DNA-Based Electrodes and Computational Approaches on the Intercalation Study of Antitumoral Drugs
Abstract
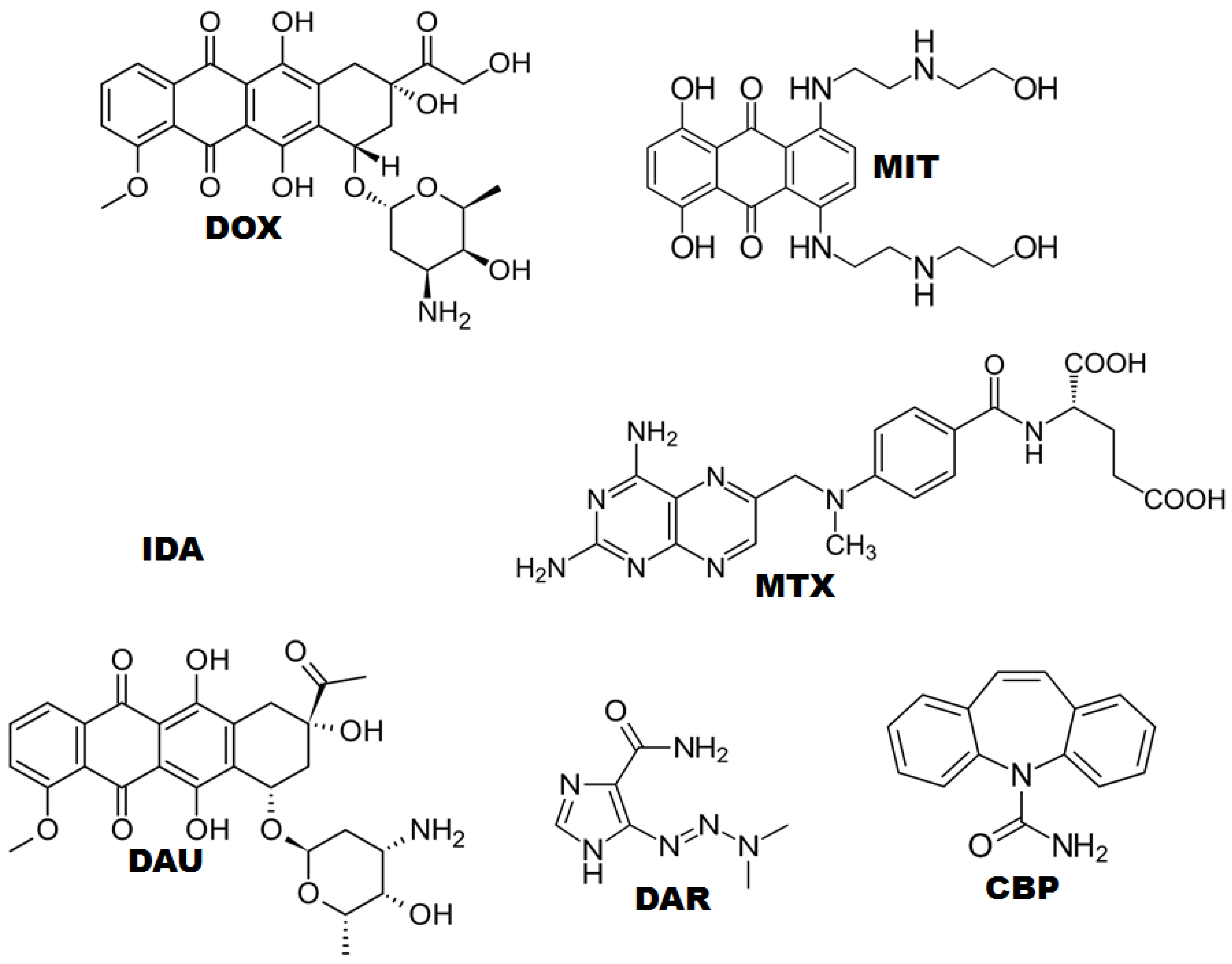
:1. Introduction
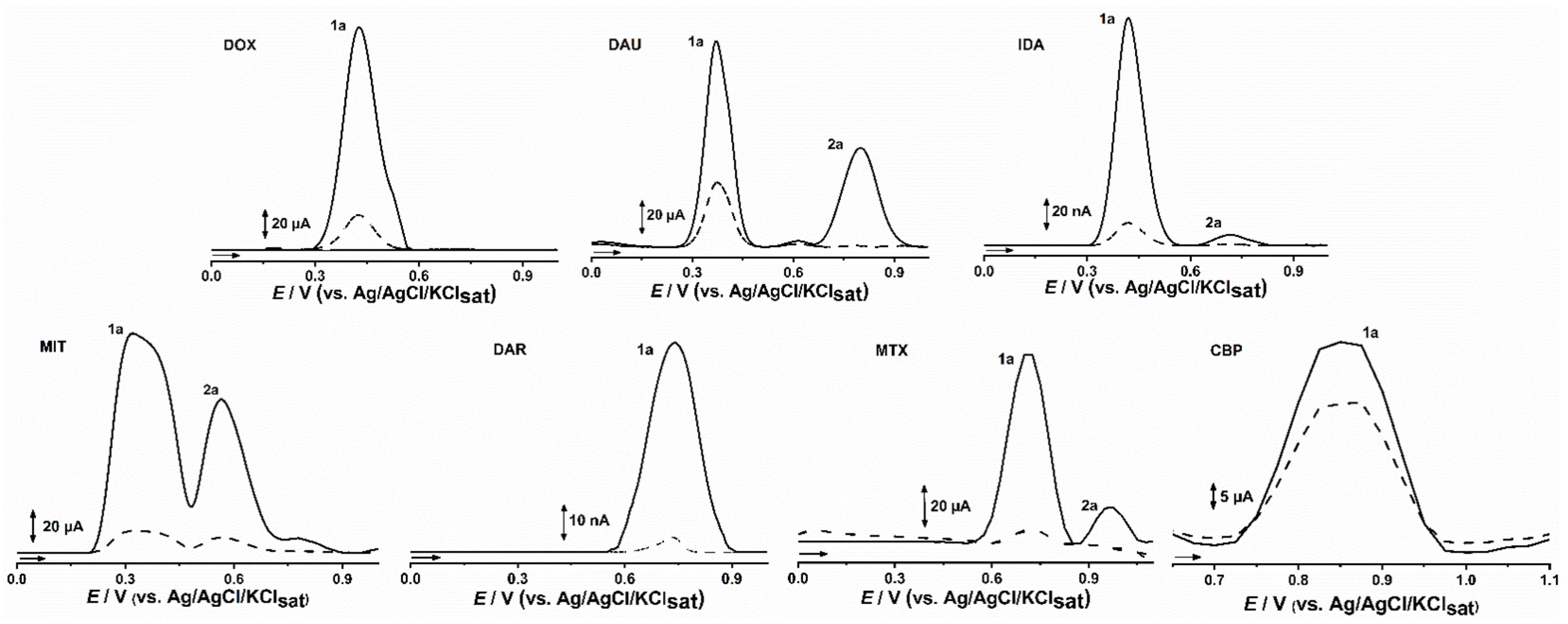
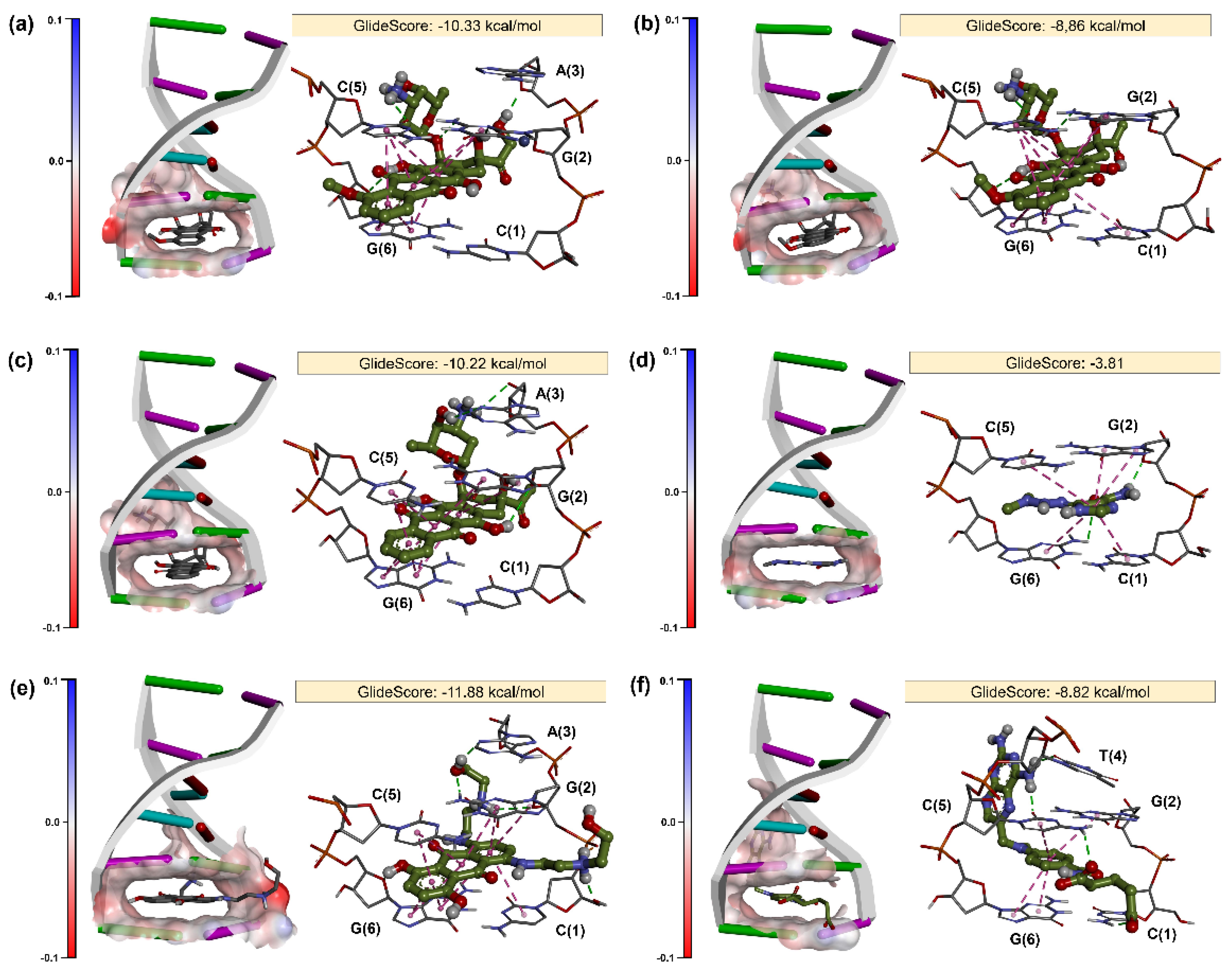
2. Results
2.1. CPDE for Voltammetric Evaluation of Anticancer Drugs
2.2. Molecular Docking
3. Materials and Methods
3.1. Reagents and Solutions
3.2. Electrochemical Assays
3.2.1. Carbon Paste Electrodes (CPE and CPDE)
3.2.2. Voltammetric Evaluation of Anticancer Drug Uptake
3.3. Molecular Docking
3.3.1. Ligand Preparation
3.3.2. DNA Preparation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Findik, M.; Bingol, H.; Erdem, A. Sensors and Actuators: B. Chemical Electrochemical Detection of Interaction between Daunorubicin and DNA by Hybrid Nanoflowers Modified Graphite Electrodes. Sens. Actuators B Chem. 2020, 329, 129120. [Google Scholar] [CrossRef]
- Li, D.; Xu, Y.; Fan, L.; Shen, B.; Ding, X.; Yuan, R.; Li, X.; Chen, W. Target-Driven Rolling Walker Based Electrochemical Biosensor for Ultrasensitive Detection of Circulating Tumor DNA Using Doxorubicin@tetrahedron-Au Tags. Biosens. Bioelectron. 2020, 148, 111826. [Google Scholar] [CrossRef]
- Diculescu, V.C.; Chiorcea-Paquim, A.M.; Oliveira-Brett, A.M. Applications of a DNA-Electrochemical Biosensor. TrAC-Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Pontinha, A.D.R.; Jorge, S.M.A.; Chiorcea Paquim, A.M.; Diculescu, V.C.; Oliveira-Brett, A.M. In Situ Evaluation of Anticancer Drug Methotrexate-DNA Interaction Using a DNA-Electrochemical Biosensor and AFM Characterization. Phys. Chem. Chem. Phys. 2011, 13, 5227–5234. [Google Scholar] [CrossRef] [PubMed]
- Ozluer, C.; Kara, H.E.S. In Vitro DNA Binding Studies of Anticancer Drug Idarubicin Using Spectroscopic Techniques. J. Photochem. Photobiol. B Biol. 2014, 138, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Charak, S.; Mehrotra, R. Structural Investigation of Idarubicin-DNA Interaction: Spectroscopic and Molecular Docking Study. Int. J. Biol. Macromol. 2013, 60, 213–218. [Google Scholar] [CrossRef]
- Bhattacharyya, J.; Basu, A.; Suresh Kumar, G. Intercalative Interaction of the Anticancer Drug Mitoxantrone with Double Stranded DNA: A Calorimetric Characterization of the Energetics. J. Chem. Thermodyn. 2014, 75, 45–51. [Google Scholar] [CrossRef]
- Krzak, A.; Swiech, O.; Majdecki, M.; Bilewicz, R. Complexing Daunorubicin with β-Cyclodextrin Derivative Increases Drug Intercalation into DNA. Electrochim. Acta 2017, 247, 139–148. [Google Scholar] [CrossRef]
- Radi, A.E.; Eissa, A.; Nassef, H.M. Voltammetric and Spectroscopic Studies on the Binding of the Antitumor Drug Dacarbazine with DNA. J. Electroanal. Chem. 2014, 717–718, 24–28. [Google Scholar] [CrossRef]
- Skalová, Š.; Langmaier, J.; Barek, J.; Vyskočil, V.; Navrátil, T. Doxorubicin Determination Using Two Novel Voltammetric Approaches: A Comparative Study. Electrochim. Acta 2020, 330, 135180. [Google Scholar] [CrossRef]
- Ribeiro, J.A.; Pereira, C.M.; Silva, F. Electrochemistry of the Interaction between Bioactive Drugs Daunorubicin and Dopamine and DNA at a Water/Oil Interface. Electrochim. Acta 2015, 180, 687–694. [Google Scholar] [CrossRef]
- Eda Satana Kara, H. Redox Mechanism of Anticancer Drug Idarubicin and In-Situ Evaluation of Interaction with DNA Using an Electrochemical Biosensor. Bioelectrochemistry 2014, 99, 17–23. [Google Scholar] [CrossRef]
- Ahmad, I.; Ahmad, M. Dacarbazine as a Minor Groove Binder of DNA: Spectroscopic, Biophysical and Molecular Docking Studies. Int. J. Biol. Macromol. 2015, 79, 193–200. [Google Scholar] [CrossRef]
- Deepa, S.; Swamy, B.E.K.; Pai, K.V. A Surfactant SDS Modified Carbon Paste Electrode as an Enhanced and Effective Electrochemical Sensor for the Determination of Doxorubicin and Dacarbazine Its Applications: A Voltammetric Study. J. Electroanal. Chem. 2020, 879, 114748. [Google Scholar] [CrossRef]
- Agarwal, S.; Jangir, D.K.; Mehrotra, R. Spectroscopic Studies of the Effects of Anticancer Drug Mitoxantrone Interaction with Calf-Thymus DNA. J. Photochem. Photobiol. B Biol. 2013, 120, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Rafique, B.; Khalid, A.M.; Akhtar, K.; Jabbar, A. Interaction of Anticancer Drug Methotrexate with DNA Analyzed by Electrochemical and Spectroscopic Methods. Biosens. Bioelectron. 2013, 44, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Kurbanoglu, S.; Dogan-Topal, B.; Hlavata, L.; Labuda, J.; Ozkan, S.A.; Uslu, B. Electrochemical Investigation of an Interaction of the Antidepressant Drug Aripiprazole with Original and Damaged Calf Thymus DsDNA. Electrochim. Acta 2015, 169, 233–240. [Google Scholar] [CrossRef]
- Aydoǧdu, G.; Günendi, G.; Zeybek, D.K.; Zeybek, B.; Pekyardimci, Ş. A Novel Electrochemical DNA Biosensor Based on Poly-(5-Amino-2-Mercapto-1, 3,4-Thiadiazole) Modified Glassy Carbon Electrode for the Determination of Nitrofurantoin. Sens. Actuators B Chem. 2014, 197, 211–219. [Google Scholar] [CrossRef]
- Tomé, L.I.N.; Marques, N.V.; Diculescu, V.C.; Oliveira-Brett, A.M. In Situ DsDNA-Bevacizumab Anticancer Monoclonal Antibody Interaction Electrochemical Evaluation. Anal. Chim. Acta 2015, 898, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Diculescu, V.C.; Oliveira-Brett, A.M. In Situ Electrochemical Evaluation of DsDNA Interaction with the Anticancer Drug Danusertib Nitrenium Radical Product Using the DNA-Electrochemical Biosensor. Bioelectrochemistry 2016, 107, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Lopes, I.C.; Carlos, S.; Oliveira, B.; Oliveira-brett, A.M. In Situ Electrochemical Evaluation of Anticancer Drug Temozolomide and Its Metabolites-DNA Interaction. Anal. Bioanal. Chem. 2013, 405, 3783–3790. [Google Scholar] [CrossRef] [PubMed]
- Buoro, R.M.; Lopes, I.C.; Diculescu, V.C.; Serrano, S.H.P.; Lemos, L.; Oliveira-Brett, A.M. In Situ Evaluation of Gemcitabine-DNA Interaction Using a DNA-Electrochemical Biosensor. Bioelectrochemistry 2014, 99, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Issaad, F.Z.; Tomé, L.I.N.; Marques, N.V.; Mouats, C.; Diculescu, V.C.; Oliveira-Brett, A.M. Bevacizumab Anticancer Monoclonal Antibody: Native and Denatured Redox Behaviour. Electrochim. Acta 2016, 206, 246–253. [Google Scholar] [CrossRef]
- Carlos, B.; Oliveira, S.; Oliveira-Brett, A.M. DNA-Electrochemical Biosensors: AFM Surface Characterisation and Application to Detection of In Situ Oxidative Damage to DNA. Comb. Chem. High Throughput Screen. 2012, 13, 628–640. [Google Scholar] [CrossRef]
- Satana, H.E.; Pontinha, A.D.R.; Diculescu, V.C.; Oliveira-Brett, A.M. Nucleoside Analogue Electrochemical Behaviour and in Situ Evaluation of DNA-Clofarabine Interaction. Bioelectrochemistry 2012, 87, 3–8. [Google Scholar] [CrossRef]
- Bayraktepe, D.E. A voltammetric study on drug-DNA interactions: Kinetic and thermodynamic aspects of the relations between the anticancer agent dasatinib and ds-DNA using a pencil lead graphite electrode. Microchem. J. 2020, 157, 104946. [Google Scholar] [CrossRef]
- Wanga, S.; Penga, T.; Yang, C.F. Electrochemical determination of interaction parameters for DNA and mitoxantrone in an irreversible redox process. Biophys. Chem. 2003, 104, 239–248. [Google Scholar] [CrossRef]
- Chen, J.; Fu, B.; Liu, T.; Yan, Z.; Li, K. A Graphene Oxide-DNA Electrochemical Sensor Based on Glassy Carbon Electrode for Sensitive Determination of Methotrexate. Electroanalysis 2018, 30, 288–295. [Google Scholar] [CrossRef]
- Alizadeh, M.; Azar, P.A.; Mozaffari, S.A.; Karimi-Maleh, H.; Tamaddon, A.-M. A DNA Based Biosensor Amplified With ZIF-8/Ionic Liquid Composite for Determination of Mitoxantrone Anticancer Drug: An Experimental/Docking Investigation. Front. Chem. 2020, 8, 814. [Google Scholar] [CrossRef]
- Hahn, Y.; Lee, H.Y. Electrochemical Behavior and Square Wave Voltammetric Determination of Doxorubicin Hydrochloride. Arch. Pharmacal Res. 2004, 27, 31–34. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A Software Program for PK(a) Prediction and Protonation State Generation for Drug-like Molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Watts, K.S.; Dalal, P.; Murphy, R.B.; Sherman, W.; Friesner, R.A.; Shelley, J.C. ConfGen: A Conformational Search Method for Efficient Generation of Bioactive Conformers. J. Chem. Inf. Model. 2010, 50, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.J.; Jenkins, T.C.; Neidle, S. Crystal Structure of a Pentamidine-Oligonucleotide Complex: Implications for DNA-Binding Properties. Biochemistry 1992, 31, 7104–7109. [Google Scholar] [CrossRef] [PubMed]
- Canals, A.; Purciolas, M.; Aymamí, J.; Coll, M. The Anticancer Agent Ellipticine Unwinds DNA by Intercalative Binding in an Orientation Parallel to Base Pairs. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 1009–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of PKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauria, A.; Barraja, P.; Dattolo, G.; Almerico, A.M. DNA Minor Groove Binders: An Overview on Molecular Modeling and QSAR Approaches. Curr. Med. Chem. 2007, 14, 2136–2160. [Google Scholar] [CrossRef]
| Drugs (I/μA) | |||||||
|---|---|---|---|---|---|---|---|
| Time (Seconds) | MIT | MTX | IDA | DAU | DOX | DAR | CBP |
| 444.481 g/mol | 454.44 g/mol | 497.500 g/mol | 527.52 g/mol | 543.52 g/mol | 182.187 g/mol | 236.269 g/mol | |
| 10 | 18 | 15 | 19 | 19 | 20 | 7 | 5 |
| 60 | 41 | 36 | 25 | 27 | 32 | 19 | 9 |
| 300 | 74 | 63 | 53 | 48 | 54 | 33 | 16 |
| ΔIt300-t10 | 56 | 48 | 34 | 29 | 34 | 26 | 11 |
| ΔICPDE-CPE | 11 | 10 | 5.0 | 4.7 | 3.8 | 5.6 | 1.4 |
| Glid Score | −11.9 | −8.9 | −10.3 | −8.9 | −10.3 | −3.8 | 0 |
| IC50 (μM) | 1.6 * | 0.6 * | 0.03 ** | 0.03 ** | 0.2 * | 20 ** | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, E.S.B.; Macêdo, I.Y.L.d.; Silva, G.N.d.M.e.; de Carvalho e Silva, A.; Gil, H.P.V.; Neves, B.J.; Gil, E.d.S. DNA-Based Electrodes and Computational Approaches on the Intercalation Study of Antitumoral Drugs. Molecules 2021, 26, 7623. https://doi.org/10.3390/molecules26247623
Rodrigues ESB, Macêdo IYLd, Silva GNdMe, de Carvalho e Silva A, Gil HPV, Neves BJ, Gil EdS. DNA-Based Electrodes and Computational Approaches on the Intercalation Study of Antitumoral Drugs. Molecules. 2021; 26(24):7623. https://doi.org/10.3390/molecules26247623
Chicago/Turabian StyleRodrigues, Edson Silvio Batista, Isaac Yves Lopes de Macêdo, Giovanna Nascimento de Mello e Silva, Arthur de Carvalho e Silva, Henric Pietro Vicente Gil, Bruno Junior Neves, and Eric de Souza Gil. 2021. "DNA-Based Electrodes and Computational Approaches on the Intercalation Study of Antitumoral Drugs" Molecules 26, no. 24: 7623. https://doi.org/10.3390/molecules26247623
APA StyleRodrigues, E. S. B., Macêdo, I. Y. L. d., Silva, G. N. d. M. e., de Carvalho e Silva, A., Gil, H. P. V., Neves, B. J., & Gil, E. d. S. (2021). DNA-Based Electrodes and Computational Approaches on the Intercalation Study of Antitumoral Drugs. Molecules, 26(24), 7623. https://doi.org/10.3390/molecules26247623






