Desert Environments Facilitate Unique Evolution of Biosynthetic Potential in Streptomyces
Abstract
1. Introduction
2. Results and Discussion
2.1. Taxonomic Identification and Phylogenetic Position of Strain SAJ15
2.2. Description of Draft Genome of Streptomyces sp. SAJ15
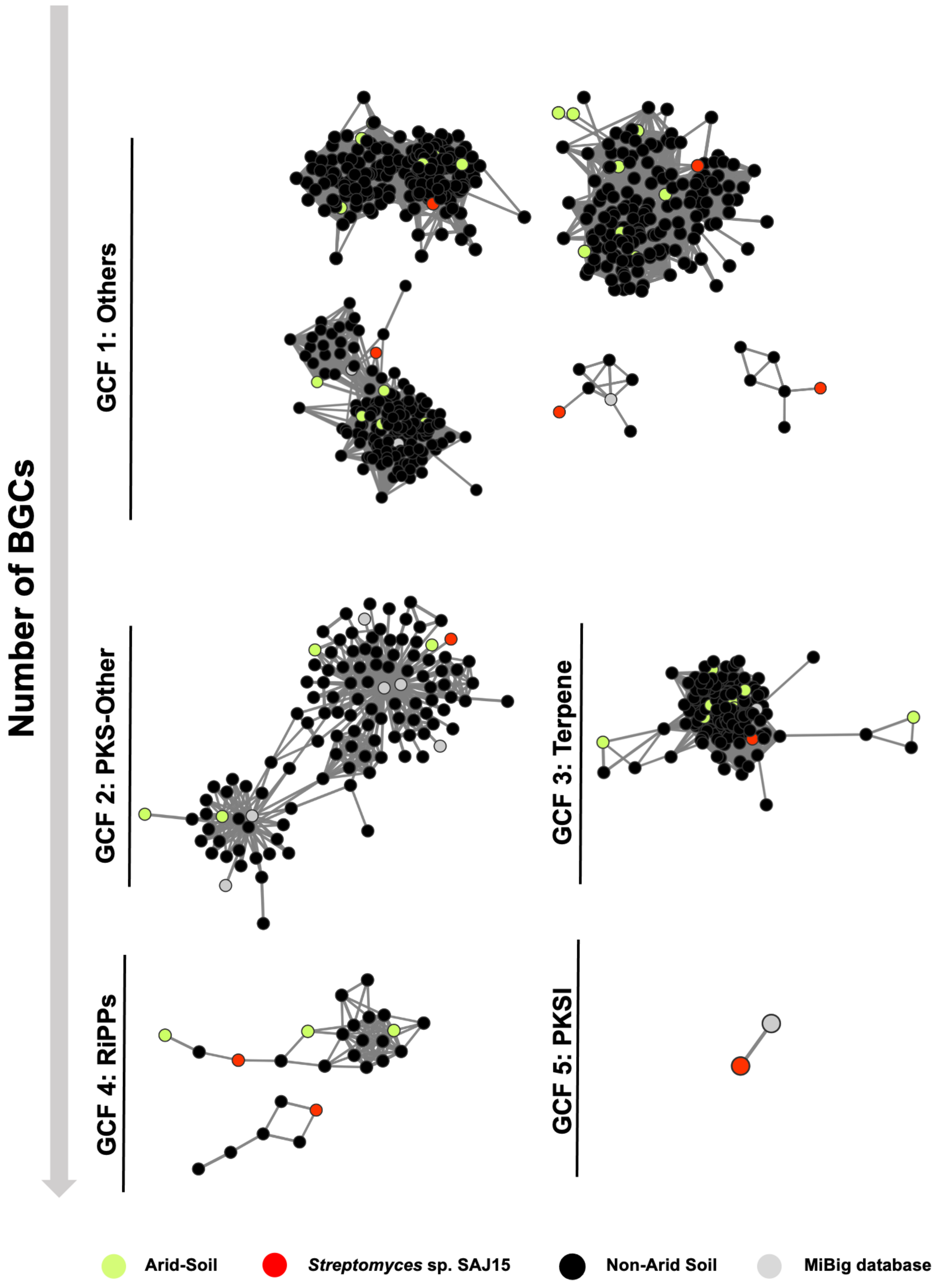
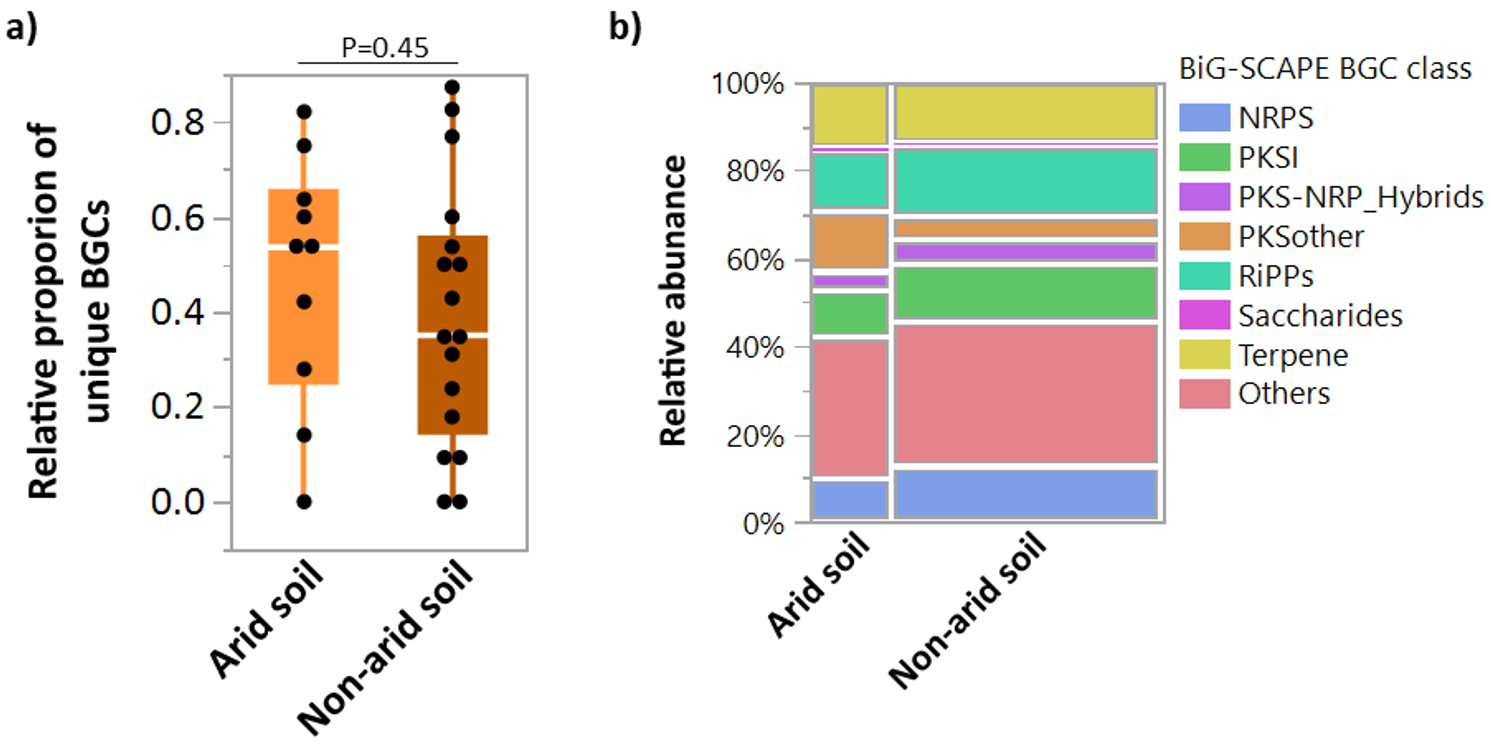
2.3. Genome Mining of Novel Biosynthetic Gene Clusters in SAJ15
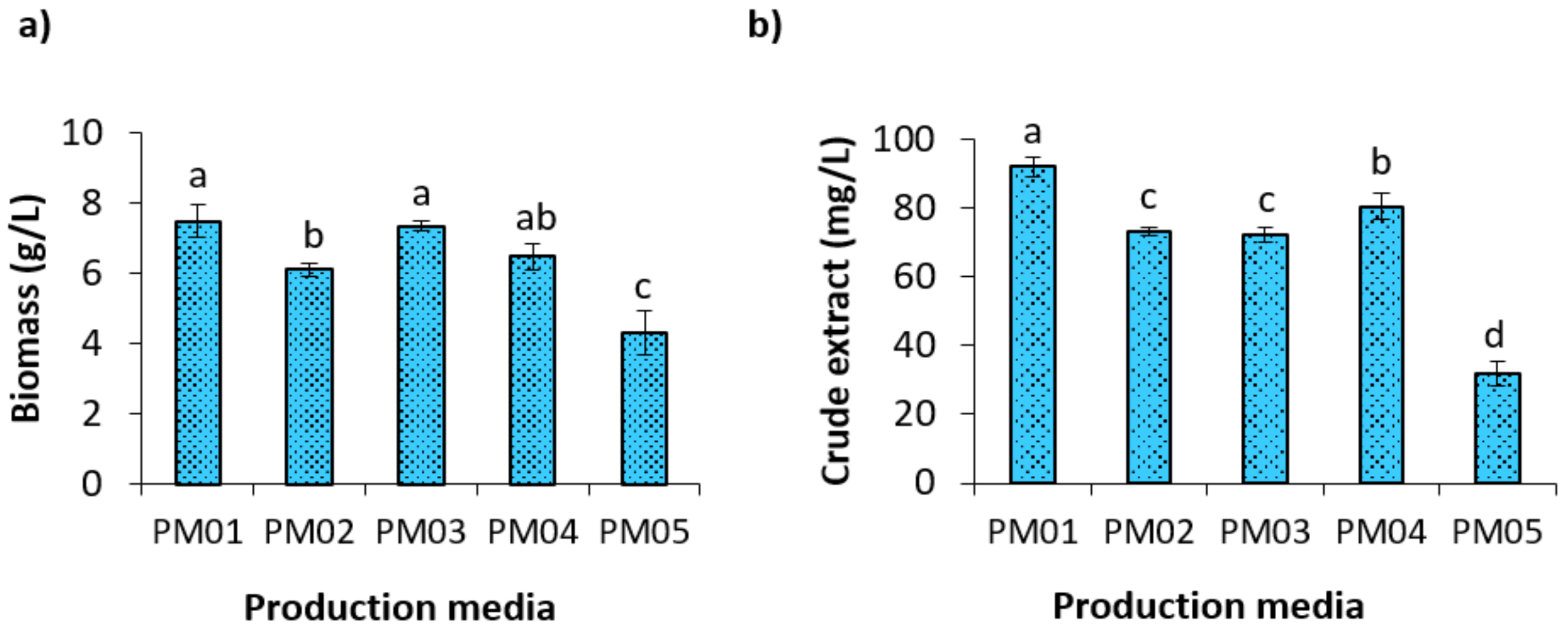
2.4. HPLC(PDA)-Guided Selection of Fermentation Medium
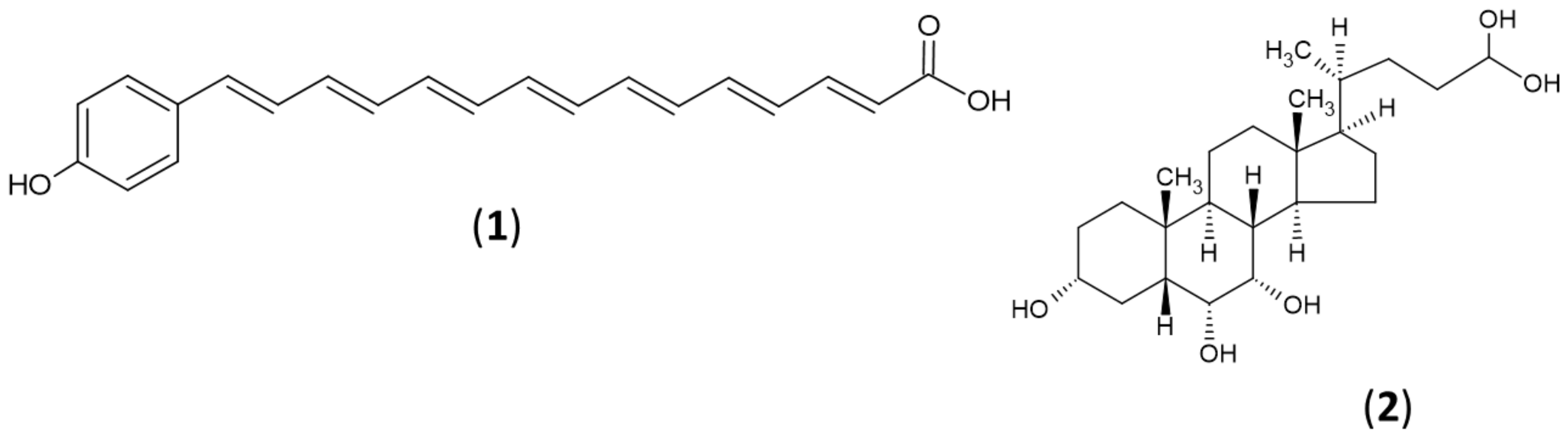
2.5. Secondary Metabolites Profiling Using HR-LC-MS
2.6. Integration of Metabolite Profile with BGCs
3. Materials and Methods
3.1. Strain and Culture Condition
3.2. 16S rDNA-Based Strain Identification
3.3. Genome Sequencing, Assembly, and Annotation
3.4. Genome Mining of Novel Biosynthetic Gene Clusters in SAJ15
3.5. Determination of Growth and Metabolite Production
3.5.1. HPLC-PDA Analysis
3.5.2. HR-LC-MS Analysis
3.5.3. HR-LC-MS Data Mapping to BGCs of Strain SAJ15
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Procopio, R.E.; Silva, I.R.; Martins, M.K.; Azevedo, J.L.; Araujo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.B.; Balachandran, L. Antibacterial agents from actinomycetes—A review. Front. Biosci. 2012, 4, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef]
- Mohammadipanah, F.; Wink, J. Actinobacteria from Arid and Desert Habitats: Diversity and Biological Activity. Front. Microbiol. 2015, 6, 1541. [Google Scholar] [CrossRef]
- Masand, M.; Sivakala, K.K.; Menghani, E.; Thinesh, T.; Anandham, R.; Sharma, G.; Sivakumar, N.; Jebakumar, S.R.D.; Jose, P.A. Biosynthetic Potential of Bioactive Streptomycetes Isolated From Arid Region of the Thar Desert, Rajasthan (India). Front. Microbiol. 2018, 9, 687. [Google Scholar] [CrossRef]
- Jose, P.A.; Jebakumar, S.R.D. Diverse Actinomycetes from Indian Coastal Solar Salterns—A Resource for Antimicrobial Screening. J. Pure Appl. Microbiol. 2013, 7, 2569–2575. [Google Scholar]
- Guo, X.; Liu, N.; Li, X.; Ding, Y.; Shang, F.; Gao, Y.; Ruan, J.; Huang, Y. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol. 2015, 81, 3086–3103. [Google Scholar] [CrossRef]
- Jose, P.A.; Jha, B. Intertidal marine sediment harbours Actinobacteria with promising bioactive and biosynthetic potential. Sci. Rep. 2017, 7, 10041. [Google Scholar] [CrossRef]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep Sea Actinomycetes and Their Secondary Metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef]
- Pajares, S.; Escalante, A.E.; Noguez, A.M.; Garcia-Oliva, F.; Martinez-Piedragil, C.; Cram, S.S.; Eguiarte, L.E.; Souza, V. Spatial heterogeneity of physicochemical properties explains differences in microbial composition in arid soils from Cuatro Cienegas, Mexico. Peerj 2016, 4, e2459. [Google Scholar] [CrossRef] [PubMed]
- Sivakala, K.K.; Jose, P.A.; Anandham, R.; Thinesh, T.; Jebakumar, S.R.D.; Samaddar, S.; Chatterjee, P.; Sivakumar, N.; Sa, T. Spatial Physiochemical and Metagenomic Analysis of Desert Environment. J. Microbiol. Biotechnol. 2018, 28, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Neilson, J.W.; Kushwaha, P.; Maier, R.M.; Barberán, A. Life-history strategies of soil microbial communities in an arid ecosystem. ISME J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Upadhyay, D.J.; Mosker, E.; Sussmuth, R.; Gupta, R.K. Culturable bioactive actinomycetes from the Great Indian Thar Desert. Ann. Microbiol. 2015, 65, 1901–1914. [Google Scholar] [CrossRef]
- Arocha-Garza, H.F.; Canales-Del Castillo, R.; Eguiarte, L.E.; Souza, V.; De la Torre-Zavala, S. High diversity and suggested endemicity of culturable Actinobacteria in an extremely oligotrophic desert oasis. Peerj 2017, 5, e3247. [Google Scholar] [CrossRef] [PubMed]
- Pait, I.G.U.; Kitani, S.; Roslan, F.W.; Ulanova, D.; Arai, M.; Ikeda, H.; Nihira, T. Discovery of a new diol-containing polyketide by heterologous expression of a silent biosynthetic gene cluster from Streptomyces lavendulae FRI-5. J. Ind. Microbiol. Biotechnol. 2018, 45, 77–87. [Google Scholar] [CrossRef]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Ruckert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jha, B. New Dimensions of Research on Actinomycetes: Quest for Next Generation Antibiotics. Front. Microbiol. 2016, 7, 1295. [Google Scholar] [CrossRef]
- Harrison, J.; Studholme, D.J. Recently published Streptomyces genome sequences. Microb. Biotechnol. 2014, 7, 373–380. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Alt, S.; Bibb, M.J. Next Generation Sequencing of Actinobacteria for the Discovery of Novel Natural Products. Mar. Drugs 2016, 14, 78. [Google Scholar] [CrossRef]
- Yang, D.; Teng, Q.; Dong, L.B.; Crnovcic, I.; Huang, T.; Ge, H.; Shen, B. Genome Mining of Streptomyces mobaraensis DSM40847 as a Bleomycin Producer Providing a Biotechnology Platform To Engineer Designer Bleomycin Analogues. Org. Lett. 2017, 19, 1386–1389. [Google Scholar]
- Sun, C.L.; Yang, Z.J.; Zhang, C.Y.; Liu, Z.Y.; He, J.Q.; Liu, Q.; Zhang, T.Y.; Ju, J.H.; Ma, J.Y. Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of Its Biosynthetic Gene Cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Kaweewan, I.; Komaki, H.; Hemmi, H.; Hoshino, K.; Hosaka, T.; Isokawa, G.; Oyoshi, T.; Kodani, S. Isolation and structure determination of a new cytotoxic peptide, curacozole, from Streptomyces curacoi based on genome mining. J. Antibiot. 2019, 72, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Katsuyama, Y.; Onaka, H.; Fujie, M.; Satoh, N.; Shin-Ya, K.; Ohnishi, Y. Production of a Novel Amide-Containing Polyene by Activating a Cryptic Biosynthetic Gene Cluster in Streptomyces sp. MSC090213JE08. Chembiochem 2016, 17, 1464–1471. [Google Scholar] [CrossRef]
- Cruz-Morales, P.; Ramos-Aboites, H.E.; Licona-Cassani, C.; Selem-Mojica, N.; Mejia-Ponce, P.M.; Souza-Saldivar, V.; Barona-Gomez, F. Actinobacteria phylogenomics, selective isolation from an iron oligotrophic environment and siderophore functional characterization, unveil new desferrioxamine traits. FEMS Microbiol. Ecol. 2017, 93, fix086. [Google Scholar] [CrossRef]
- Dorador, C.; Meneses, D.; Urtuvia, V.; Demergasso, C.; Vila, I.; Witzel, K.-P.; Imhoff, J.F. Diversity ofBacteroidetesin high-altitude saline evaporitic basins in northern Chile. J. Geophys. Res. Biogeosci. 2009, 114. [Google Scholar] [CrossRef]
- Busarakam, K.; Bull, A.T.; Girard, G.; Labeda, D.P.; van Wezel, G.P.; Goodfellow, M. Streptomyces leeuwenhoekii sp. nov. the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie Leeuwenhoek 2014, 105, 849–861. [Google Scholar] [CrossRef]
- Elsayed, S.S.; Trusch, F.; Deng, H.; Raab, A.; Prokes, I.; Busarakam, K.; Asenjo, J.A.; Andrews, B.A.; van West, P.; Bull, A.T.; et al. Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert. J. Org. Chem. 2015, 80, 10252–10260. [Google Scholar] [CrossRef]
- Rateb, M.E.; Houssen, W.E.; Arnold, M.; Abdelrahman, M.H.; Deng, H.; Harrison, W.T.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Ferguson, G.; et al. Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J. Nat. Prod. 2011, 74, 1491–1499. [Google Scholar] [CrossRef]
- Castro, J.F.; Razmilic, V.; Gomez-Escribano, J.P.; Andrews, B.; Asenjo, J.; Bibb, M. The ‘gifted’ actinomycete Streptomyces leeuwenhoekii. Antonie Leeuwenhoek 2018, 111, 1433–1448. [Google Scholar] [CrossRef]
- Benaud, N.; Edwards, R.J.; Amos, T.G.; D’Agostino, P.M.; Gutiérrez-Chávez, C.; Montgomery, K.; Nicetic, I.; Ferrari, B.C. Antarctic desert soil bacteria exhibit high novel natural product potential, evaluated through long-read genome sequencing and comparative genomics. Environ. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sujarit, K.; Kudo, T.; Ohkuma, M.; Pathom-Aree, W.; Lumyong, S. Streptomyces palmae sp. nov. isolated from oil palm (Elaeis guineensis) rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Archer, S.D.; Boyle, R.H.; Lacap-Bugler, D.C.; Belnap, J.; Pointing, S.B. Niche Filtering of Bacteria in Soil and Rock Habitats of the Colorado Plateau Desert, Utah, USA. Front. Microbiol 2016, 7, 1489. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J. Genome Update. Let the consumer beware: Streptomyces genome sequence quality. Microb. Biotechnol. 2016, 9, 3–7. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Koberl, M.; White, R.A., 3rd; Erschen, S.; El-Arabi, T.F.; Jansson, J.K.; Berg, G. Draft Genome Sequence of Streptomyces sp. Strain Wb2n-11, a Desert Isolate with Broad-Spectrum Antagonism against Soilborne Phytopathogens. Genome Announc. 2015, 3, e00860-15. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Sadeghi, A.; Soltani, B.M.; Nekouei, M.K.; Jouzani, G.S.; Mirzaei, H.H.; Sadeghizadeh, M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. [Google Scholar] [CrossRef]
- Czech, L.; Hoppner, A.; Kobus, S.; Seubert, A.; Riclea, R.; Dickschat, J.S.; Heider, J.; Smits, S.H.J.; Bremer, E. Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis. Sci. Rep. 2019, 9, 364. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoneyama, K.; Shiraishi, S.; Watanabe, A.; Takahashi, N. Chemical Structures of New Piericidins Produced by Streptomyces-Pactum. Agric. Biol. Chem. 1977, 41, 855–862. [Google Scholar] [CrossRef]
- Urakawa, A.; Sasaki, T.; Yoshida, K.; Otani, T.; Lei, Y.; Yun, W. IT-143-A and B, novel piericidin-group antibiotics produced by Streptomyces sp. J. Antibiot. 1996, 49, 1052–1055. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Munoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Antonio, T.; Bellao, C.; Correa, T.; Cavallieri, A.P.; Badino, A.C.; Araujo, M.L.G.D. Evaluation of Different Media for the Production of Cephalosporins by Streptomyces clavuligerus ATCC 27064. Braz. Arch. Biol. Technol. 2012, 55, 819–825. [Google Scholar] [CrossRef][Green Version]
- Rateb, M.E.; Yu, Z.; Yan, Y.; Yang, D.; Huang, T.; Vodanovic-Jankovic, S.; Kron, M.A.; Shen, B. Medium optimization of Streptomyces sp. 17944 for tirandamycin B production and isolation and structural elucidation of tirandamycins H, I and J. J. Antibiot. 2014, 67, 127–132. [Google Scholar] [CrossRef]
- Masurekar, P.S. Nutritional and engineering aspects of microbial process development. Prog. Drug Res. 2008, 65, 293–328. [Google Scholar]
- Elibol, M. Optimization of medium composition for actinorhodin production by Streptomyces coelicolor A3(2) with response surface methodology. Process. Biochem. 2004, 39, 1057–1062. [Google Scholar] [CrossRef]
- Montuschi, P.; Ciabattoni, G. Bronchodilating drugs for chronic obstructive pulmonary disease: Current status and future trends. J. Med. Chem. 2015, 58, 4131–4164. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, W.; Zhu, Y.; Zhang, Q.; Tian, X.; Zhang, S.; Zhang, C. Elucidating hydroxylation and methylation steps tailoring piericidin A1 biosynthesis. Org. Lett. 2014, 16, 736–739. [Google Scholar] [CrossRef]
- Senges, C.H.R.; Al-Dilaimi, A.; Marchbank, D.H.; Wibberg, D.; Winkler, A.; Haltli, B.; Nowrousian, M.; Kalinowski, J.; Kerr, R.G.; Bandow, J.E. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. USA 2018, 115, 2490–2495. [Google Scholar] [CrossRef]
- Maansson, M.; Vynne, N.G.; Klitgaard, A.; Nybo, J.L.; Melchiorsen, J.; Nguyen, D.D.; Sanchez, L.M.; Ziemert, N.; Dorrestein, P.C.; Andersen, M.R.; et al. An Integrated Metabolomic and Genomic Mining Workflow To Uncover the Biosynthetic Potential of Bacteria. mSystems 2016, 1, e00028-15. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zacchetti, B.; Ram, A.F.; van Wezel, G.P.; Claessen, D.; Hae Choi, Y. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci. Rep. 2015, 5, 10868. [Google Scholar] [CrossRef] [PubMed]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Ravel, J. Methods for In Silico Prediction of Microbial Polyketide and Nonribosomal Peptide Biosynthetic Pathways from DNA Sequence Data. In Complex Enzymes in Microbial Natural Product Biosynthesis, Part A: Overview Articles and Peptides; Elsevier: Amsterdam, The Netherlands, 2009; pp. 181–217. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivakala, K.K.; Gutiérrez-García, K.; Jose, P.A.; Thinesh, T.; Anandham, R.; Barona-Gómez, F.; Sivakumar, N. Desert Environments Facilitate Unique Evolution of Biosynthetic Potential in Streptomyces. Molecules 2021, 26, 588. https://doi.org/10.3390/molecules26030588
Sivakala KK, Gutiérrez-García K, Jose PA, Thinesh T, Anandham R, Barona-Gómez F, Sivakumar N. Desert Environments Facilitate Unique Evolution of Biosynthetic Potential in Streptomyces. Molecules. 2021; 26(3):588. https://doi.org/10.3390/molecules26030588
Chicago/Turabian StyleSivakala, Kunjukrishnan Kamalakshi, Karina Gutiérrez-García, Polpass Arul Jose, Thangadurai Thinesh, Rangasamy Anandham, Francisco Barona-Gómez, and Natesan Sivakumar. 2021. "Desert Environments Facilitate Unique Evolution of Biosynthetic Potential in Streptomyces" Molecules 26, no. 3: 588. https://doi.org/10.3390/molecules26030588
APA StyleSivakala, K. K., Gutiérrez-García, K., Jose, P. A., Thinesh, T., Anandham, R., Barona-Gómez, F., & Sivakumar, N. (2021). Desert Environments Facilitate Unique Evolution of Biosynthetic Potential in Streptomyces. Molecules, 26(3), 588. https://doi.org/10.3390/molecules26030588






_Barona-Gomez.png)
