Characterization of Signalling Pathways That Link Apoptosis and Autophagy to Cell Death Induced by Estrone Analogues Which Reversibly Depolymerize Microtubules
Abstract
1. Introduction
2. Results
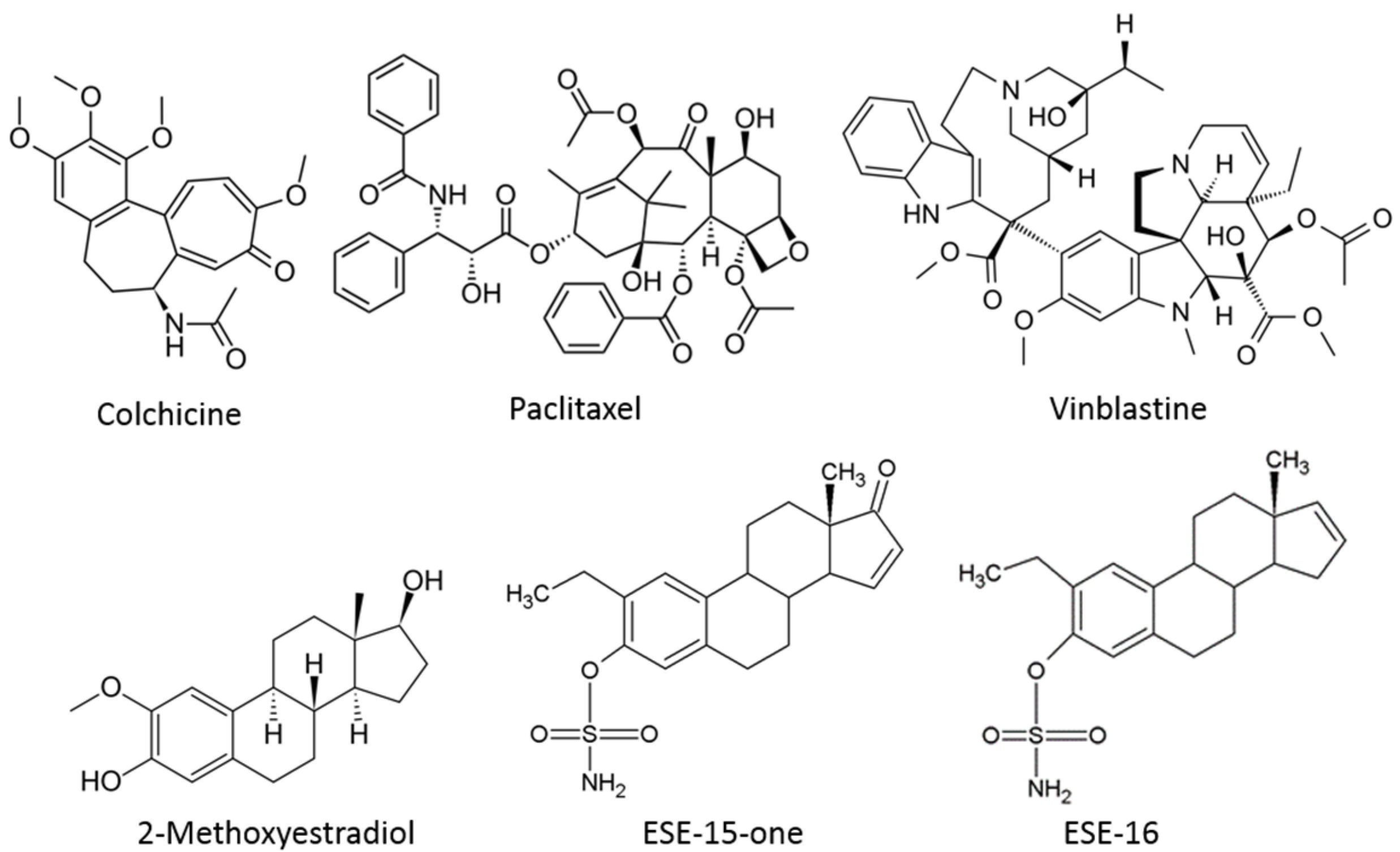
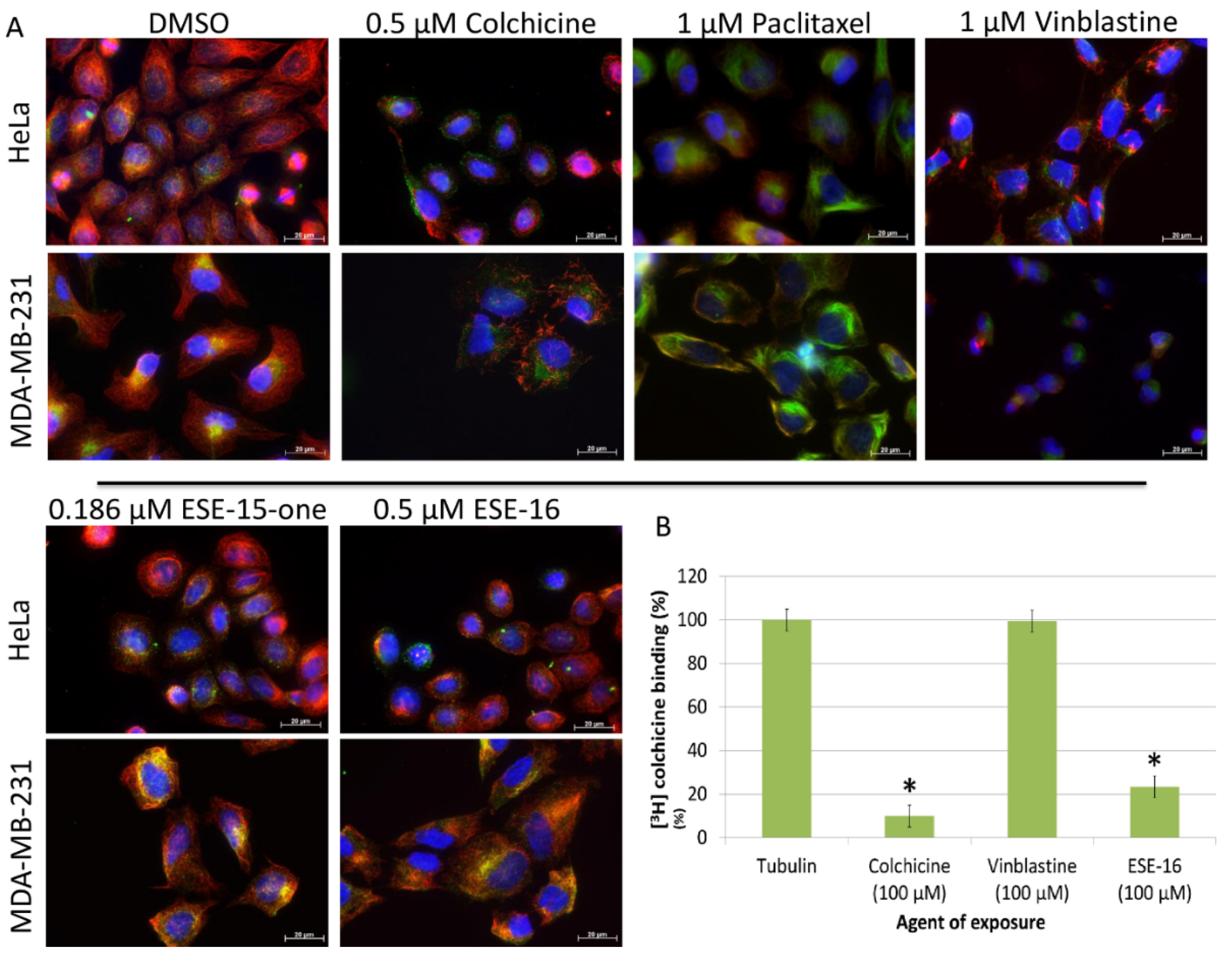
2.1. ESE-15-one and ESE-16 Effect Microtubules by Binding to the Colchicine Binding Site
2.2. The Microtubule Depolymerizing Effects of the 2-ME Analogues Are Reversible
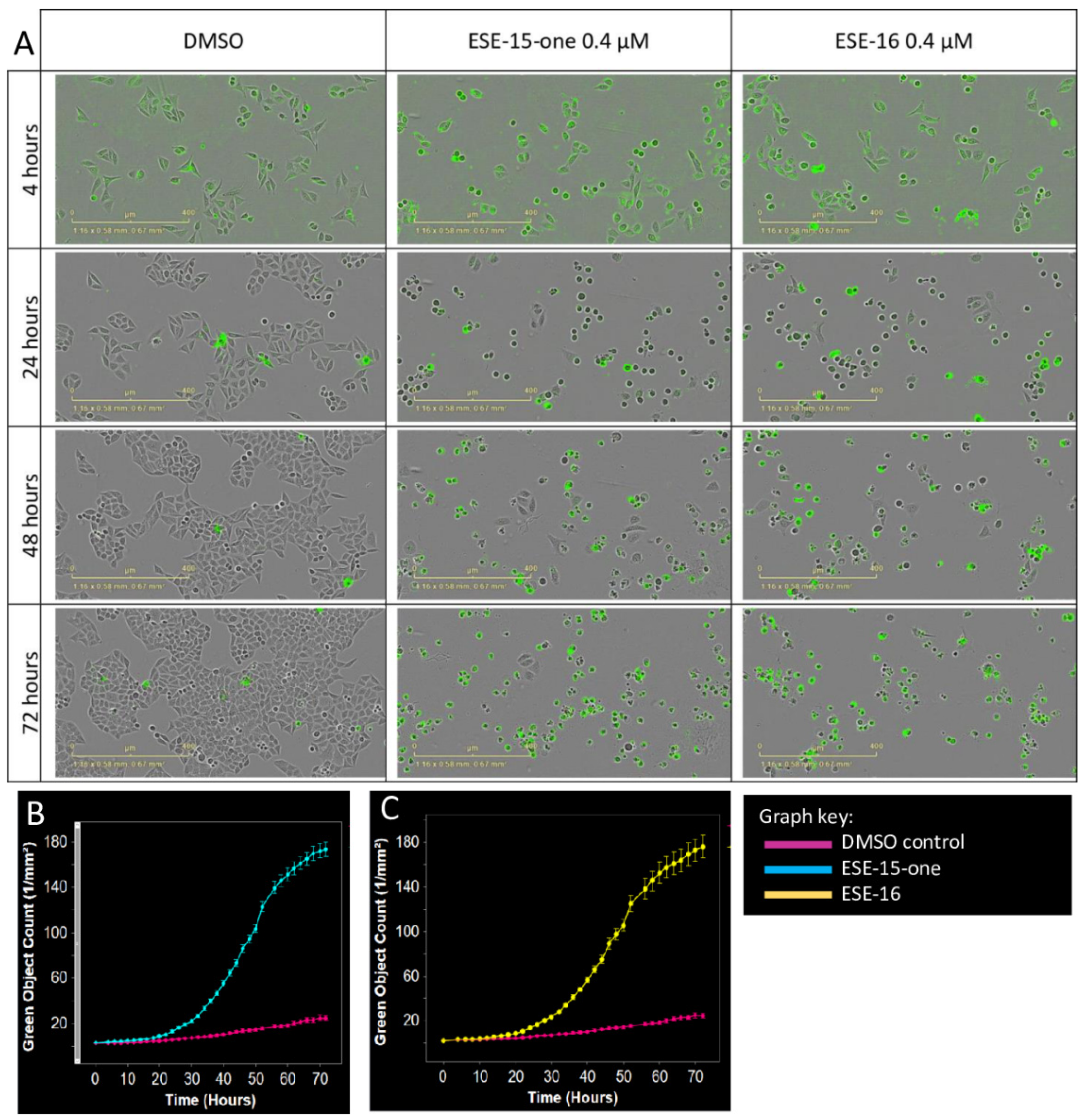
2.3. Kinetics of Cell Death Differ Slightly in Response to ESE-15-one and ESE-16 Exposure
2.4. Changes in the Mitochondrial Transmembrane Potential Are Not Detected after a 2-h Drug-Exposure
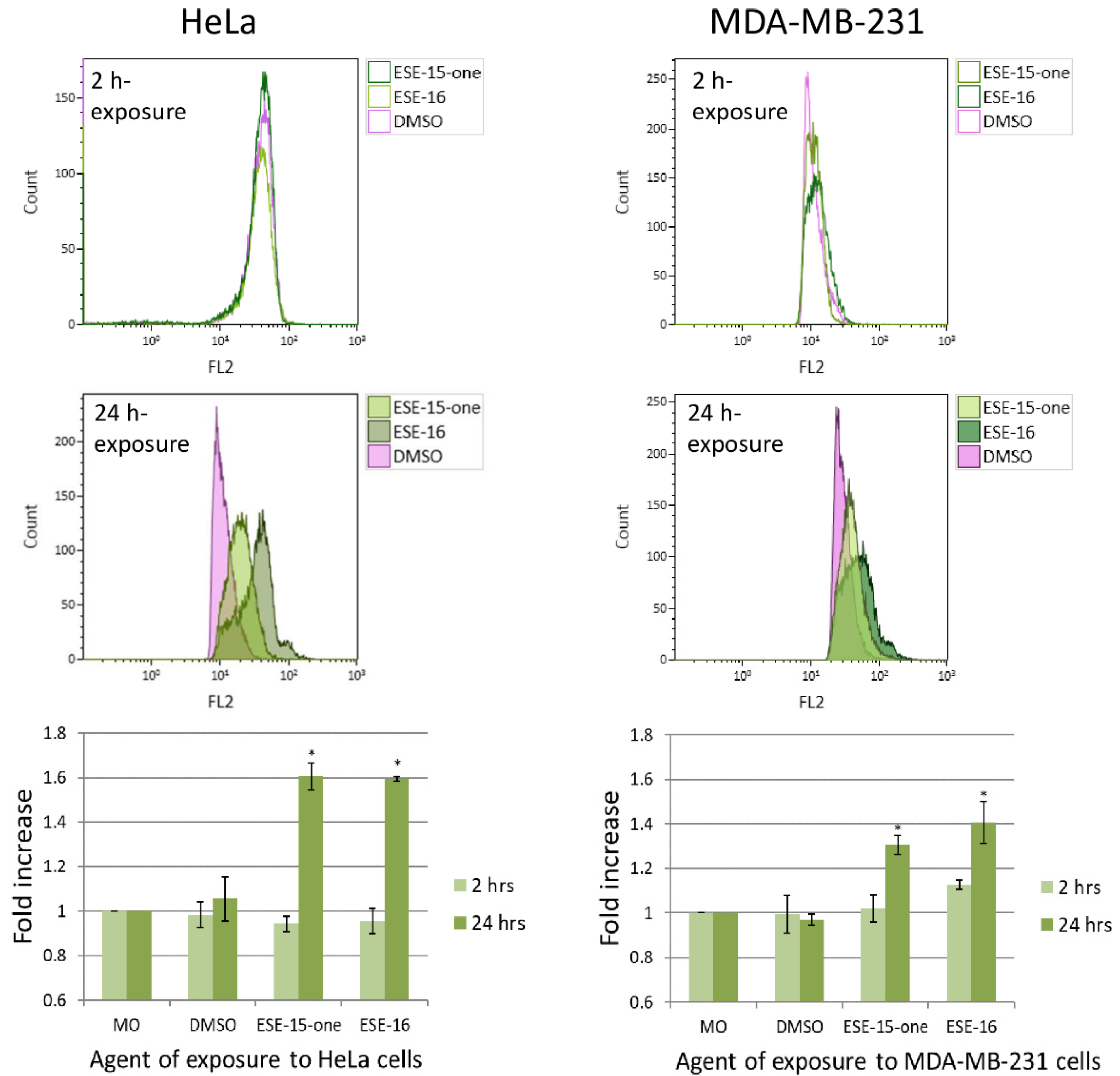
2.5. Superoxide Production Is Increased by ESE-15-one and ESE-16-Exposure in Both Cell Lines
2.6. The 2-ME Analogues Cause a Prolonged Transit through Metaphase and Induced a G2/M Block
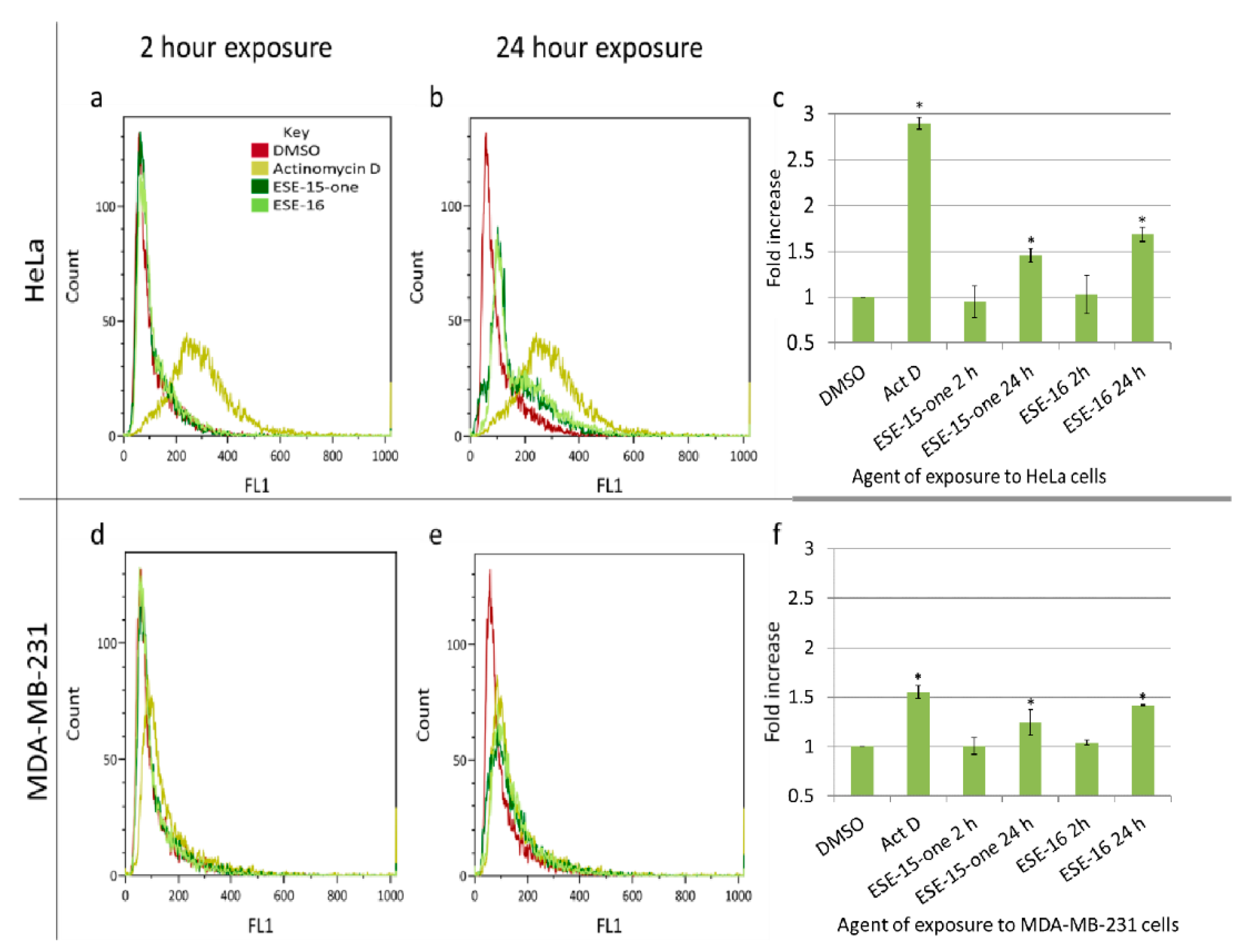
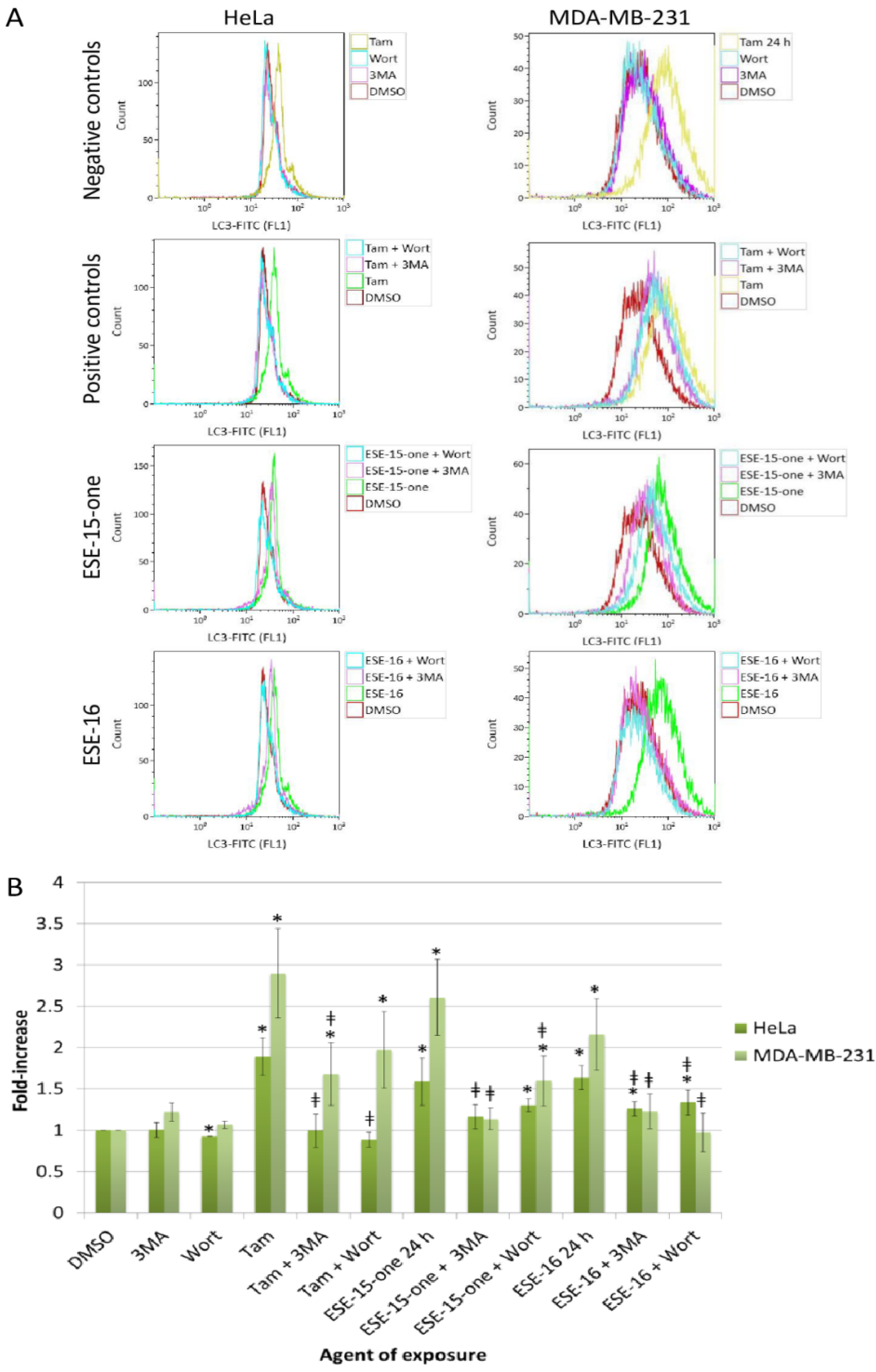
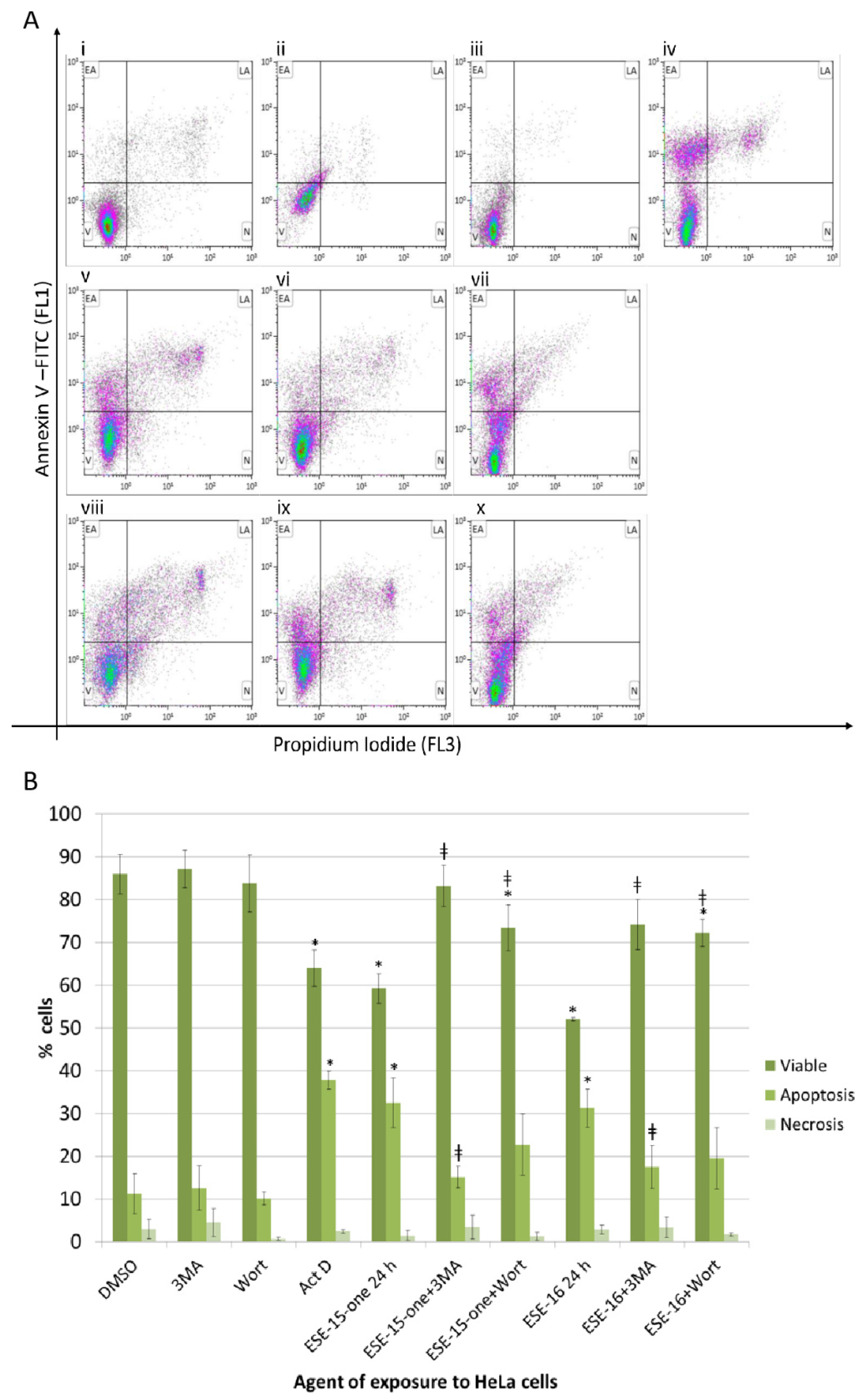
2.7. The Estrone Analogues Increase LC3B Expression and Induce Apoptosis, a Phenomenon Which Is Partially Attenuated if Autophagy Is Blocked
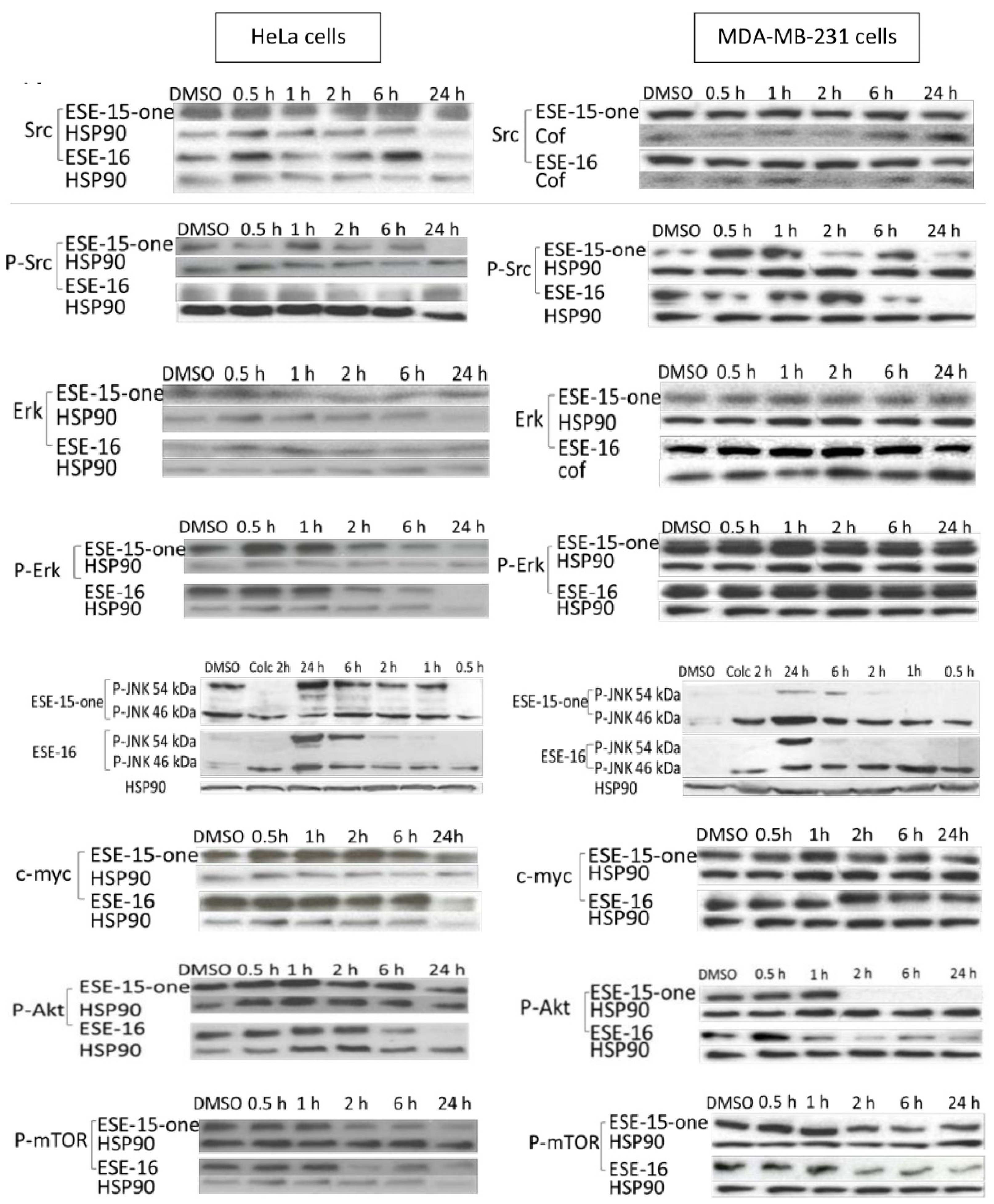
2.8. Signal Transduction Pathways: RPPA and Western Blot Analyses Highlight Signalling Involved in Cell Cycle Progression, Autophagy and Apoptosis
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Culture Methods and Chemicals
4.2. Tubulin Competitive Binding Assay: Tritiated [3H] Colchicine Binding
4.3. Immunofluorescence Microscopy
4.4. Reversibility of Microtubule Toxicity Effects
4.5. Kinetics of Cytotoxicity
4.6. Changes in the Mitochondrial Transmembrane Potential: Flow Cytometric Quantification Utilizing the MitoCaptureTM Mitochondrial Apoptosis Detection Kit
4.7. Generation of Reactive Oxygen Species: Flow Cytometric Quantification of Superoxide Molecules
4.8. Time-Lapse Imaging
4.9. Cell Cycle Analysis
4.10. Apoptosis Assay
4.11. Autophagic Vacuole Quantification
4.12. Inhibition of Autophagy
4.13. Reverse Phase Protein Array Analyses
4.14. Western Blots
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Nigg, E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001, 2, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, E.; Sinnappan, S.; Munoz, M.A.; Kavallaris, M. ENMD-1198, a new analogue of 2-methoxyestradiol, displays both antiangiogenic and vascular-disrupting properties. Mol. Cancer Ther. 2010, 9, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, S.; Knipling, L.; Sackett, D.L.; Wolff, J. Localization of the colchicine-binding site of tubulin. Proc. Natl. Acad. Sci. USA 1993, 90, 11598–11602. [Google Scholar] [CrossRef]
- Gigant, B.; Wang, C.; Ravelli, R.B.G.; Roussi, F.; Steinmetz, M.O.; Curmi, P.A.; Sobel, A.; Knossow, M. Structural basis for the regulation of tubulin by vinblastine. Nature 2005, 435, 519–522. [Google Scholar] [CrossRef]
- Matson, D.R.; Stukenberg, P.T. Spindle Poisons and Cell Fate: A Tale of Two Pathways. Mol. Interv. 2011, 11, 141–150. [Google Scholar] [CrossRef]
- Riedl, S.J.; Salvesen, G.S. The apoptosome: Signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 2007, 8, 405–413. [Google Scholar] [CrossRef]
- Mooberry, S.L. Mechanism of action of 2-methoxyestradiol: New developments. Drug Resist. Updat. 2003, 6, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Tkaczuk, K.R.; Yared, J.A. Update on taxane development: New analogs and new formulations. Drug Des. Dev. Ther. 2012, 6, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A.; Kamath, K. How do microtubule-targeted drugs work? An overview. Curr. Cancer Drug Targets 2007, 7, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Da, J.; Landstrom, M.; Ulmsten, U.; Fu, X. Antiproliferative activity and toxicity of 2-methoxyestradiol in cervical cancer xenograft mice. Int. J. Gynecol. Cancer 2005, 15, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Tevaarwerk, A.J.; Holen, K.D.; Alberti, D.B.; Sidor, C.; Arnott, J.; Quon, C.; Wilding, G.; Liu, G. Phase I Trial of 2-Methoxyestradiol NanoCrystal Dispersion in Advanced Solid Malignancies. Clin. Cancer Res. 2009, 15, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, N.J.; Sarkar, M.A.; Venitz, J.; Figg, W.D. 2-Methoxyestradiol, a promising anticancer agent. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2003, 23, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, C.J.; Liu, G.; Yiannoutsos, C.; Kolesar, J.; Horvath, D.; Staab, M.J.; Fife, K.; Armstrong, V.; Treston, A.; Sidor, C.; et al. A Phase II Multicenter, Randomized, Double-Blind, Safety Trial Assessing the Pharmacokinetics, Pharmacodynamics, and Efficacy of Oral 2-Methoxyestradiol Capsules in Hormone-Refractory Prostate Cancer. Clin. Cancer Res. 2005, 11, 6625–6633. [Google Scholar] [CrossRef]
- Mabjeesh, N.J.; Escuin, D.; Lavallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 2003, 3, 363–375. [Google Scholar] [CrossRef]
- Van Zijl, C.; Lottering, M.; Steffens, F.; Joubert, A.M. In vitro effects of 2-methoxyestradiol on MCF-12A and MCF-7 cell growth, morphology and mitotic spindle formation. Cell Biochem. Funct. 2008, 26, 632–642. [Google Scholar] [CrossRef]
- D’Amato, R.J.; Lin, C.M.; Flynn, E.; Folkman, J.; Hamel, E. 2-Methoxyestradiol, an endogenous mammalian metabolite, inhibits tubulin polymerization by interacting at the colchicine site. Proc. Natl. Acad. Sci. USA 1994, 91, 3964–3968. [Google Scholar] [CrossRef]
- Mooberry, S.L. New insights into 2-methoxyestradiol, a promising antiangiogenic and antitumor agent. Curr. Opin. Oncol. 2003, 15, 425–430. [Google Scholar] [CrossRef]
- Stander, A.; Joubert, F.; Joubert, A.M. Docking, Synthesis, and in vitro Evaluation of Antimitotic Estrone Analogs. Chem. Biol. Drug Des. 2011, 77, 173–181. [Google Scholar] [CrossRef]
- Theron, A.E.; Prudent, R.; Nolte, E.; Bout, I.V.D.; Punchoo, R.; Marais, S.; Du Toit, P.; Hlophe, Y.; Van Papendorp, D.; Lafanechère, L.; et al. Novel in silico-designed estradiol analogues are cytotoxic to a multidrug-resistant cell line at nanomolar concentrations. Cancer Chemother. Pharmacol. 2015, 75, 431–437. [Google Scholar] [CrossRef]
- Chiche, J.; Ilc, K.; Laferrière, J.; Trottier, E.; Dayan, F.; Mazure, N.M.; Brahimi-Horn, M.C.; Pouysségur, J. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009, 69, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Desai, K.G.H.; Mallery, S.R.; Schwendeman, S.P. Effect of formulation parameters on 2-methoxyestradiol release from injectable cylindrical poly(dl-lactide-co-glycolide) implants. Eur. J. Pharm. Biopharm. 2008, 70, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Marais, S.; Mqoco, T.; Stander, A.; van Papendorp, D.; Joubert, A. The in vitro effects of a sulphamoylated derivative of 2-methoxyestradiol on cell number, morphology and alpha-tubulin disruption in cervical adenocarcinoma (HeLa) cells. Biomed. Res. 2012, 23, 357–362. [Google Scholar]
- Theron, A.E.; Nolte, E.M.; Lafanechère, L.; Joubert, A.M. Molecular crosstalk between apoptosis and autophagy induced by a novel 2-methoxyestradiol analogue in cervical adenocarcinoma cells. Cancer Cell Int. 2013, 13, 87. [Google Scholar] [CrossRef]
- Edsall, A.B.; Mohanakrishnan, A.K.; Yang, D.; Fanwick, P.E.; Hamel, E.; Hanson, A.D.; Agoston, G.E.; Cushman, M. Effects of Altering the Electronics of 2-Methoxyestradiol on Cell Proliferation, on Cytotoxicity in Human Cancer Cell Cultures, and on Tubulin Polymerization. J. Med. Chem. 2004, 47, 5126–5139. [Google Scholar] [CrossRef]
- Visagie, M.; Michelle, H.; Theron, A.; Anne, E.; Mqoco, T.; Thandi, V.; Vieira, W.A.; Prudent, R.; Martinez, A.; Lafanechère, L.; et al. Sulphamoylated 2-Methoxyestradiol Analogues Induce Apoptosis in Adenocarcinoma Cell Lines. PLoS ONE 2013, 8, e71935. [Google Scholar] [CrossRef]
- Nkandeu, D.S.; Mqoco, T.; Visagie, M.H.; Stander, B.A.; Wolmarans, E.; Cronjé, M.J.; Joubert, A.M. In vitro changes in mitochondrial potential, aggresome formation and caspase activity by a novel 17-β-estradiol analogue in breast adenocarcinoma cells. Cell Biochem. Funct. 2013, 31, 566–574. [Google Scholar] [CrossRef]
- De Forges, H.; Bouissou, A.; Perez, F. Interplay between microtubule dynamics and intracellular organization. Int. J. Biochem. Cell Biol. 2012, 44, 266–274. [Google Scholar] [CrossRef]
- Rovini, A.; Savry, A.; Braguer, D.; Carré, M. Microtubule-targeted agents: When mitochondria become essential to chemotherapy. Biochim. Biophys. Acta BBA Bioenerg. 2011, 1807, 679–688. [Google Scholar] [CrossRef]
- Poillet-Perez, L.; Despouy, G.; Delage-Mourroux, R.; Boyer-Guittaut, M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015, 4, 184–192. [Google Scholar] [CrossRef]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, T.; Roth, J.A. Superinduction of wild-type p53 protein after 2-methoxyestradiol treatment of Ad5p53-transduced cells induces tumor cell apoptosis. Oncogene 1998, 17, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Li, S.; Westermarck, J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008, 22, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Minemoto, Y.; Lin, A. c-Jun N-Terminal Protein Kinase 1 (JNK1), but Not JNK2, Is Essential for Tumor Necrosis Factor Alpha-Induced c-Jun Kinase Activation and Apoptosis. Mol. Cell. Biol. 2004, 24, 10844–10856. [Google Scholar] [CrossRef] [PubMed]
- Dreskin, S.C.; Thomas, G.W.; Dale, S.N.; Heasley, L.E. Isoforms of Jun Kinase Are Differentially Expressed and Activated in Human Monocyte/Macrophage (THP-1) Cells. J. Immunol. 2001, 166, 5646–5653. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Prudent, R.; Vassal-Stermann, É.; Nguyen, C.-H.; Mollaret, M.; Viallet, J.; Desroches-Castan, A.; Martinez, A.; Barette, C.; Pillet, C.; Valdameri, G.; et al. Azaindole derivatives are inhibitors of microtubule dynamics, with anti-cancer and anti-angiogenic activities. Br. J. Pharmacol. 2013, 168, 673–685. [Google Scholar] [CrossRef]
- Polioudaki, H.; Kastrinaki, M.-C.; Papadaki, H.A.; Theodoropoulos, P.A. Microtubule-interacting drugs induce moderate and reversible damage to human bone marrow mesenchymal stem cells. Cell Prolif. 2009, 42, 434–447. [Google Scholar] [CrossRef]
- Blajeski, A.L.; Phan, V.A.; Kottke, T.J.; Kaufmann, S.H. G(1) and G(2) cell-cycle arrest following microtubule depolymerization in human breast cancer cells. J. Clin. Investig. 2002, 110, 91–99. [Google Scholar] [CrossRef]
- Currier, A.W.; Kolb, E.A.; Gorlick, R.G.; Roth, M.E.; Gopalakrishnan, V.; Sampson, V.B. p27/Kip1 functions as a tumor suppressor and oncoprotein in osteosarcoma. Sci. Rep. 2019, 9, 6161. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Gottlieb, R.A.; Andres, A.M.; Sin, J.; Taylor, D.P. Untangling autophagy measurements: All fluxed up. Circ. Res. 2015, 116, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Wolmarans, E.; Mqoco, T.; Thandi, V.; Stander, A.; Nkandeu, S.D.; Sippel, K.H.; McKenna, R.; Joubert, A.M. Novel estradiol analogue induces apoptosis and autophagy in esophageal carcinoma cells. Cell. Mol. Biol. Lett. 2014, 19, 98–115. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.I.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [PubMed]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992. [Google Scholar] [CrossRef]
- Bonet-Ponce, L.; Saez-Atienzar, S.; Da Casa, C.; Sancho-Pelluz, J.; Barcia, J.M.; Martinez-Gil, N.; Nava, E.; Jordan, J.; Romero, F.J.; Galindo, M.F. Rotenone Induces the Formation of 4-Hydroxynonenal Aggresomes. Role of ROS-Mediated Tubulin Hyperacetylation and Autophagic Flux Disruption. Mol. Neurobiol. 2015, 53, 6194–6208. [Google Scholar] [CrossRef]
- Viola, G.; Bortolozzi, R.; Hamel, E.; Moro, S.; Brun, P.; Castagliuolo, I.; Ferlin, M.G.; Basso, G. MG-2477, a new tubulin inhibitor, induces autophagy through inhibition of the Akt/mTOR pathway and delayed apoptosis in A549 cells. Biochem. Pharmacol. 2012, 83, 16–26. [Google Scholar] [CrossRef]
- Xie, B.; Zhao, H.-C.; Yao, S.-K.; Zhuo, D.-X.; Lv, D.-C.; Wu, C.-L.; Ma, D.-L.; Gao, C.; Shu, X.-M.; Ai, Z.-L. Autophagy inhibition enhances etoposide-induced cell death in human hepatoma G2 cells. Int. J. Mol. Med. 2011, 27, 599–606. [Google Scholar] [CrossRef]
- Qadir, M.A.; Kwok, B.; Dragowska, W.H.; To, K.H.; Le, D.; Bally, M.B.; Gorski, S.M. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 2008, 112, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Desai, B.N.; Myers, B.R.; Schreiber, S.L. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2002, 99, 4319–4324. [Google Scholar] [CrossRef] [PubMed]
- Shanware, N.P.; Bray, K.; Abraham, R.T. The PI3K, Metabolic, and Autophagy Networks: Interactive Partners in Cellular Health and Disease. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 89–106. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Calnan, D.R.; Brunet, A. The FoxO code. Oncogene 2008, 27, 2276–2288. [Google Scholar] [CrossRef]
- Tang, H.; Lee, M.; Budak, M.T.; Pietras, N.; Hittinger, S.; Vu, M.; Khuong, A.; Hoang, C.D.; Hussain, S.N.A.; Levine, S.; et al. Intrinsic apoptosis in mechanically ventilated human diaphragm: Linkage to a novel Fos/FoxO1/Stat3-Bim axis. FASEB J. 2011, 25, 2921–2936. [Google Scholar] [CrossRef]
- Park, S.-J.; Sohn, H.-Y.; Yoon, J.; Park, S.I. Down-regulation of FoxO-dependent c-FLIP expression mediates TRAIL-induced apoptosis in activated hepatic stellate cells. Cell. Signal. 2009, 21, 1495–1503. [Google Scholar] [CrossRef]
- Alessi, D.R.; Kozlowski, M.T.; Weng, Q.-P.; Morrice, N.; Avruch, J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol. 1998, 8, 69–81. [Google Scholar] [CrossRef]
- Chung, J.; Grammar, T.C.; Lemon, K.P.; Kazlauskas, A.; Blenis, J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nat. Cell Biol. 1994, 370, 71–75. [Google Scholar] [CrossRef]
- Hidalgo, M.; Rowinsky, E.K. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 2000, 19, 6680–6686. [Google Scholar] [CrossRef]
- Grewe, M.; Gansauge, F.; Schmid, R.M.; Adler, G.; Seufferlein, T. Regulation of cell growth and cyclin D1 expression by the constitutively active FRAP-p70s6K pathway in human pancreatic cancer cells. Cancer Res. 1999, 59, 3581–3587. [Google Scholar] [PubMed]
- Seufferlein, T.; Rozengurt, E. Rapamycin inhibits constitutive p70s6k phosphorylation, cell proliferation, and colony formation in small cell lung cancer cells. Cancer Res. 1996, 56, 3895–3897. [Google Scholar] [PubMed]
- Oh, W.J.; Jacinto, E. mTOR complex 2 signaling and functions. Cell Cycle 2011, 10, 2305–2316. [Google Scholar] [CrossRef] [PubMed]
- Datan, E.; Shirazian, A.; Benjamin, S.; Matassov, D.; Tinari, A.; Malorni, W.; Lockshin, R.A.; Garcia-Sastre, A.; Zakeri, Z. mTOR/p70S6K signaling distinguishes routine, maintenance-level autophagy from autophagic cell death during influenza A infection. Virology 2014, 175–190. [Google Scholar] [CrossRef]
- Betz, C.; Stracka, D.; Prescianotto-Baschong, C.; Frieden, M.; Demaurex, N.; Hall, M.N. mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA 2013, 110, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Patergnani, S.; Missiroli, S.; Marchi, S.; Giorgi, C. Mitochondria-Associated Endoplasmic Reticulum Membranes Microenvironment: Targeting Autophagic and Apoptotic Pathways in Cancer Therapy. Front. Oncol. 2015, 5, 173. [Google Scholar] [CrossRef]
- Lee, S.T.; Lee, J.Y.; Han, C.R.; Kim, Y.H.; Jun, D.Y.; Taub, D.; Kim, Y. Dependency of 2-methoxyestradiol-induced mitochondrial apoptosis on mitotic spindle network impairment and prometaphase arrest in human Jurkat T cells. Biochem. Pharmacol. 2015, 94, 257–269. [Google Scholar] [CrossRef]
- Chauhan, D.; Hideshima, T.; Podar, K.; Mitsiades, C.; Munshi, N.C.; Kharbanda, S.; Anderson, K.C.; Li, G.; Mitsiades, N. JNK-dependent Release of Mitochondrial Protein, Smac, during Apoptosis in Multiple Myeloma (MM) Cells. J. Biol. Chem. 2003, 278, 17593–17596. [Google Scholar] [CrossRef]
- Liu, J.; Lin, A. Role of JNK activation in apoptosis: A double-edged sword. Cell Res. 2005, 15, 36–42. [Google Scholar] [CrossRef]
- Larroquecadoso, P.; Swiader, A.; Ingueneau, C.; Negresalvayre, A.; Elbaz, M.; Reyland, M.E.; Salvayre, R.; Vindis, C. Role of protein kinase C δ in ER stress and apoptosis induced by oxidized LDL in human vascular smooth muscle cells. Cell Death Dis. 2013, 4, e520. [Google Scholar] [CrossRef]
- Cagnol, S.; Chambard, J.-C. ERK and cell death: Mechanisms of ERK-induced cell death—Apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Forman, H.J. Redox signaling and the MAP kinase pathways. BioFactors 2003, 17, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life 2006, 58, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.; Blaukat, A.; Fu, X.; Heldin, N.-E.; Landström, M. Mechanisms for 2-methoxyestradiol-induced apoptosis of prostate cancer cells. FEBS Lett. 2002, 531, 141–151. [Google Scholar] [CrossRef]
- Fukui, M.; Zhu, B.T. Mechanism of 2-methoxyestradiol-induced apoptosis and growth arrest in human breast cancer cells. Mol. Carcinog. 2009, 48, 66–78. [Google Scholar] [CrossRef]
- Wong, C.H.; Iskandar, K.B.; Yadav, S.K.; Hirpara, J.L.; Loh, T.; Pervaiz, S. Simultaneous Induction of Non-Canonical Autophagy and Apoptosis in Cancer Cells by ROS-Dependent ERK and JNK Activation. PLoS ONE 2010, 5, e9996. [Google Scholar] [CrossRef]
- Paturle-Lafanechere, L.; Edde, B.; Denoulet, P.; Van Dorsselaer, A.; Mazarguil, H.; Le Caer, J.P.; Wehland, J.; Job, D. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry 1991, 30, 10523–10528. [Google Scholar] [CrossRef]
- Paturle-Lafanechère, L.; Manier, M.; Trigault, N.; Pirollet, F.; Mazarguil, H.; Job, D. Accumulation of delta 2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J. Cell Sci. 1994, 107, 1529–1543. [Google Scholar]
- Paturle, L.; Wehland, J.; Margolis, R.L.; Job, D.; Lafanechère, L. Complete separation of tyrosinated, detyrosinated, and nontyrosinatable brain tubulin subpopulations using affinity chromatography. Biochemistry 1989, 28, 2698–2704. [Google Scholar] [CrossRef]
- Troncale, S.; Barbet, A.; Coulibaly, L.; Henry, E.; He, B.; Barillot, E.; Dubois, T.; Hupé, P.; De Koning, L. NormaCurve: A SuperCurve-Based Method That Simultaneously Quantifies and Normalizes Reverse Phase Protein Array Data. PLoS ONE 2012, 7, e38686. [Google Scholar] [CrossRef] [PubMed]


| Antibody name | Pathway(s) | HeLa | MDA-MB-231 | ||
|---|---|---|---|---|---|
| ESE-15-One | ESE-16 | ESE-15-One | ESE-16 | ||
| Beclin-1 | Autophagy, apoptosis | - | - | ↑1–2 h | ↑1–2 h |
| Puma (C-Term) | Autophagy, apoptosis | - | - | ↑ 0.5–6 h | ↑ 0.5–6 h |
| TRAIL | Apoptosis | ? Mild ↑ 0.5-6 h | ? | mild ↑ over 24 h | mild ↑ over 24 h |
| P-PAK1 (S144) | Cytoskeleton, cell cycle, apoptosis | ↑ 1–6 h | - | ↑ 1–6 h | - |
| P-Rb (S807/811) | Checkpoint, cell cycle | ↑↑ over 24 h | ↑↑ over 24 h | ↑ over 24 h | ↑ over 24 h |
| P-p27 KIP1 (T198) | Checkpoint, cell cycle | ↑↑ over 24 h | ↑↑ over 24 h | ↑↑ over 24 h | ↑↑ over 24 h |
| p15 INK4B | Cell cycle | ↑ up to 2 h then plateau | ↑ up to 2 h then plateau | ↑ up to 2 h then plateau | ↑ up to 2 h then plateau |
| Akt | PI3K pathway | ? Mild ↑ 0.5–2 h | - | ? Mild ↑ 0.5–2 h | - |
| P-PTEN (S380/T382/383) | PI3K pathway | ? Mild ↑ 16–24 h | ? Mild ↑ 16–24 h | - | - |
| FOXO1 (C29H4) | PI3K pathway | ↓↓ over 24 h | ↓↓ over 24 h | mild ↓ over 24h | mild ↓ over 24h |
| PI3 Kinase p110 β-subunit | PI3K pathway | - | - | ↑ 1–6 h | ↑1–6 h |
| P-Akt (S473) (193H12) | PI3K pathway | ↓ over 24 h | ↓ over 24 h | - | - |
| P-Akt substrate (RXRXX S/T) | PI3K pathway | ↑ up to 2 h | ↑ up to 2 h | ↑ 1–2 h | ↑ 1–2 h |
| P-mTOR (S2448) | PI3K pathway | ↑ 0.5–6h, then ↓ | ↑ 0.5–6h, then ↓ | ↑ 0.5–6h, then ↓ | ↑ 0.5–6h, then ↓ |
| P-p70 S6 kinase (T421/S424) | PI3K pathway | ↑ 12–24 h | ↑ 12–24 h | Mild ↑ 12–24 h | Mild ↑ 12–24 h |
| P-Stat3 (S727) | JAK/STAT signalling | - | - | Mild ↑ 0.5–6 h | Mild ↑ 0.5–6 h |
| P-Smad3 (S423/425) | TGF-β signalling | ? ↑1–6 h | ? ↑ 1–6 h | ↑ 1–6 h | ↑ 1–6 h |
| Proteins apparently not affected by compound exposure | P-p53 (S15), β-catenin (6B3), Dvl3, PKA C-α, P-AMPK β1 (S181), P-NF-kB p65 (S536) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mercier, A.E.; Prudent, R.; Pepper, M.S.; De Koning, L.; Nolte, E.; Peronne, L.; Nel, M.; Lafanechère, L.; Joubert, A.M. Characterization of Signalling Pathways That Link Apoptosis and Autophagy to Cell Death Induced by Estrone Analogues Which Reversibly Depolymerize Microtubules. Molecules 2021, 26, 706. https://doi.org/10.3390/molecules26030706
Mercier AE, Prudent R, Pepper MS, De Koning L, Nolte E, Peronne L, Nel M, Lafanechère L, Joubert AM. Characterization of Signalling Pathways That Link Apoptosis and Autophagy to Cell Death Induced by Estrone Analogues Which Reversibly Depolymerize Microtubules. Molecules. 2021; 26(3):706. https://doi.org/10.3390/molecules26030706
Chicago/Turabian StyleMercier, Anne E., Renaud Prudent, Michael S. Pepper, Leanne De Koning, Elsie Nolte, Lauralie Peronne, Marcel Nel, Laurence Lafanechère, and Anna M. Joubert. 2021. "Characterization of Signalling Pathways That Link Apoptosis and Autophagy to Cell Death Induced by Estrone Analogues Which Reversibly Depolymerize Microtubules" Molecules 26, no. 3: 706. https://doi.org/10.3390/molecules26030706
APA StyleMercier, A. E., Prudent, R., Pepper, M. S., De Koning, L., Nolte, E., Peronne, L., Nel, M., Lafanechère, L., & Joubert, A. M. (2021). Characterization of Signalling Pathways That Link Apoptosis and Autophagy to Cell Death Induced by Estrone Analogues Which Reversibly Depolymerize Microtubules. Molecules, 26(3), 706. https://doi.org/10.3390/molecules26030706








