Erucic Acid-Rich Yellow Mustard Oil Improves Insulin Resistance in KK-Ay Mice
Abstract
:1. Introduction
2. Results
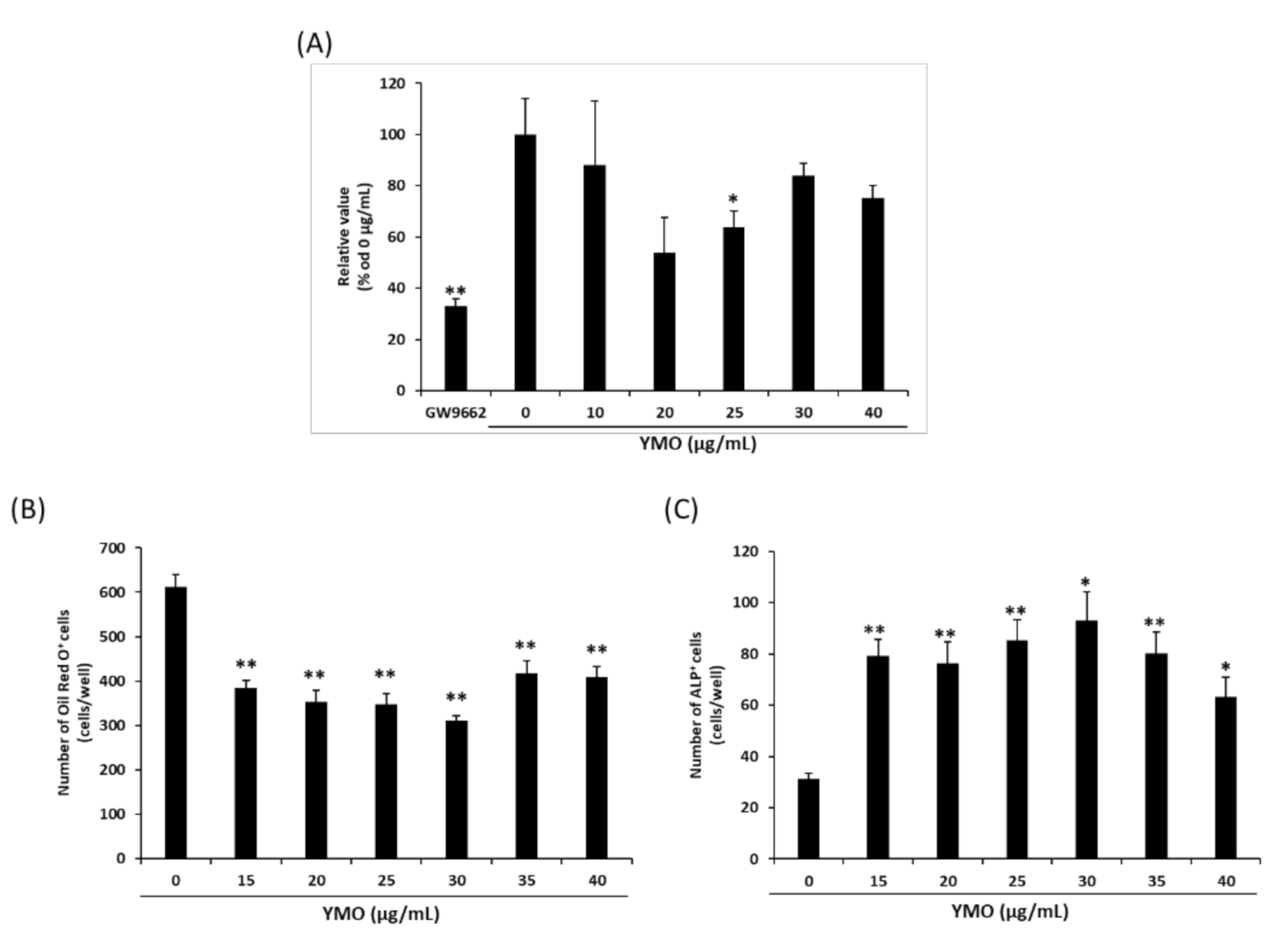
2.1. YMO Suppressed PPARγ Transcriptional Activity and Differentiation of MSCs into Adipocytes
2.2. YMO Suppressed Fat Accumulation in KK-Ay Mice
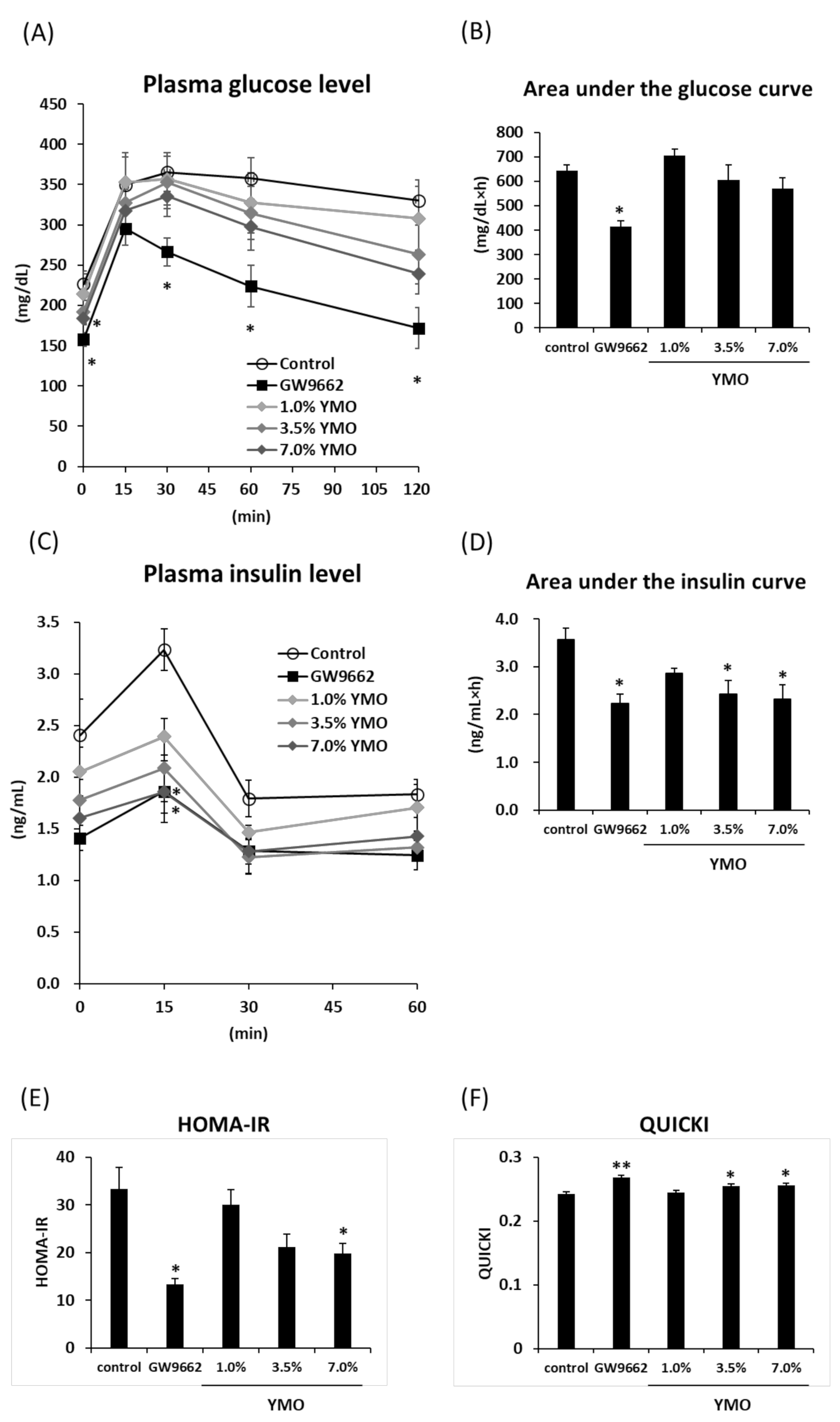
2.3. YMO Improved Insulin Resistance in KK-Ay Mice
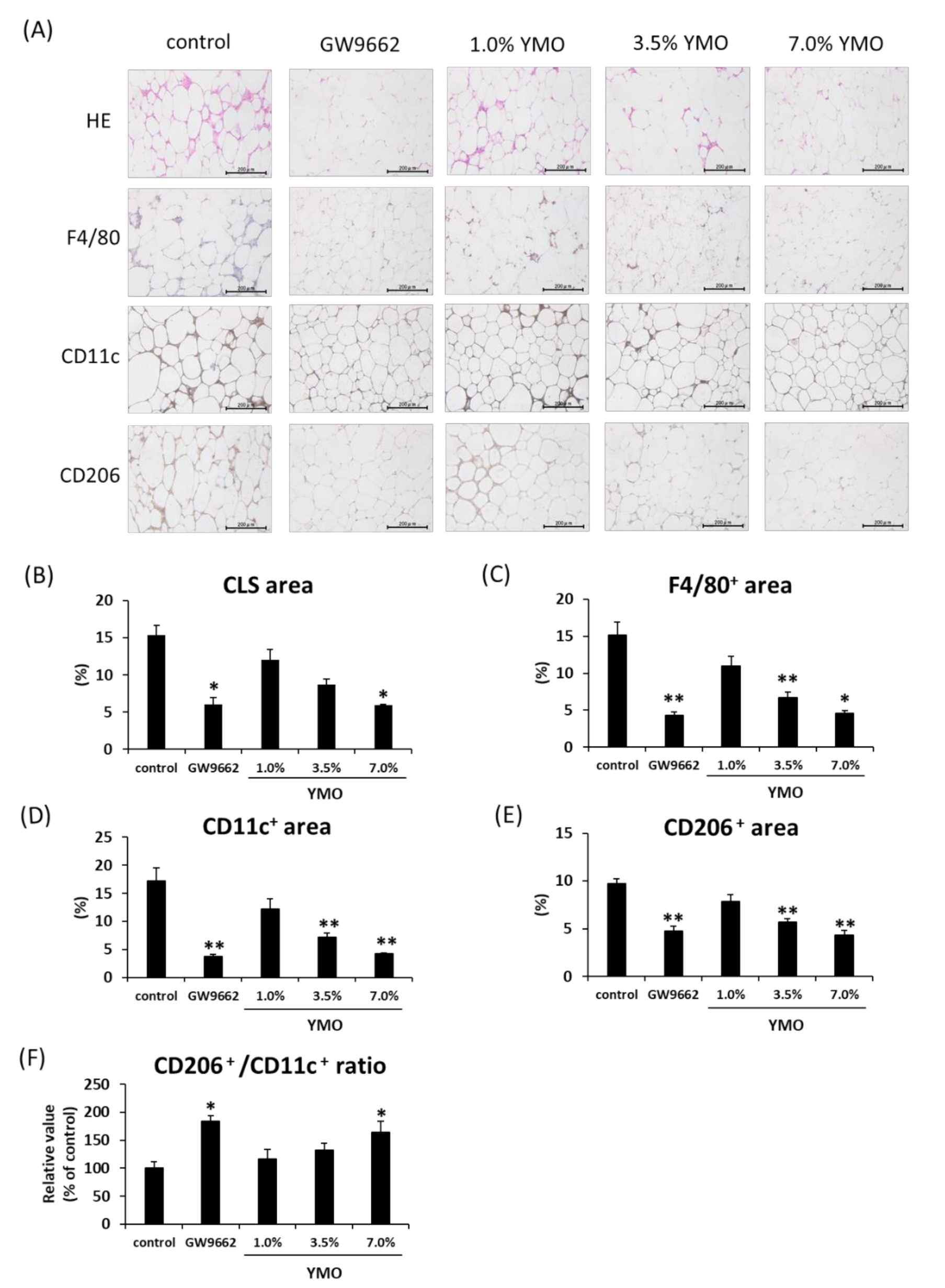
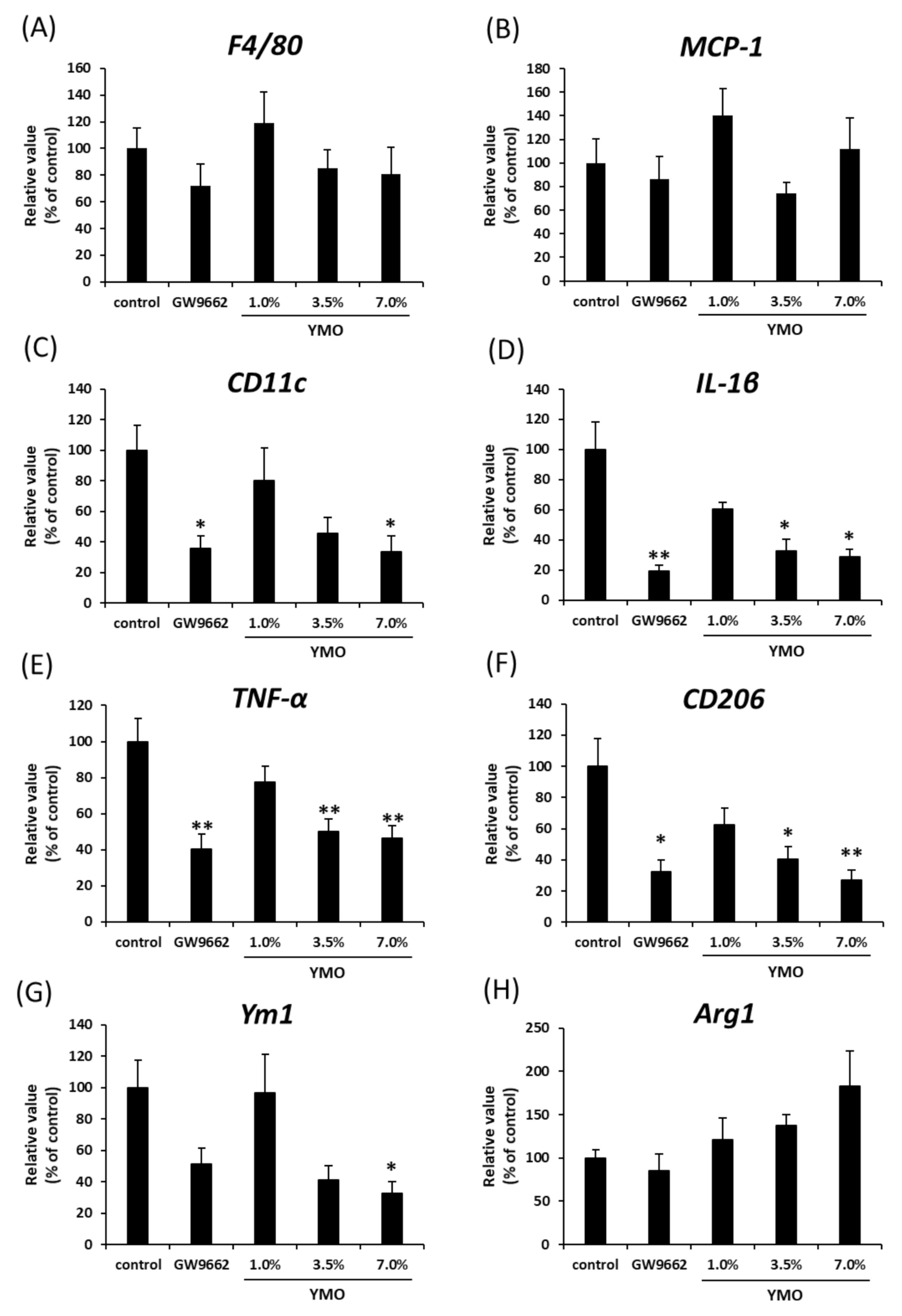
2.4. YMO Suppressed Infiltration of Macrophages into Adipose Tissue in KK-Ay Mice
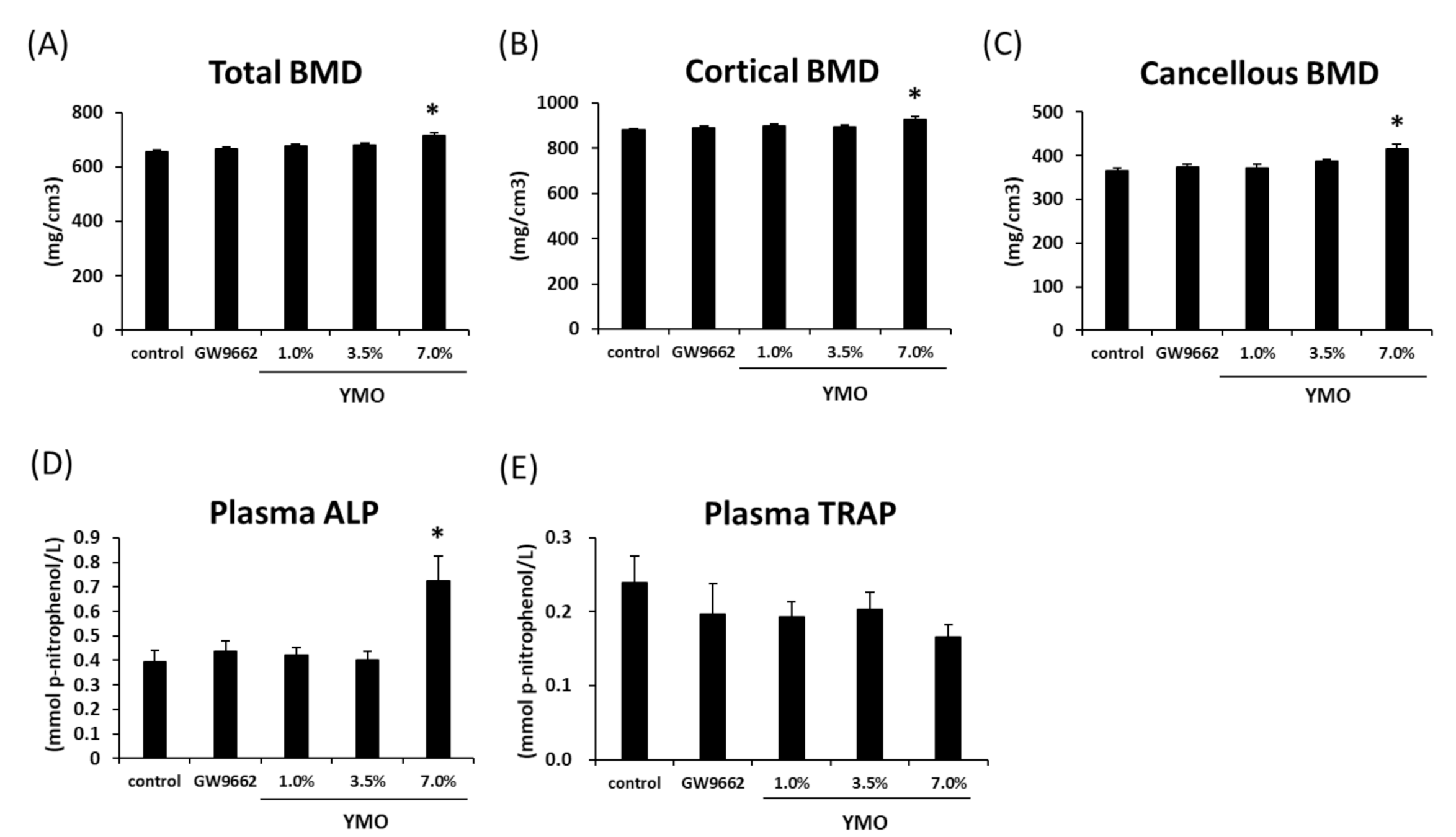
2.5. YMO Increased Bone Mineral Density (BMD) and Osteoblastic Bone Formation Marker
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Hydrolyzation of YMO
4.3. Fatty Acid Analysis of YMO
4.4. PPARγ Luciferase Reporter Assay
4.5. Culture of C3H10T1/2 Cells
4.6. Oil Red O and ALP Staining
4.7. Animal Experiments
4.8. OGTT
4.9. Quantitative Real-Time PCR Analysis
4.10. Histological Analysis of Adipose Tissues
4.11. Analysis of Bone Mineral Density (BMD)
4.12. Measurement of Plasma Bone Metabolic Markers
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Astrup, A.; Finer, N. Redefining Type 2 Diabetes: ’Diabesity’ or ’Obesity Dependent Diabetes Mellitus’? Obes. Rev. 2000, 1, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Salans, L.B.; Cushman, S.W.; Weismann, R.E. Studies of human adipose tissue, Adipose cell size and number in nonobese and obese patients. J. Clin. Invest. 1973, 52, 929–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siersbaek, R.; Nielsen, R.; Mandrup, S. PPARγ in adipocyte differentiation and metabolism--novel insights from genome-wide studies. FEBS Lett. 2010, 584, 3242–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picard, F.; Auwerx, J. PPARγ and glucose homeostasis. Annu. Rev. Nutr. 2002, 22, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.C.; Louche, K.; Nibbelink, M.; Prunet, B.; Bross, A.; Desbazeille, M.; Dacquet, C.; Renard, P.; Casteilla, L.; Pénicaud, L. Fenofibrate prevents Rosiglitazone-induced body weight gain in ob/ob mice. Int. J. Obes. (Lond.) 2005, 29, 864–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.A.; Weinstein, R.S.; Stewart, S.A.; Parfitt, A.M.; Manolagas, S.C.; Jilka, R.L. Rosiglitazone causes bone loss in mice by suppressing osteoblast differentiation and bone formation. Endocrinology 2005, 146, 226–235. [Google Scholar] [CrossRef] [Green Version]
- Sottile, V.; Seuwen, K.; Kneissel, M. Enhanced marrow adipogenesis and bone resorption in estrogen-deprived rats treated with the PPARgamma agonist BRL49653 (rosiglitazone). Calcif. Tissue Int. 2005, 75, 329–337. [Google Scholar] [CrossRef]
- Jones, J.R.; Barrick, C.; Kim, K.A.; Lindner, J.; Blondeau, B.; Fujimoto, Y.; Shiota, M.; Kesterson, R.A.; Kahn, B.B.; Magnuson, M.A. Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 6207–6212. [Google Scholar] [CrossRef] [Green Version]
- Gumbilai, V.; Ebihara, K.; Aizawa-Abe, M.; Ebihara, C.; Zhao, M.; Yamamoto, Y.; Mashimo, T.; Hosoda, K.; Serikawa, T.; Nakao, K. Fat Mass Reduction With Adipocyte Hypertrophy and Insulin Resistance in Heterozygous PPARγ Mutant Rats. Diabetes 2016, 65, 2954–2965. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, T.; Kamon, J.; Waki, H.; Murakami, K.; Motojima, K.; Komeda, K.; Ide, T.; Kubota, N.; Terauchi, Y.; Tobe, K.; et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. J. Biol. Chem. 2001, 276, 41245–41254. [Google Scholar] [CrossRef] [Green Version]
- Nakano, R.; Kurosaki, E.; Yoshida, S.; Yokono, M.; Shimaya, A.; Maruyama, T.; Shibasaki, M. Antagonism of peroxisome proliferator-activated receptor γ prevents high-fat diet-induced obesity in vivo. Biochem. Pharmacol. 2006, 72, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Waki, H.; Kamon, J.; Murakami, K.; Motojima, K.; Komeda, K.; Miki, H.; Kubota, N.; Terauchi, Y.; Tsuchida, A.; et al. Inhibition of RXR and PPARγ ameliorates diet-induced obesity and type 2 diabetes. J. Clin. Investig. 2001, 108, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Duque, G.; Li, W.; Vidal, C.; Bermeo, S.; Rivas, D.; Henderson, J. Pharmacological inhibition of PPARγ increases osteoblastogenesis and bone mass in male C57BL/6 mice. J. Bone Miner. Res. 2013, 28, 639–648. [Google Scholar] [CrossRef]
- David, V.; Martin, A.; Lafage-Proust, M.H.; Malaval, L.; Peyroche, S.; Jones, D.B.; Vico, L.; Guignandon, A. Mechanical loading down-regulates peroxisome proliferator-activated receptor γ in bone marrow stromal cells and favors osteoblastogenesis at the expense of adipogenesis. Endocrinology 2007, 148, 2553–2562. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Dohi, H.; Egashira, Y.; Hirai, S. Erucic acid derived from rosemary regulates differentiation of mesenchymal stem cells into osteoblasts/adipocytes via suppression of peroxisome proliferator-activated receptor γ transcriptional activity. Phytother. Res. 2020, 34, 1358–1366. [Google Scholar] [CrossRef]
- Ishaq, M.; Razi, R.; Khan, S.A. Exploring genotypic variations for improved oil content and healthy fatty acids composition in rapeseed (Brassica napus L.). J. Sci. Food Agric. 2017, 97, 1924–1930. [Google Scholar] [CrossRef]
- Wendlinger, C.; Hammann, S.; Vetter, W. Various concentrations of erucic acid in mustard oil and mustard. Food Chem. 2014, 153, 393–397. [Google Scholar] [CrossRef]
- Ciubota-Rosie, C.; Macoveanu, M.; Fernàndez, C.M.; Ramos, M.J.; Pérez, A.; Moreno, A. Sinapis alba seed as a prospective biodiesel source. Biomass Bioenergy 2013, 53, 83–90. [Google Scholar] [CrossRef]
- Tzen, J.; Cao, Y.; Laurent, P.; Ratnayake, C.; Huang, A. Lipids, Proteins, and Structure of Seed Oil Bodies from Diverse Species. Plant Physiol. 1993, 101, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Ducy, P.; Starbuck, M.; Priemel, M.; Shen, J.; Pinero, G.; Geoffroy, V.; Amling, M.; Karsenty, G.A. Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999, 13, 1025–1036. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Dong, Y.; Huang, X.; Li, M.; Qin, L.; Ren, Y.; Guo, F.; Chen, A.; Huang, S. Decreased osteoclastogenesis, osteoblastogenesis and low bone mass in a mouse model of type 2 diabetes. Mol. Med. Rep. 2014, 10, 1935–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takasawa, K.; Kubota, N.; Terauchi, Y.; Kadowaki, T. Impact of increased PPARγ activity in adipocytes in vivo on adiposity, insulin sensitivity and the effects of rosiglitazone treatment. Endocr. J. 2008, 55, 767–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuno, A.; Tamemoto, H.; Tobe, K.; Ueki, K.; Mori, Y.; Iwamoto, K.; Umesono, K.; Akanuma, Y.; Fujiwara, T.; Horikoshi, H.; et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J. Clin. Investig. 1998, 101, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Jin, D.; Chen, X. Luteolin enhances insulin sensitivity via activation of PPARγ transcriptional activity in adipocytes. J. Nutr. Biochem. 2010, 21, 941–947. [Google Scholar] [CrossRef]
- Takahashi, N.; Goto, T.; Taimatsu, A.; Egawa, K.; Katoh, S.; Kusudo, T.; Sakamoto, T.; Ohyane, C.; Lee, J.Y.; Kim, Y.I.; et al. Bixin regulates mRNA expression involved in adipogenesis and enhances insulin sensitivity in 3T3-L1 adipocytes through PPARγ activation. Biochem. Biophys. Res. Commun. 2009, 390, 1372–1376. [Google Scholar] [CrossRef]
- Kennedy, A.; Chung, S.; LaPoint, K.; Fabiyi, O.; McIntosh, M.K. Trans-10, cis-12 conjugated linoleic acid antagonizes ligand-dependent PPARγ activity in primary cultures of human adipocytes. J. Nutr. 2008, 138, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Platt, I.D.; El-Sohemy, A. Regulation of osteoblast and adipocyte differentiation from human mesenchymal stem cells by conjugated linoleic acid. J. Nutr. Biochem. 2009, 20, 956–964. [Google Scholar] [CrossRef]
- Smith, R.H.; Powell, G.L. The critical micelle concentration of some physiologically important fatty acyl-coenzyme A’s as a function of chain length. Arch. Biochem. Biophys. 1986, 244, 357–360. [Google Scholar] [CrossRef]
- Kassem, M.; Philippe, F.; Isabelle, W. Adipose tissue in obesity-related inflammation and insulin resistance: Cells, cytokines, and chemokines. ISRN Inflamm. 2013. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.; Kim, C.S.; Kwon, B.S.; Kawada, T. Mesenteric Adipose Tissue-Derived Monocyte Chemoattractant Protein-1 Plays a Crucial Role in Adipose Tissue Macrophage Migration and Activation in Obese Mice. Obesity 2006, 14, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Kamei, N.; Tobe, K.; Suzuki, R.; Ohsugi, M.; Watanabe, T.; Kubota, N.; Ohtsuka-Kowatari, N.; Kumagai, K.; Sakamoto, K.; Kobayashi, M.; et al. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem. 2006, 281, 26602–26614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisaka, S.; Usui, I.; Bukhari, A.; Ikutani, M.; Oya, T.; Kanatani, Y.; Tsuneyama, K.; Nagai, Y.; Takatsu, K.; Urakaze, M.; et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes 2009, 58, 2574–2582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, A.A.; Reid Bolus, W.; Hasty, A.H. A decade of progress in adipose tissue macrophage biology. Immunol. Rev. 2014, 262, 134–152. [Google Scholar] [CrossRef] [Green Version]
- Cinti, S.; Mitchell, G.; Barbatelli, G.; Murano, I.; Ceresi, E.; Faloia, E.; Wang, S.; Fortier, M.; Greenberg, A.S.; Obin, M.S. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res. 2005, 46, 2347–2355. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.H.; Banks, A.S.; Estall, J.L.; Kajimura, S.; Boström, P.; Laznik, D.; Ruas, J.L.; Chalmers, M.J.; Kamenecka, T.M.; Blüher, M.; et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature 2010, 466, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.H.; Banks, A.S.; Kamenecka, T.M.; Busby, S.A.; Chalmers, M.J.; Kumar, N.; Kuruvilla, D.S.; Shin, Y.; He, Y.; Bruning, J.B.; et al. Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature 2011, 477, 477–481. [Google Scholar] [CrossRef]
- Banks, A.S.; McAllister, F.E.; Camporez, J.P.; Zushin, P.J.; Jurczak, M.J.; Laznik-Bogoslavski, D.; Shulman, G.I.; Gygi, S.P.; Spiegelman, B.M. An ERK/Cdk5 axis controls the diabetogenic actions of PPARγ. Nature 2015, 517, 391–395. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zhou, Q.; Zhong, F.; Guo, S.; Hao, X.; Li, C.; Wang, W.; Chen, N. 15-Deoxy-Δ12,14-Prostaglandin J2 Modulates Lipopolysaccharide-Induced Chemokine Expression by Blocking Nuclear Factor-κB Activation via Peroxisome Proliferator Activated Receptor-γ-Independent Mechanism in Renal Tubular Epithelial Cells. Nephron. Exp. Nephrol. 2013, 123, 1–10. [Google Scholar] [CrossRef]
- Choi, S.S.; Kim, E.S.; Jung, J.E.; Marciano, D.P.; Jo, A.; Koo, J.Y.; Choi, S.Y.; Yang, Y.R.; Jang, H.J.; Kim, E.K.; et al. PPARγ Antagonist Gleevec Improves Insulin Sensitivity and Promotes the Browning of White Adipose Tissue. Diabetes 2016, 65, 829–839. [Google Scholar] [CrossRef] [Green Version]
- Shui, C.; Spelsberg, T.C.; Riggs, B.L.; Khosla, S. Changes in Runx2/Cbfa1 expression and activity during osteoblastic differentiation of human bone marrow stromal cells. J. Bone Miner. Res. 2003, 18, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Hinoi, E.; Fujimori, S.; Wang, L.; Hojo, H.; Uno, K.; Yoneda, Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J. Biol. Chem. 2006, 281, 18015–18024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Léotoing, L.; Davicco, M.J.; Lebecque, P.; Wittrant, Y.; Coxam, V. The flavonoid fisetin promotes osteoblasts differentiation through Runx2 transcriptional activity. Mol. Nutr. Food Res. 2014, 58, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, M.H.; Wozney, J.M.; Cho, J.Y.; Ryoo, H.M. Bone morphogenetic protein-2-induced alkaline phosphatase expression is stimulated by Dlx5 and repressed by Msx2. J. Biol. Chem. 2004, 279, 50773–50780. [Google Scholar] [CrossRef] [Green Version]
- Felthaus, O.; Gosau, M.; Klein, S.; Prantl, L.; Reichert, T.E.; Schmalz, G.; Morsczeck, C. Dexamethasone-related osteogenic differentiation of dental follicle cells depends on ZBTB16 but not Runx2. Cell Tissue Res. 2014, 357, 695–705. [Google Scholar] [CrossRef]
- Beekman, K.M.; Veldhuis-Vlug, A.G.; van der Veen, A.; den Heijer, M.; Maas, M.; Kerckhofs, G.; Parac-Vogt, T.N.; Bisschop, P.H.; Bravenboer, N. The effect of PPARγ inhibition on bone marrow adipose tissue and bone in C3H/HeJ mice. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E96–E105. [Google Scholar] [CrossRef]
- Wronski, T.J.; Cintrón, M.; Dann, L.M. Temporal relationship between bone loss and increased bone turnover in ovariectomized rats. Calcif. Tissue Int. 1988, 43, 179–183. [Google Scholar] [CrossRef]
- Leslie, W.D.; Rubin, M.R.; Schwartz, A.V.; Kanis, J.A. Type 2 diabetes and bone. J. Bone Miner. Res. 2012, 27, 2231–2237. [Google Scholar] [CrossRef]
- Wallace, T.M.; Levy, J.C.; Matthews, D.R. Use and abuse of HOMA modeling. Diabetes Care 2004, 27, 1487–1495. [Google Scholar] [CrossRef] [Green Version]
- Muniyappa, R.; Lee, S.; Chen, H.; Quon, M.J. Current approaches for assessing insulin sensitivity and resistance in vivo: Advantages, limitations, and appropriate usage. Am. J. Physiol. Endocrinol. Metab. 2008, 294, E15–E26. [Google Scholar] [CrossRef] [Green Version]
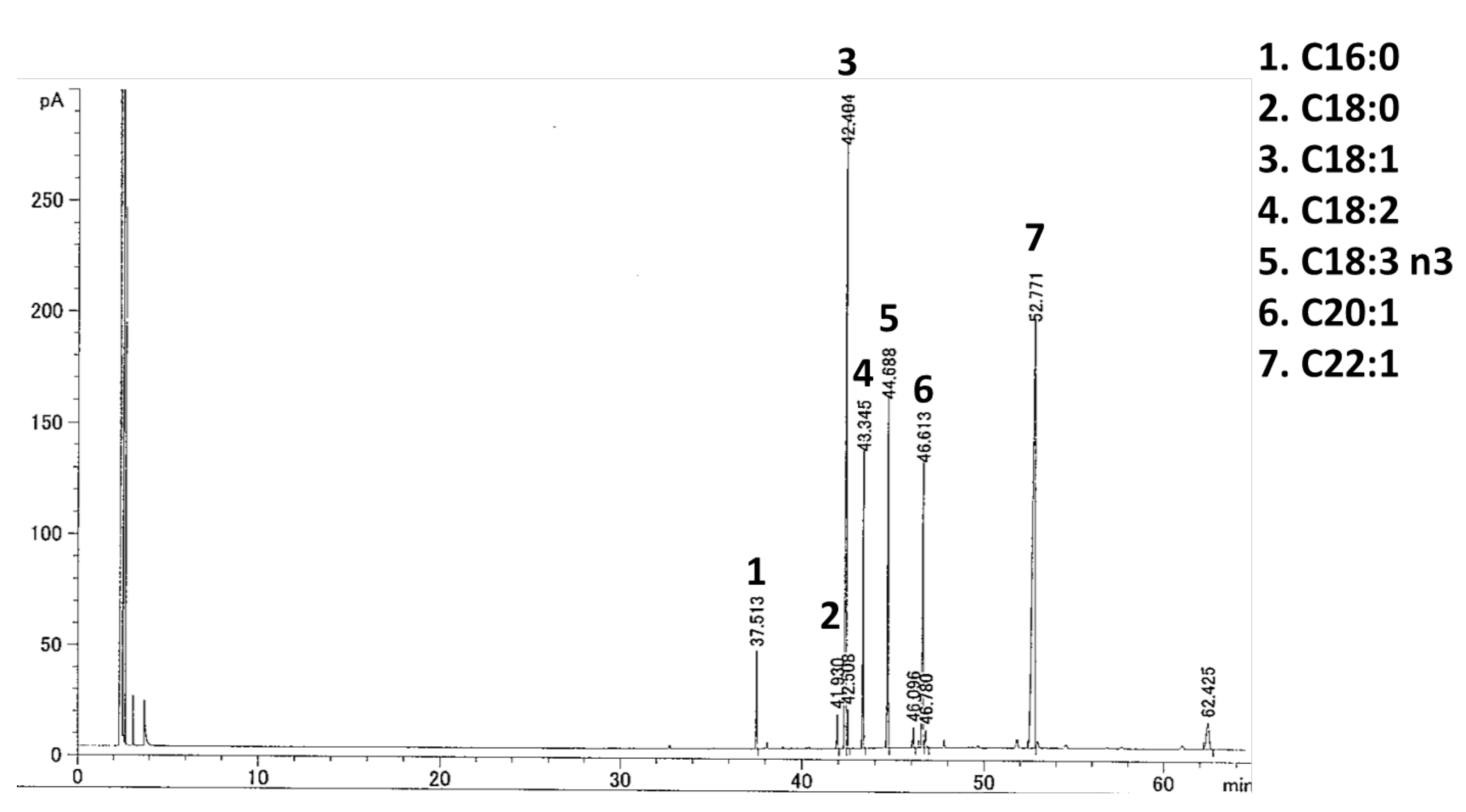


| Fatty Acid (%) | YMO |
|---|---|
| C16:0 | 2.61 |
| C18:0 | 1.05 |
| C18:1 | 24.35 |
| C18:2 | 8.92 |
| C18:3 n3 | 10.49 |
| C20:1 | 10.88 |
| C22:1 | 36.97 |
| Others | 4.73 |
| Control | GW9662 | 1.0% YMO | 3.5% YMO | 7.0% YMO | |
|---|---|---|---|---|---|
| Final body weight (g) | 42.29 ± 0.70 | 40.16 ± 0.95 | 41.98 ± 1.36 | 39.59 ± 0.68 * | 41.41 ± 0.84 |
| Subcutaneous adipose tissue (g) | 1.62 ± 0.08 | 1.41 ± 0.05 | 1.63 ± 0.10 | 1.21 ± 0.10 * | 1.47 ± 0.07 |
| Perirenal adipose tissue (g) | 0.91 ± 0.09 | 0.94 ± 0.07 | 0.88 ± 0.10 | 0.66 ± 0.06 | 0.59 ± 0.08 * |
| Mesenteric adipose tissue (g) | 1.09 ± 0.04 | 0.99 ± 0.05 | 1.21 ± 0.03 | 0.96 ± 0.06 | 1.12 ± 0.05 |
| Epididymal adipose tissue (g) | 1.04 ± 0.04 | 0.98 ± 0.03 | 1.03 ± 0.03 | 0.96 ± 0.04 | 1.05 ± 0.06 |
| Genes | Accession Number | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|---|
| PPARγ | NM_001127330 | GGAGATCTCCAGTGATATCGACCA | ACGGCTTCTACGGATCGAAACT |
| aP2 | NM_001122948 | AAGACAGCTCCTCCTCGAAGGTT | TGACCAAATCCCCATTTACGC |
| LPL | NM_008509 | GCCCAGCAACATTATCCAGT | GGTCAGACTTCCTGCTACGC |
| Runx2 | DQ065175 | CCCAGCCACTTTACCTACA | TATGGAGTGCTGCTGGTCTG |
| Osterix | AF184902 | ACTCATCCCTAATGGCTCGTG | GGTAGGGAGCTGGGTTAAGG |
| ALP | BC065175 | GCTGATCATTCCCACGTTTT | CTGGGCCTGGTAGTTGTTGT |
| Col1 | NM_007742.4 | GAGCGGAGAGTACTGGATCG | GCTTCTTTTCCTTGGGGTTC |
| F4/80 | X93328.1 | TTTCCTCGCCTGCTTCTTC | CCCCGTCTCTGTATTCAACC |
| MCP-1 | NM_011333.3 | AGGTCCCTGTCATGCTTCTG | TCTGGACCCATTCCTTCTTG |
| CD11c | NM_007482 | TGGGTTTGTTTCTTGTCTTG | GCCTGTGTGATAGCCACATTT |
| IL-1β | NM_008361.4 | GCCCATCCTCTGTGACTCAT | AGGCCACAGGTATTTTGTCG |
| TNF-α | NM_013693 | ACACTCAGATCATCTTCTCAAAATTCG | GTGTGGGTGAGGAGCACGTAGT |
| CD206 | NM_008625 | GCGCTGCGTGGACGCTCTAA | ACAGGGTGACGGAAGCCCAGT |
| Ym1 | NM_009892 | AGAAGGGAGTTTCAAACCTGGT | GTCTTGCTCATGTGTGTAAGTGA |
| Arg1 | NM_007482 | CAGTTGGAAGCATCTCTGGC | GTGAGCATCCACCCAAATGAC |
| 36B4 | NM_007475.5 | TGTGTGTCTGCAGARCGGGTAC | CTTTGGCGGGATTAGTCGAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, A.; Ishizaki, M.; Kimira, Y.; Egashira, Y.; Hirai, S. Erucic Acid-Rich Yellow Mustard Oil Improves Insulin Resistance in KK-Ay Mice. Molecules 2021, 26, 546. https://doi.org/10.3390/molecules26030546
Takahashi A, Ishizaki M, Kimira Y, Egashira Y, Hirai S. Erucic Acid-Rich Yellow Mustard Oil Improves Insulin Resistance in KK-Ay Mice. Molecules. 2021; 26(3):546. https://doi.org/10.3390/molecules26030546
Chicago/Turabian StyleTakahashi, Asako, Mayu Ishizaki, Yoshifumi Kimira, Yukari Egashira, and Shizuka Hirai. 2021. "Erucic Acid-Rich Yellow Mustard Oil Improves Insulin Resistance in KK-Ay Mice" Molecules 26, no. 3: 546. https://doi.org/10.3390/molecules26030546
APA StyleTakahashi, A., Ishizaki, M., Kimira, Y., Egashira, Y., & Hirai, S. (2021). Erucic Acid-Rich Yellow Mustard Oil Improves Insulin Resistance in KK-Ay Mice. Molecules, 26(3), 546. https://doi.org/10.3390/molecules26030546







