Associates, Inc.: St. Louis, MO, USA, 2013. [Google Scholar]
- Clark, M.; Cramer, R.D.; Van Opdenbosch, N. Validation of the General Purpose Tripos 5.2 Force Field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Morishita, D.; Takami, M.; Yoshikawa, S.; Katayama, R.; Sato, S.; Kukimoto-Niino, M.; Umehara, T.; Shirouzu, M.; Sekimizu, K.; Yokoyama, S.; et al. Cell-Permeable Carboxyl-Terminal P27 Kip1 Peptide Exhibits Anti-Tumor Activity by Inhibiting Pim-1 Kinase. J. Biol. Chem. 2011, 286, 2681–2688. [Google Scholar] [CrossRef]
- Jones, G.; Willet, P.; Glen, R.C. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes. Dassault Systèmes BIOVIA, Discovery Studio Visualizer, v19.1.0.18287; Dassault Systèmes: San Diego, CA, USA, 2019. [Google Scholar]
- Ponte, A.L.; Ribeiro-Fleury, T.; Chabot, V.; Gouilleux, F.; Langonné, A.; Hérault, O.; Charbord, P.; Domenech, J. Granulocyte-Colony-Stimulating Factor Stimulation of Bone Marrow Mesenchymal Stromal Cells Promotes CD34+ Cell Migration Via a Matrix Metalloproteinase-2-Dependent Mechanism. Stem Cells Dev. 2012, 21, 3162–3172. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).