The Microfluidic Toolbox for Analyzing Exosome Biomarkers of Aging
Abstract
1. Introduction
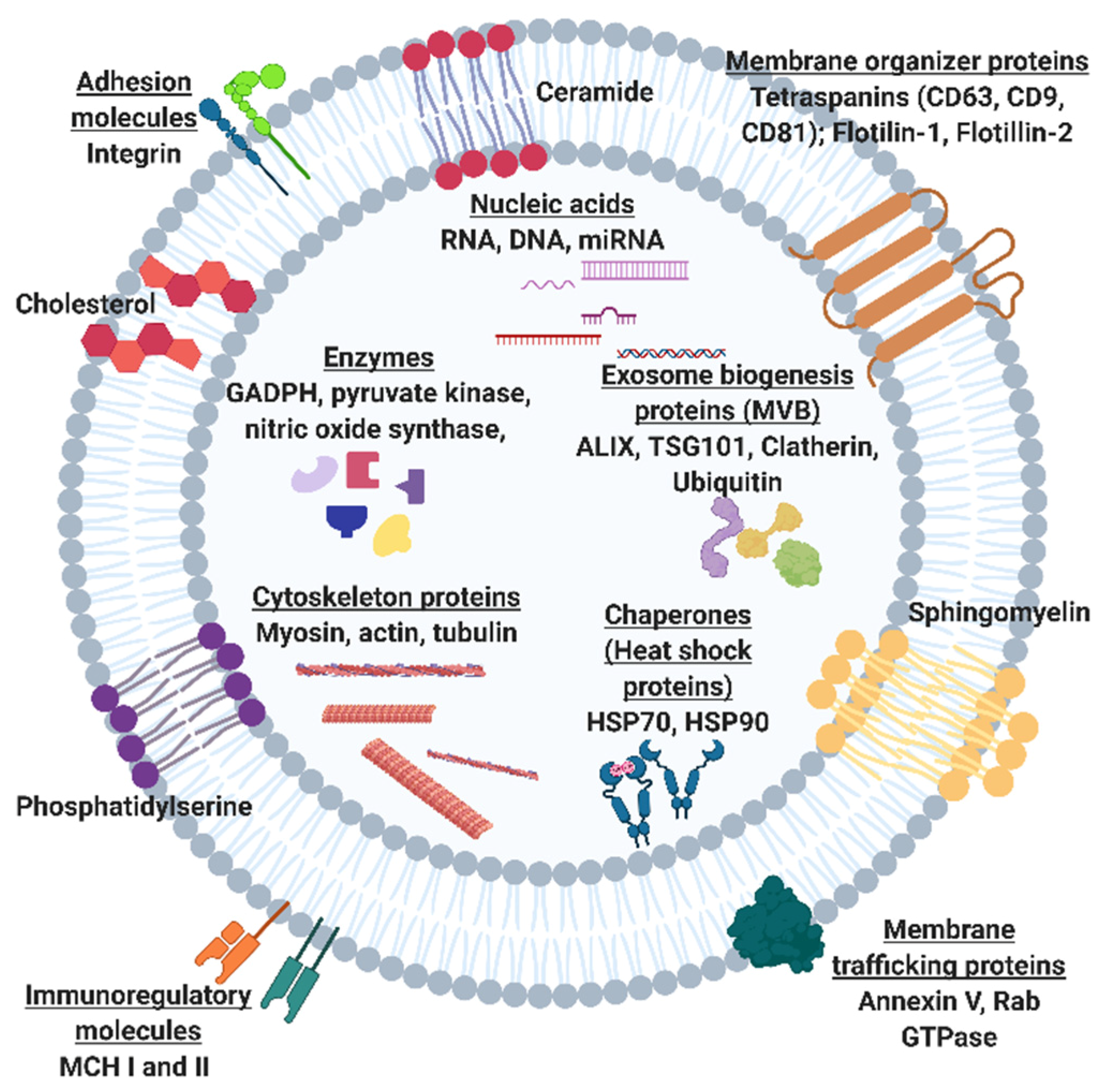
2. The Potential of Exosomal Biomarkers for Precision Medicine and Liquid Biopsies
3. Microfluidic Solutions for Exosome Isolations
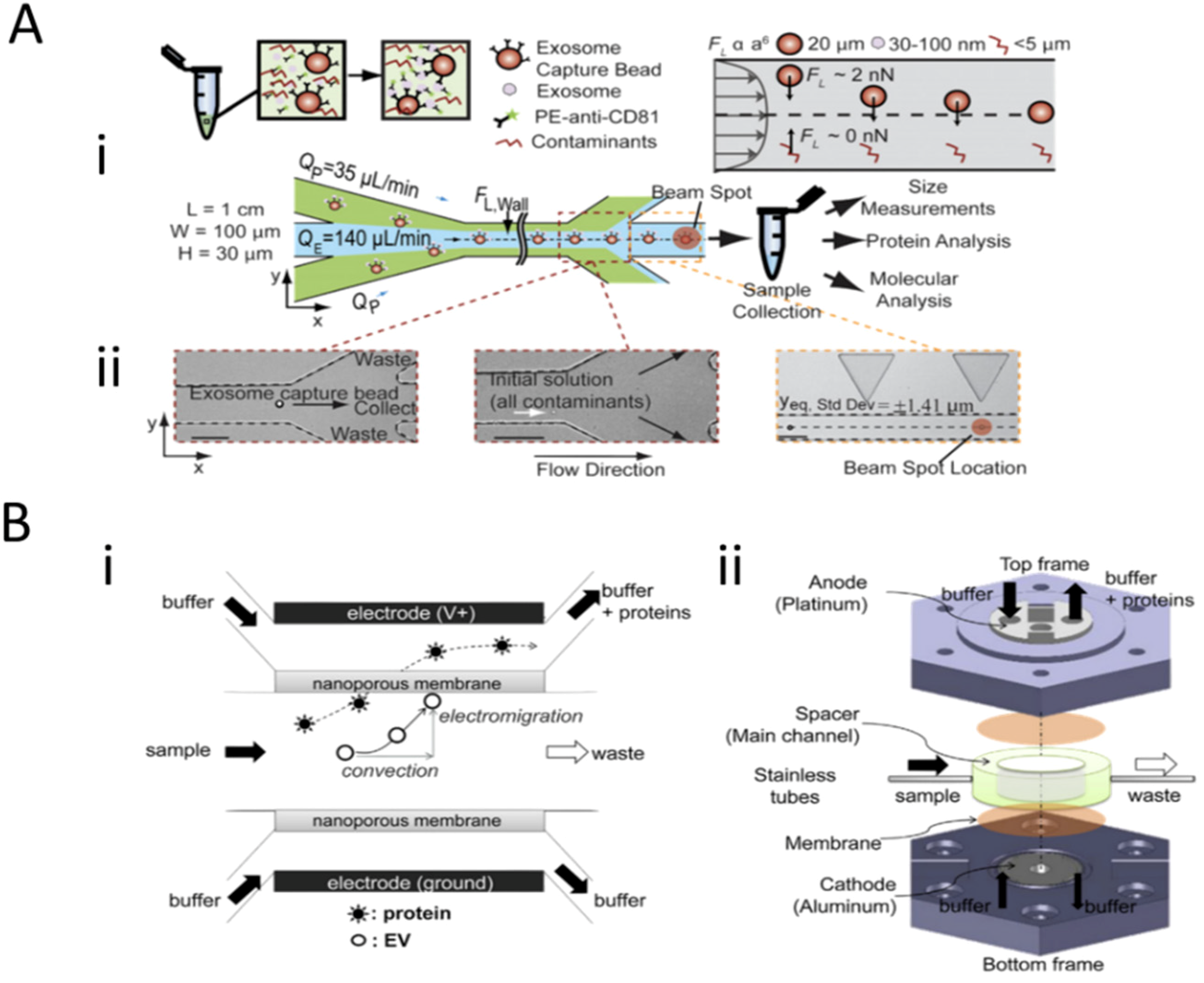
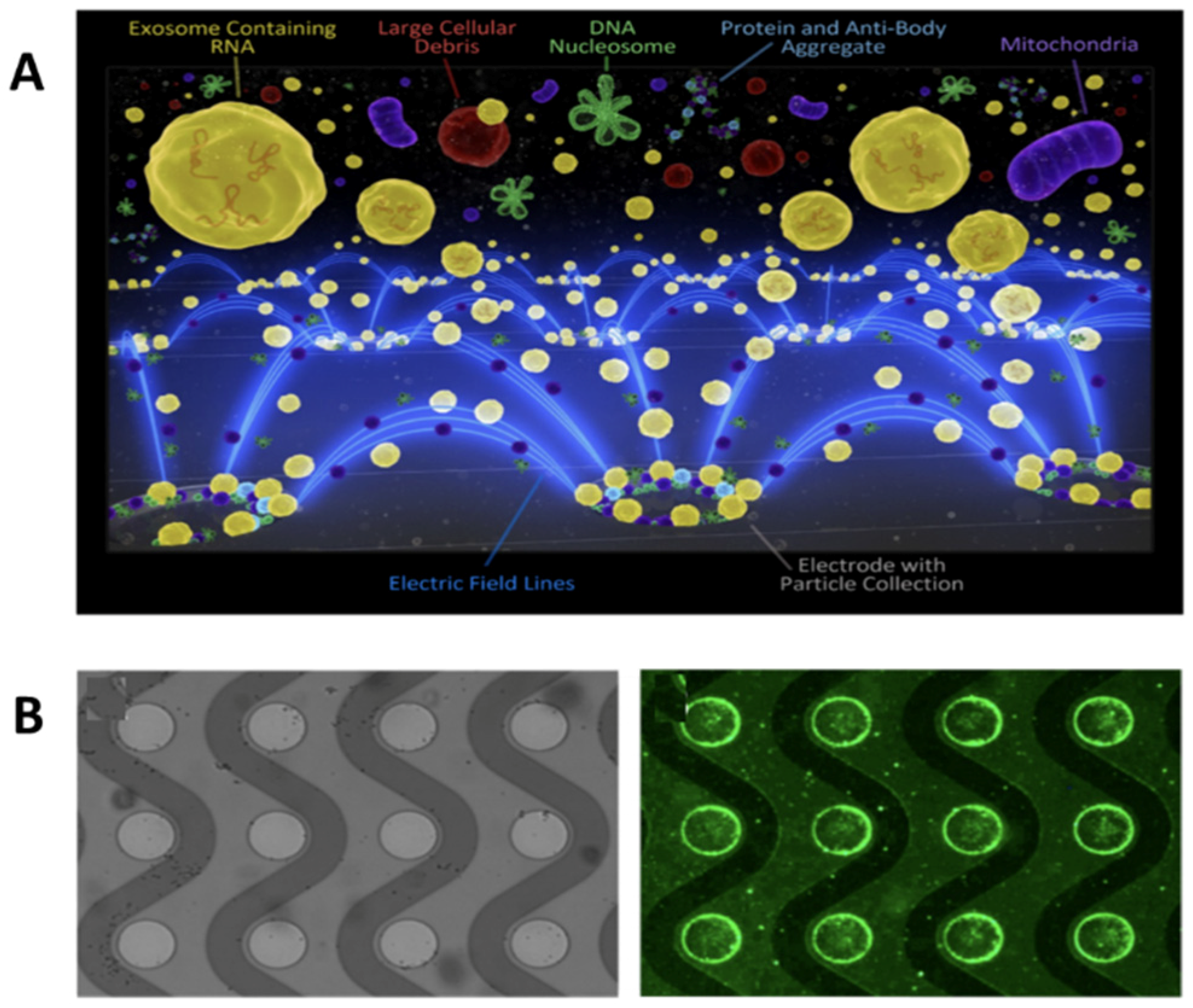
3.1. Field-Based Isolation of Exosomes
3.2. Surface-Based Isolation of Exosomes
4. Exosomal Detection Systems to Monitor Age-Associated Pathologies
4.1. Technologies for Profiling of Antigen-Specific Exosomal Biomarkers
4.1.1. Immunoassay-Based Technologies
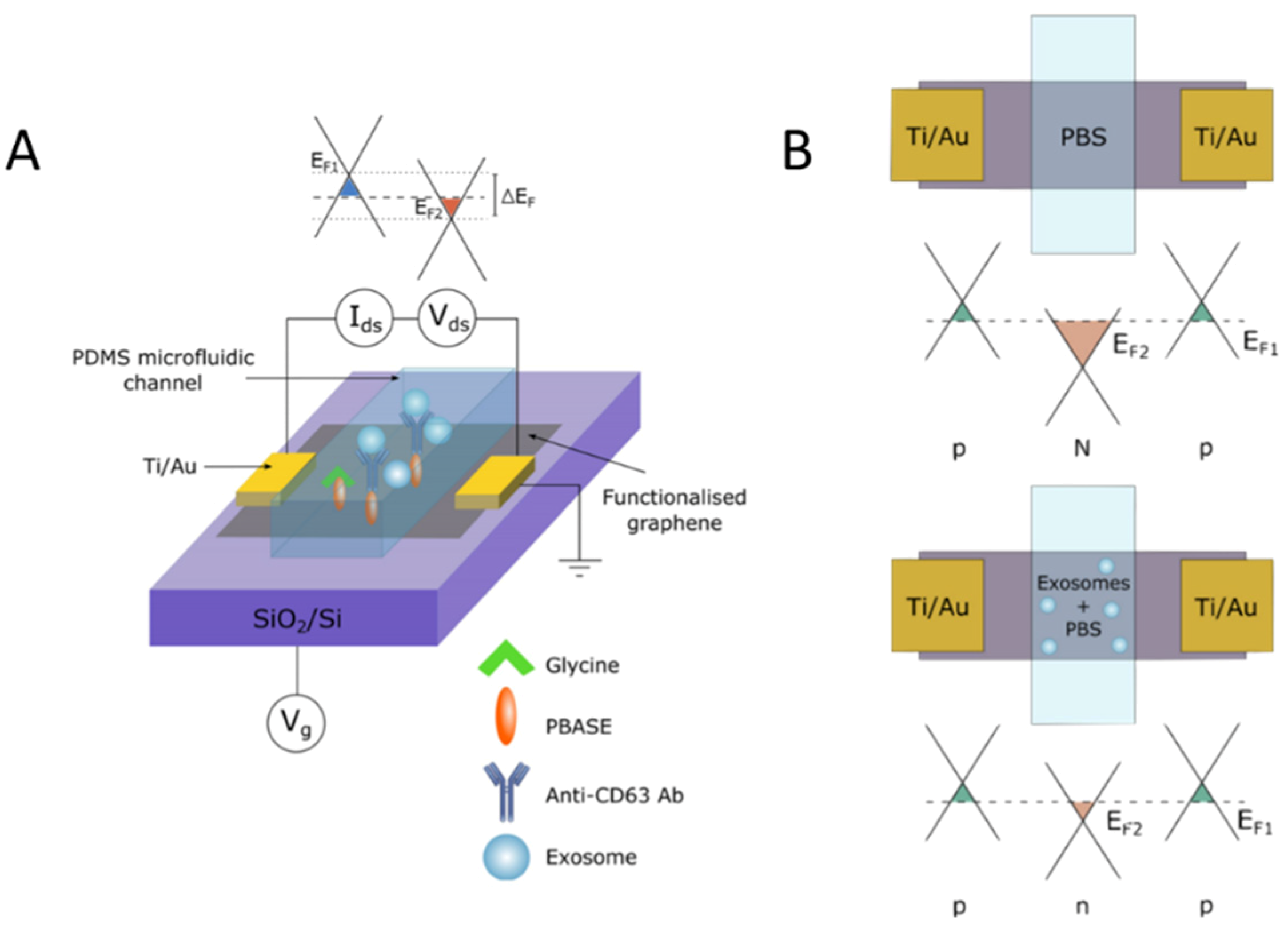
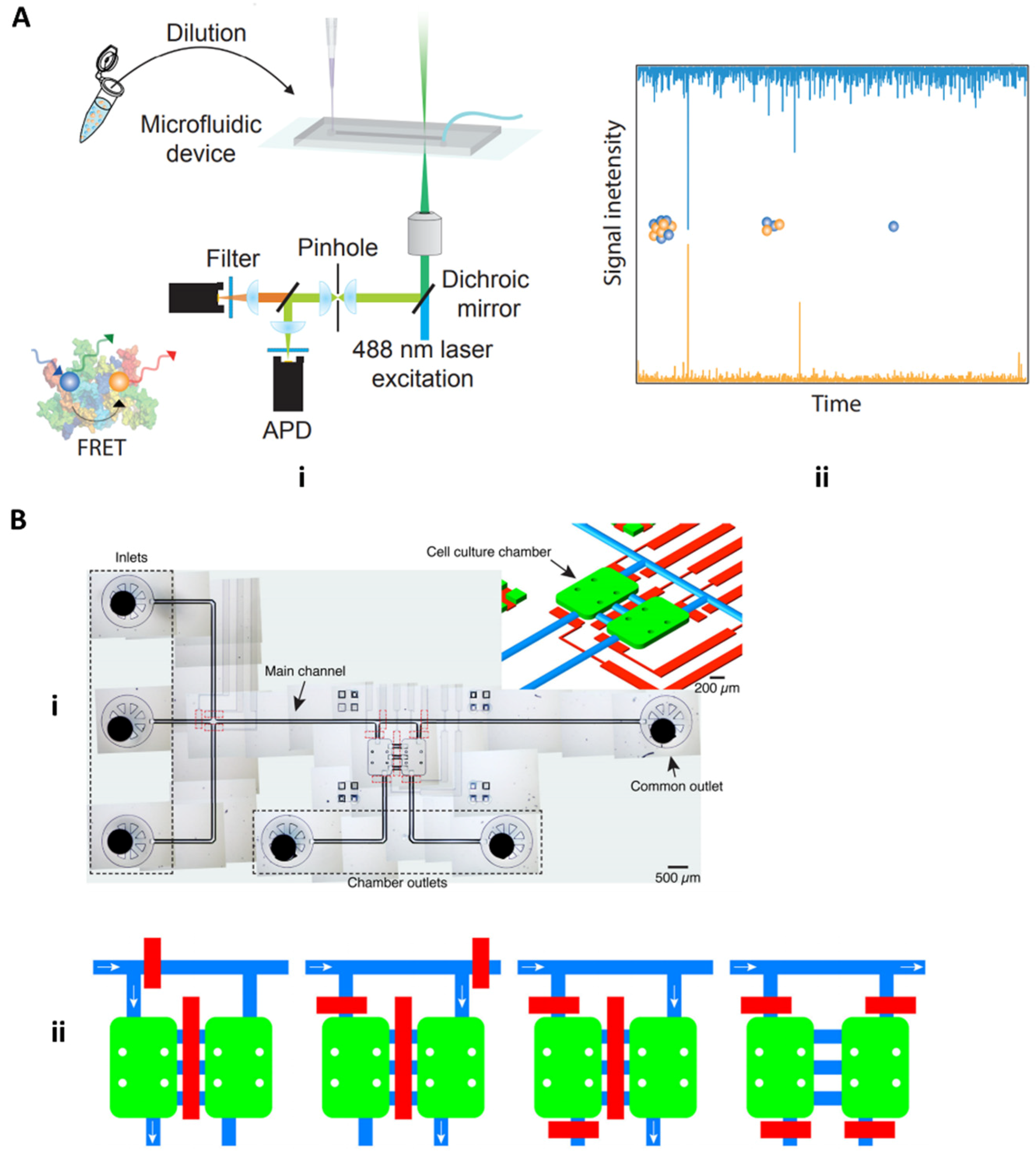
4.1.2. Fluorescence and Field-Based Technologies
4.2. Microfluidic Approaches for Screening Neurotoxic Biomarkers
4.2.1. Microfluidic Detection of Alzheimer’s Disease Biomarkers: Tau Protein and Amyloid-Beta
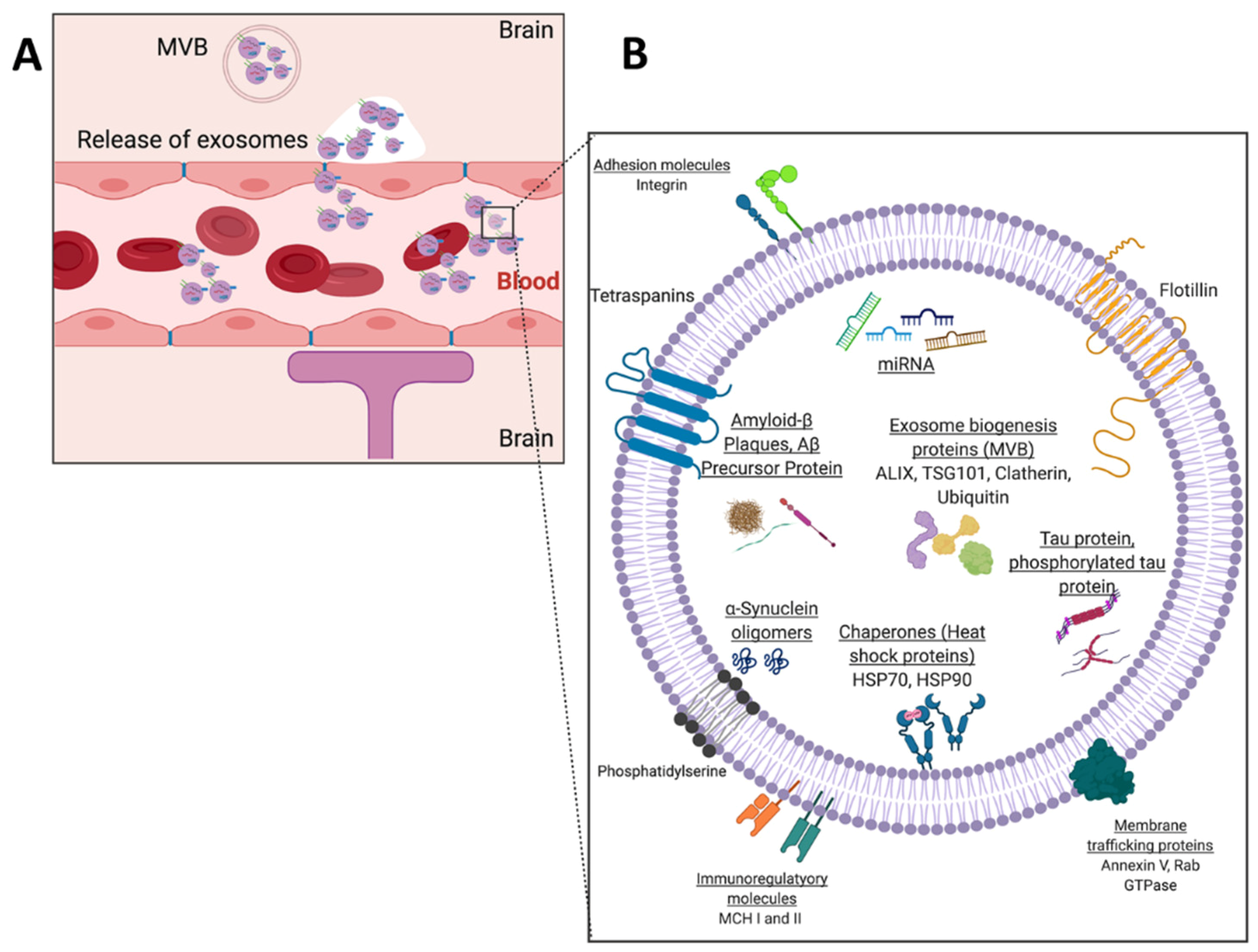
4.2.2. Opportunities to Develop Technologies for Profiling of Exosomal Cargo Biomarkers
5. Challenges to Commercialization
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Franceschi, C.; Garagnani, P.; Morsiani, C.; Conte, M.; Santoro, A.; Grignolio, A.; Monti, D.; Capri, M.; Salvioli, S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front. Med. 2018, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Shin, Y.; Chung, S.; Hwang, D.W.; Lee, D.S. Convective Exosome-Tracing Microfluidics for Analysis of Cell-Non-Autonomous Neurogenesis. Biomaterials 2017, 112, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. CtDNA and CTCs in Liquid Biopsy – Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Lowes, L.E.; Bratman, S.V.; Dittamore, R.; Done, S.; Kelley, S.O.; Mai, S.; Morin, R.D.; Wyatt, A.W.; Allan, A.L. Circulating Tumor Cells (CTC) and Cell-Free DNA (CfDNA) Workshop 2016: Scientific Opportunities and Logistics for Cancer Clinical Trial Incorporation. Int. J. Mol. Sci. 2016, 17, 1505. [Google Scholar] [CrossRef] [PubMed]
- Shaw, Gina Exosomes: Promising Targets for Liquid Biopsy. Available online: http://www.biocompare.com/Editorial-Articles/357781-Exosomes-Promising-Targets-for-Liquid-Biopsy/ (accessed on 19 May 2020).
- Lang, J.E. Advantages and Disadvantages of CtDNA vs CTC Assays: How to Move the Needle Forward towards Clinical Application; Keck Medicine of USC: Los Angeles, CA, USA, 2007. [Google Scholar]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for Exosome Isolation and Analysis: Enabling Liquid Biopsy for Personalized Medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7. [Google Scholar] [CrossRef]
- De Vrij, J.; Maas, S.L.N.; van Nispen, M.; Sena-Esteves, M.; Limpens, R.W.A.; Koster, A.J.; Leenstra, S.; Lamfers, M.L.; Broekman, M.L.D. Quantification of Nanosized Extracellular Membrane Vesicles with Scanning Ion Occlusion Sensing. Nanomed. (Lond.) 2013, 8, 1443–1458. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Zhu, Q.; Heon, M.; Zhao, Z.; He, M. Microfluidic Engineering of Exosomes: Editing Cellular Messages for Precision Therapeutics. Lab Chip 2018, 18, 1690–1703. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy Delivery Tools and Biomarkers of Diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; Bianco, M.; Nigro, A.; Primiceri, E.; Ferrara, F.; Romano, A.; Quattrini, A.; Furlan, R.; Arima, V.; Maruccio, G. Lab-on-Chip for Exosomes and Microvesicles Detection and Characterization. Sensors 2018, 18, 3175. [Google Scholar] [CrossRef] [PubMed]
- Cell Guidance Systems Exospin: Trusted Exosome Purification. Available online: https://www.cellgs.com/products/exo-spinand8482-midi-columns.html (accessed on 16 December 2020).
- System Biosciences ExoQuick Exosome Preciptiation Solution. Available online: https://systembio.com/wp-content/uploads/MANUAL_EXOQXXA-1-1.pdf (accessed on 16 December 2020).
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of Membrane Affinity-Based Method with Size-Exclusion Chromatography for Isolation of Exosome-like Vesicles from Human Plasma. J. Transl. Med. 2018, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Eskelin, K.; Poranen, M.M.; Oksanen, H.M. Asymmetrical Flow Field-Flow Fractionation on Virus and Virus-Like Particle Applications. Microorganisms 2019, 7, 555. [Google Scholar] [CrossRef]
- Thakur, A.; Qiu, G.; Ng, S.-P.; Guan, J.; Yue, J.; Lee, Y.; Wu, C.-M.L. Direct Detection of Two Different Tumor-Derived Extracellular Vesicles by SAM-AuNIs LSPR Biosensor. Biosens. Bioelectron. 2017, 94, 400–407. [Google Scholar] [CrossRef]
- Dudani, J.S.; Gossett, D.R.; Tse, H.T.K.; Lamm, R.J.; Kulkarni, R.P.; Carlo, D.D. Rapid Inertial Solution Exchange for Enrichment and Flow Cytometric Detection of Microvesicles. Biomicrofluidics 2015, 9, 014112. [Google Scholar] [CrossRef]
- Gossett, D.R.; Tse, H.T.K.; Dudani, J.S.; Goda, K.; Woods, T.A.; Graves, S.W.; Di Carlo, D. Inertial Manipulation and Transfer of Microparticles across Laminar Fluid Streams. Small 2012, 8, 2757–2764. [Google Scholar] [CrossRef]
- Cho, S.; Jo, W.; Heo, Y.; Kang, J.Y.; Kwak, R.; Park, J. Isolation of Extracellular Vesicle from Blood Plasma Using Electrophoretic Migration through Porous Membrane. Sens. Actuators B Chem. 2016, 233, 289–297. [Google Scholar] [CrossRef]
- Tao, D.; Shui, B.; Gu, Y.; Cheng, J.; Zhang, W.; Jaffrezic-Renault, N.; Song, S.; Guo, Z. Development of a Label-Free Electrochemical Aptasensor for the Detection of Tau381 and Its Preliminary Application in AD and Non-AD Patients’ Sera. Biosensors 2019, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A Visible and Colorimetric Aptasensor Based on DNA-Capped Single-Walled Carbon Nanotubes for Detection of Exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cheng, S.-B.; Cao, P.; Qiu, Q.-F.; Chen, Y.; Xie, M.; Xu, Y.; Huang, W.-H. Detection of Exosomes by ZnO Nanowires Coated Three-Dimensional Scaffold Chip Device. Biosens. Bioelectron. 2018, 122, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.-G.; Kong, M.-Q.; Zhou, S.; Sheng, Y.-F.; Wang, P.; Yu, T.; Inci, F.; Kuo, W.P.; Li, L.-J.; Demirci, U.; et al. An Integrated Double-Filtration Microfluidic Device for Isolation, Enrichment and Quantification of Urinary Extracellular Vesicles for Detection of Bladder Cancer. Sci. Rep. 2017, 7, 46224. [Google Scholar] [CrossRef] [PubMed]
- Chemically Functionalised Graphene FET Biosensor for the Label-Free Sensing of Exosomes | Scientific Reports. Available online: https://www.nature.com/articles/s41598-019-50412-9 (accessed on 14 July 2020).
- Robbins, P.D. Extracellular Vesicles and Aging. Stem Cell Investig. 2017, 4, 98. [Google Scholar] [CrossRef] [PubMed]
- D’Anca, M.; Fenoglio, C.; Serpente, M.; Arosio, B.; Cesari, M.; Scarpini, E.A.; Galimberti, D. Exosome Determinants of Physiological Aging and Age-Related Neurodegenerative Diseases. Front. Aging Neurosci. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Real Time and Label Free Profiling of Clinically Relevant Exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. ACS Nano 2017, 11, 11182–11193. [Google Scholar] [CrossRef]
- Theodoraki, M.-N.; Yerneni, S.; Gooding, W.E.; Ohr, J.; Clump, D.A.; Bauman, J.E.; Ferris, R.L.; Whiteside, T.L. Circulating Exosomes Measure Responses to Therapy in Head and Neck Cancer Patients Treated with Cetuximab, Ipilimumab, and IMRT. Oncoimmunology 2019, 8, 1593805. [Google Scholar] [CrossRef]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.-Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C.; et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane Vesicles as Conveyors of Immune Responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zeng, X.; An, Z.; Yang, Y.; Eisenbaum, M.; Gu, X.; Jornet, J.M.; Dy, G.K.; Reid, M.E.; Gan, Q.; et al. Sensitive Detection of Exosomal Proteins via a Compact Surface Plasmon Resonance Biosensor for Cancer Diagnosis. ACS Sens. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ramshani, Z.; Zhang, C.; Richards, K.; Chen, L.; Xu, G.; Stiles, B.L.; Hill, R.; Senapati, S.; Go, D.B.; Chang, H.-C. Extracellular Vesicle MicroRNA Quantification from Plasma Using an Integrated Microfluidic Device. Commun. Biol. 2019, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Go, D.B.; Atashbar, M.Z.; Ramshani, Z.; Chang, H.-C. Surface Acoustic Wave Devices for Chemical Sensing and Microfluidics: A Review and Perspective. Anal. Methods 2017, 9, 4112–4134. [Google Scholar] [CrossRef]
- Taller, D.; Richards, K.; Slouka, Z.; Senapati, S.; Hill, R.; Go, D.B.; Chang, H.-C. On-Chip Surface Acoustic Wave Lysis and Ion-Exchange Nanomembrane Detection of Exosomal RNA for Pancreatic Cancer Study and Diagnosis. Lab Chip 2015, 15, 1656–1666. [Google Scholar] [CrossRef]
- Rama, E.C.; González-García, M.B.; Costa-García, A. Competitive Electrochemical Immunosensor for Amyloid-Beta 1-42 Detection Based on Gold Nanostructurated Screen-Printed Carbon Electrodes. Sens. Actuators B Chem. 2014, 201, 567–571. [Google Scholar] [CrossRef]
- Tao, W.; Xie, Q.; Wang, H.; Ke, S.; Lin, P.; Zeng, X. Integration of a Miniature Quartz Crystal Microbalance with a Microfluidic Chip for Amyloid Beta-Aβ42 Quantitation. Sensors (Basel) 2015, 15, 25746–25760. [Google Scholar] [CrossRef]
- Yoo, Y.K.; Kim, J.; Kim, G.; Kim, Y.S.; Kim, H.Y.; Lee, S.; Cho, W.W.; Kim, S.; Lee, S.-M.; Lee, B.C.; et al. A Highly Sensitive Plasma-Based Amyloid-β Detection System through Medium-Changing and Noise Cancellation System for Early Diagnosis of the Alzheimer’s Disease. Sci. Rep. 2017, 7, 8882. [Google Scholar] [CrossRef]
- Ameri, M.; Shabaninejad, Z.; Movahedpour, A.; Sahebkar, A.; Mohammadi, S.; Hosseindoost, S.; Ebrahimi, M.S.; Savardashtaki, A.; Karimipour, M.; Mirzaei, H. Biosensors for Detection of Tau Protein as an Alzheimer’s Disease Marker. Int. J. Biol. Macromol. 2020, 162, 1100–1108. [Google Scholar] [CrossRef]
- Frost, B.; Götz, J.; Feany, M.B. Connecting the Dots Between Tau Dysfunction and Neurodegeneration. Trends Cell Biol. 2015, 25, 46–53. [Google Scholar] [CrossRef]
- Vestergaard, M.; Kerman, K.; Kim, D.-K.; Hiep, H.M.; Tamiya, E. Detection of Alzheimer’s Tau Protein Using Localised Surface Plasmon Resonance-Based Immunochip. Talanta 2008, 74, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Scarano, S.; Lisi, S.; Palladino, P.; Minunni, M. Real-Time Tau Protein Detection by Sandwich-Based Piezoelectric Biosensing: Exploring Tubulin as a Mass Enhancer. Sensors 2018, 18, 946. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, V.S.E.; Langford, T.; Sefo, L.A.-C.; Hewitt, A.W.; Verma, N. Screening for Diabetic Eye Disease among Samoan Adults: A Pilot Study. Ophthalmol. Ther. 2017, 6, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Courte, J.; Bousset, L.; Boxberg, Y.V.; Villard, C.; Melki, R.; Peyrin, J.-M. The Expression Level of Alpha-Synuclein in Different Neuronal Populations Is the Primary Determinant of Its Prion-like Seeding. Sci. Rep. 2020, 10, 4895. [Google Scholar] [CrossRef] [PubMed]
- Horrocks, M.H.; Tosatto, L.; Dear, A.J.; Garcia, G.A.; Iljina, M.; Cremades, N.; Dalla Serra, M.; Knowles, T.P.J.; Dobson, C.M.; Klenerman, D. Fast Flow Microfluidics and Single-Molecule Fluorescence for the Rapid Characterization of α-Synuclein Oligomers. Anal. Chem. 2015, 87, 8818–8826. [Google Scholar] [CrossRef]
- Fernandes, J.T.S.; Chutna, O.; Chu, V.; Conde, J.P.; Outeiro, T.F. A Novel Microfluidic Cell Co-Culture Platform for the Study of the Molecular Mechanisms of Parkinson’s Disease and Other Synucleinopathies. Front. Neurosci. 2016, 10. [Google Scholar] [CrossRef]
- Boukouris, S.; Mathivanan, S. Exosomes in Bodily Fluids Are a Highly Stable Resource of Disease Biomarkers. Proteomics Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef]
- Nik Mohamed Kamal, N.N.S.B.; Shahidan, W.N.S. Non-Exosomal and Exosomal Circulatory MicroRNAs: Which Are More Valid as Biomarkers? Front. Pharmacol. 2020, 10. [Google Scholar] [CrossRef]
- Meng, W.; He, C.; Hao, Y.; Wang, L.; Li, L.; Zhu, G. Prospects and Challenges of Extracellular Vesicle-Based Drug Delivery System: Considering Cell Source. Drug Deliv. 2020, 27, 585–598. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of Preclinical Alzheimer’s Disease by a Profile of Pathogenic Proteins in Neurally Derived Blood Exosomes: A Case-Control Study. Alzheimers Dement 2015, 11, 600–607.e1. [Google Scholar] [CrossRef]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.Y.; et al. Exosome-Associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef] [PubMed]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kügler, S.; Ikezu, T. Depletion of Microglia and Inhibition of Exosome Synthesis Halt Tau Propagation. Nat. Neurosci. 2015, 18, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Polanco, J.C.; Scicluna, B.J.; Hill, A.F.; Götz, J. Extracellular Vesicles Isolated from the Brains of RTg4510 Mice Seed Tau Protein Aggregation in a Threshold-Dependent Manner. J. Biol. Chem. 2016, 291, 12445–12466. [Google Scholar] [CrossRef] [PubMed]
- Sardar Sinha, M.; Ansell-Schultz, A.; Civitelli, L.; Hildesjö, C.; Larsson, M.; Lannfelt, L.; Ingelsson, M.; Hallbeck, M. Alzheimer’s Disease Pathology Propagation by Exosomes Containing Toxic Amyloid-Beta Oligomers. Acta Neuropathol. 2018, 136, 41–56. [Google Scholar] [CrossRef]
- Ngolab, J.; Trinh, I.; Rockenstein, E.; Mante, M.; Florio, J.; Trejo, M.; Masliah, D.; Adame, A.; Masliah, E.; Rissman, R.A. Brain-Derived Exosomes from Dementia with Lewy Bodies Propagate α-Synuclein Pathology. Acta Neuropathol. Commun. 2017, 5, 46. [Google Scholar] [CrossRef]
- Sun, J.; Wang, L.; Bao, H.; Premi, S.; Das, U.; Chapman, E.R.; Roy, S. Functional Cooperation of α-Synuclein and VAMP2 in Synaptic Vesicle Recycling. Proc. Natl. Acad. Sci. USA 2019, 116, 11113–11115. [Google Scholar] [CrossRef]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Shamout, F.; Zhu, T.; Clifton, D.A. Machine Learning for Clinical Outcome Prediction. IEEE Rev. Biomed. Eng. 2020. [Google Scholar] [CrossRef]
- Weng, W.-H. Machine Learning for Clinical Predictive Analytics. In Leveraging Data Science for Global Health; Celi, L.A., Majumder, M.S., Ordóñez, P., Osorio, J.S., Paik, K.E., Somai, M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 199–217. ISBN 978-3-030-47994-7. [Google Scholar]
- Fisher, C.K.; Smith, A.M.; Walsh, J.R. Machine Learning for Comprehensive Forecasting of Alzheimer’s Disease Progression. Sci. Rep. 2019, 9, 13622. [Google Scholar] [CrossRef]





| Criteria | Description | |
|---|---|---|
| R | Real-time connectivity | Tests are connected, and/or a reader or mobile phone is used to power the reaction and/or read the test results to give appropriate data to decision-makers |
| E | Ease of specimen collection | Tests should be designed for use with non-invasive specimens |
| A | Affordable | Tests are affordable to end-users and health systems |
| S | Sensitive | Avoid false-negatives |
| S | Specific | Avoid false-positives |
| U | User-friendly | The procedure of testing is simple with few steps and little training |
| R | Rapid and robust | Results are available for giving treatment within the first visit (15 min to 2 h); Tests can survive as stock without additional transport or storage like refrigeration |
| E | Equipment-free or simple environment | The test does not require any special equipment |
| D | Deliverable to end-users | Accessible to those who need the tests |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeCastro, J.; Littig, J.; Chou, P.P.; Mack-Onyeike, J.; Srinivasan, A.; Conboy, M.J.; Conboy, I.M.; Aran, K. The Microfluidic Toolbox for Analyzing Exosome Biomarkers of Aging. Molecules 2021, 26, 535. https://doi.org/10.3390/molecules26030535
DeCastro J, Littig J, Chou PP, Mack-Onyeike J, Srinivasan A, Conboy MJ, Conboy IM, Aran K. The Microfluidic Toolbox for Analyzing Exosome Biomarkers of Aging. Molecules. 2021; 26(3):535. https://doi.org/10.3390/molecules26030535
Chicago/Turabian StyleDeCastro, Jonalyn, Joshua Littig, Peichi Peggy Chou, Jada Mack-Onyeike, Amrita Srinivasan, Michael J. Conboy, Irina M. Conboy, and Kiana Aran. 2021. "The Microfluidic Toolbox for Analyzing Exosome Biomarkers of Aging" Molecules 26, no. 3: 535. https://doi.org/10.3390/molecules26030535
APA StyleDeCastro, J., Littig, J., Chou, P. P., Mack-Onyeike, J., Srinivasan, A., Conboy, M. J., Conboy, I. M., & Aran, K. (2021). The Microfluidic Toolbox for Analyzing Exosome Biomarkers of Aging. Molecules, 26(3), 535. https://doi.org/10.3390/molecules26030535








