Biosynthesis of Tetrapyrrole Cofactors by Bacterial Community Inhabiting Porphyrine-Containing Shale Rock (Fore-Sudetic Monocline)
Abstract
1. Introduction
2. Results
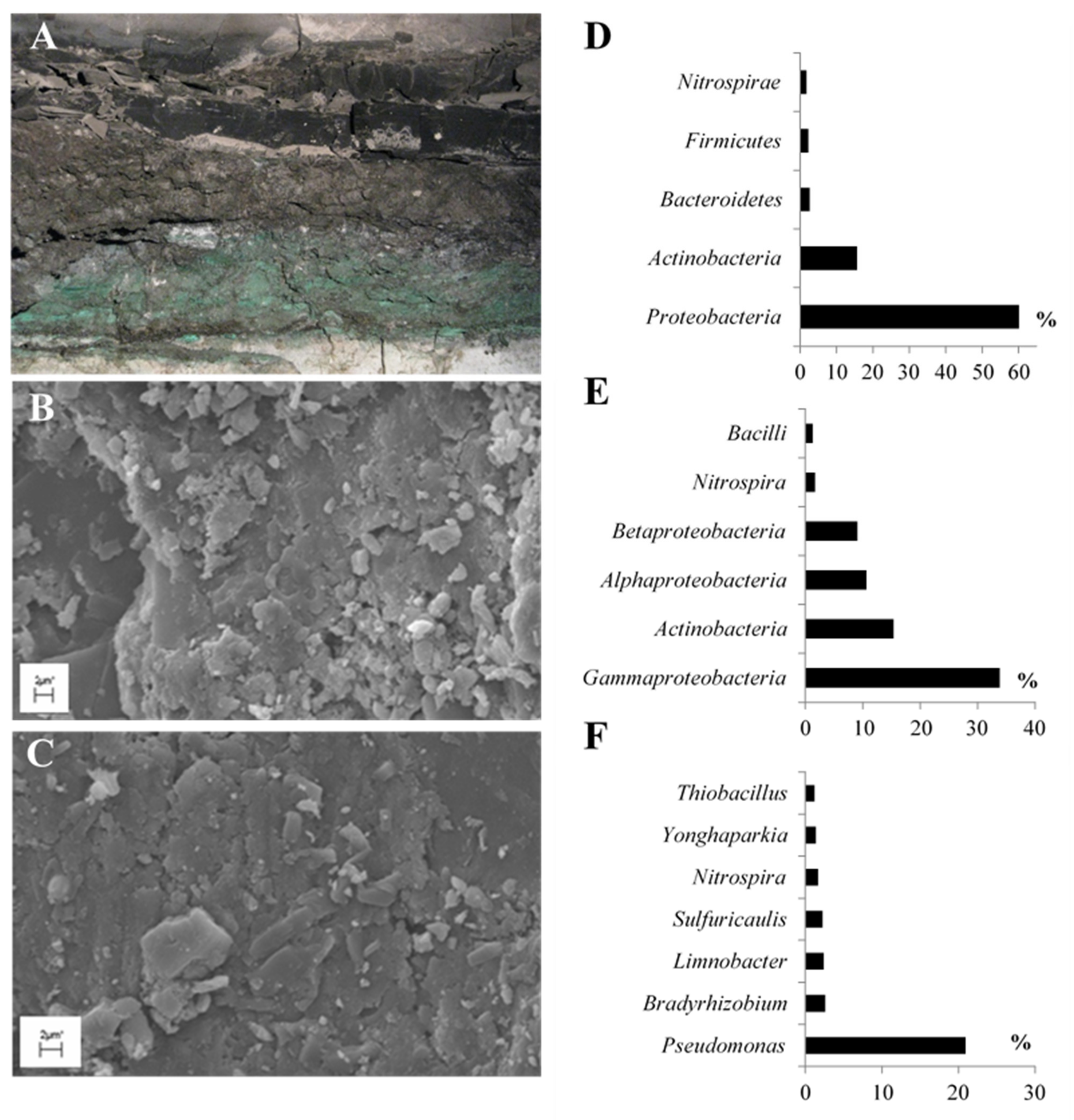
2.1. Taxonomic Diversity of BC Inhabiting Black Shale
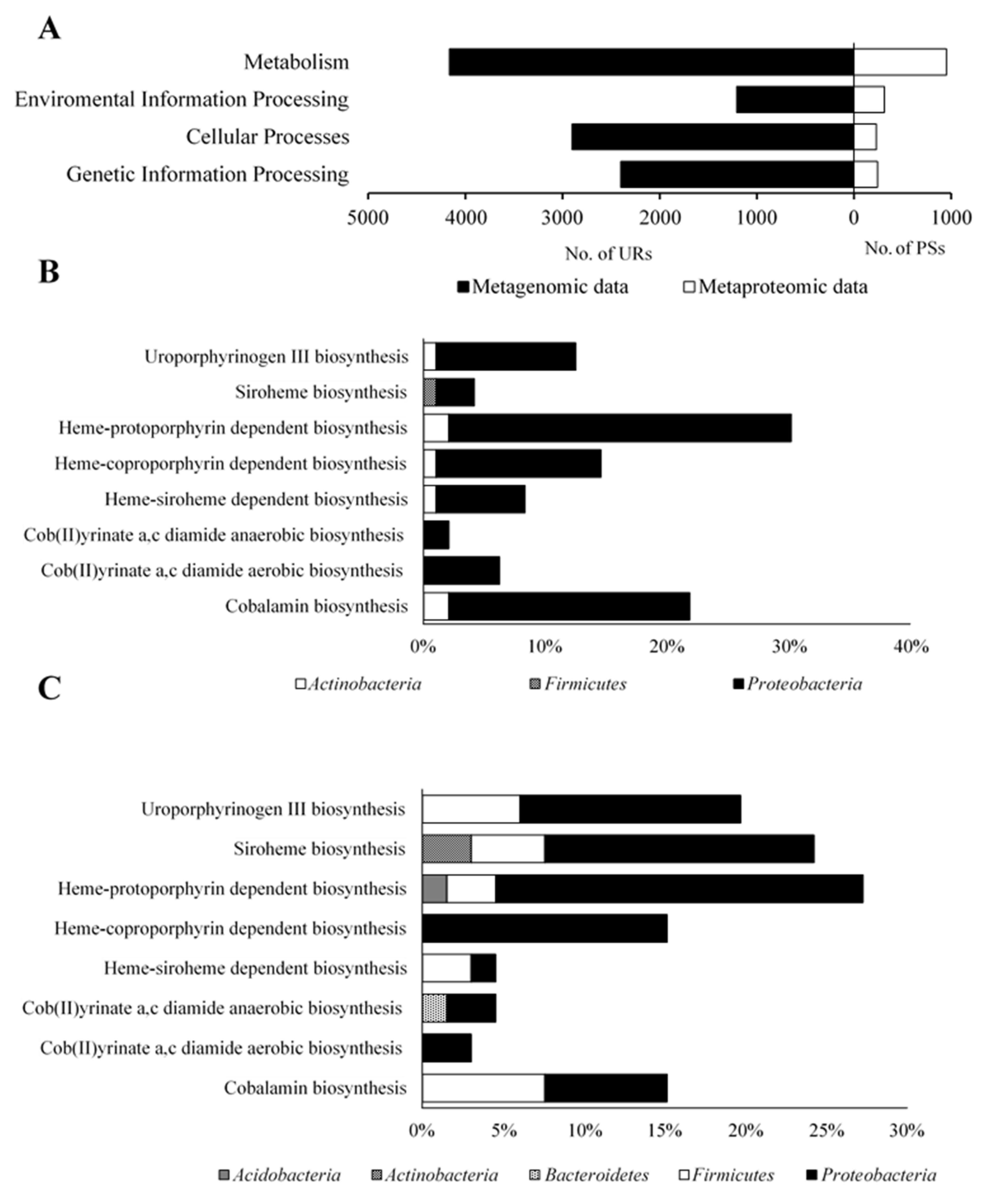
2.2. General Characteristics of the Metagenome and Metaproteome of BC
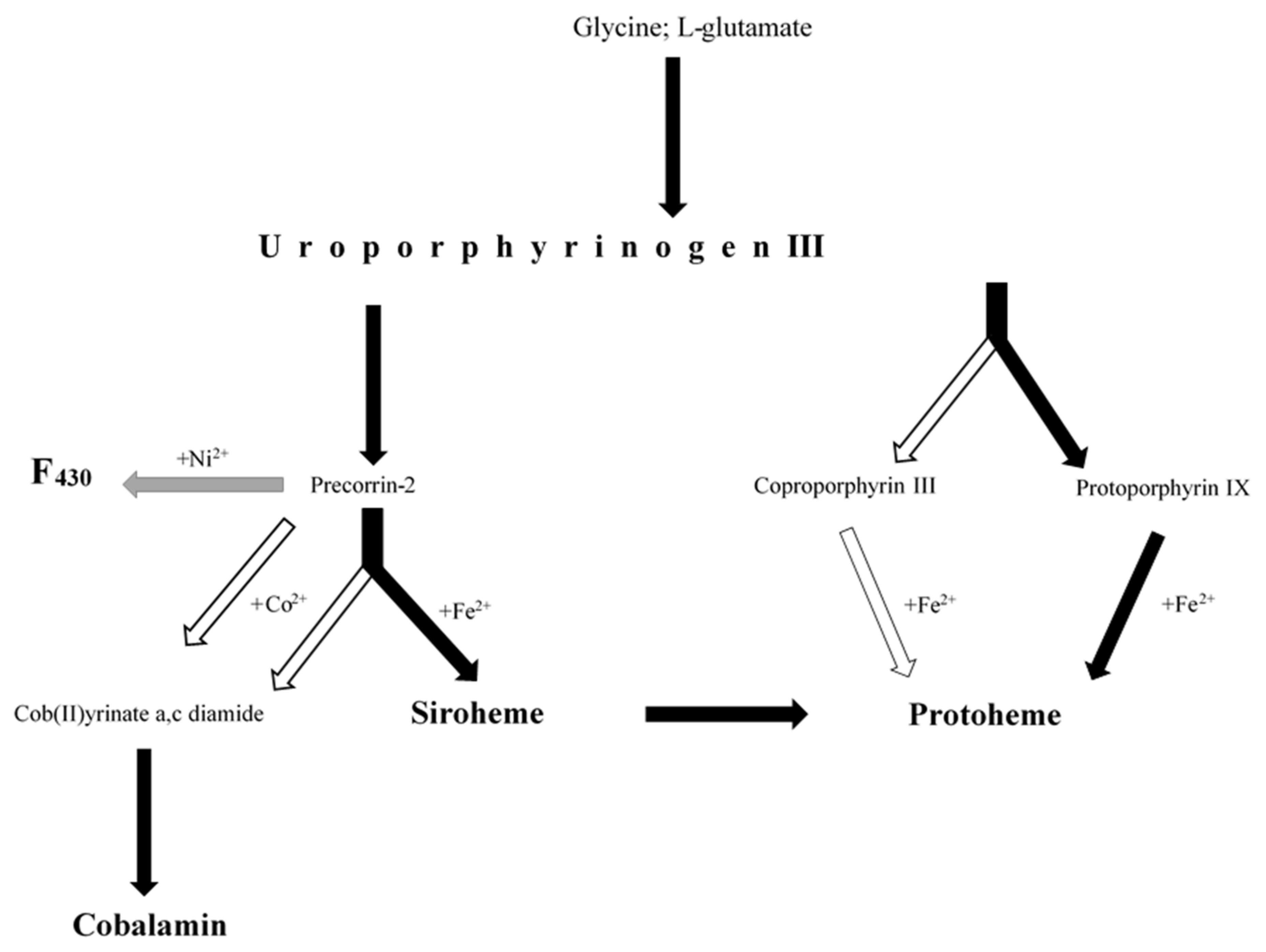
2.3. Biosynthesis of Cofactors Detected in the Studied BC
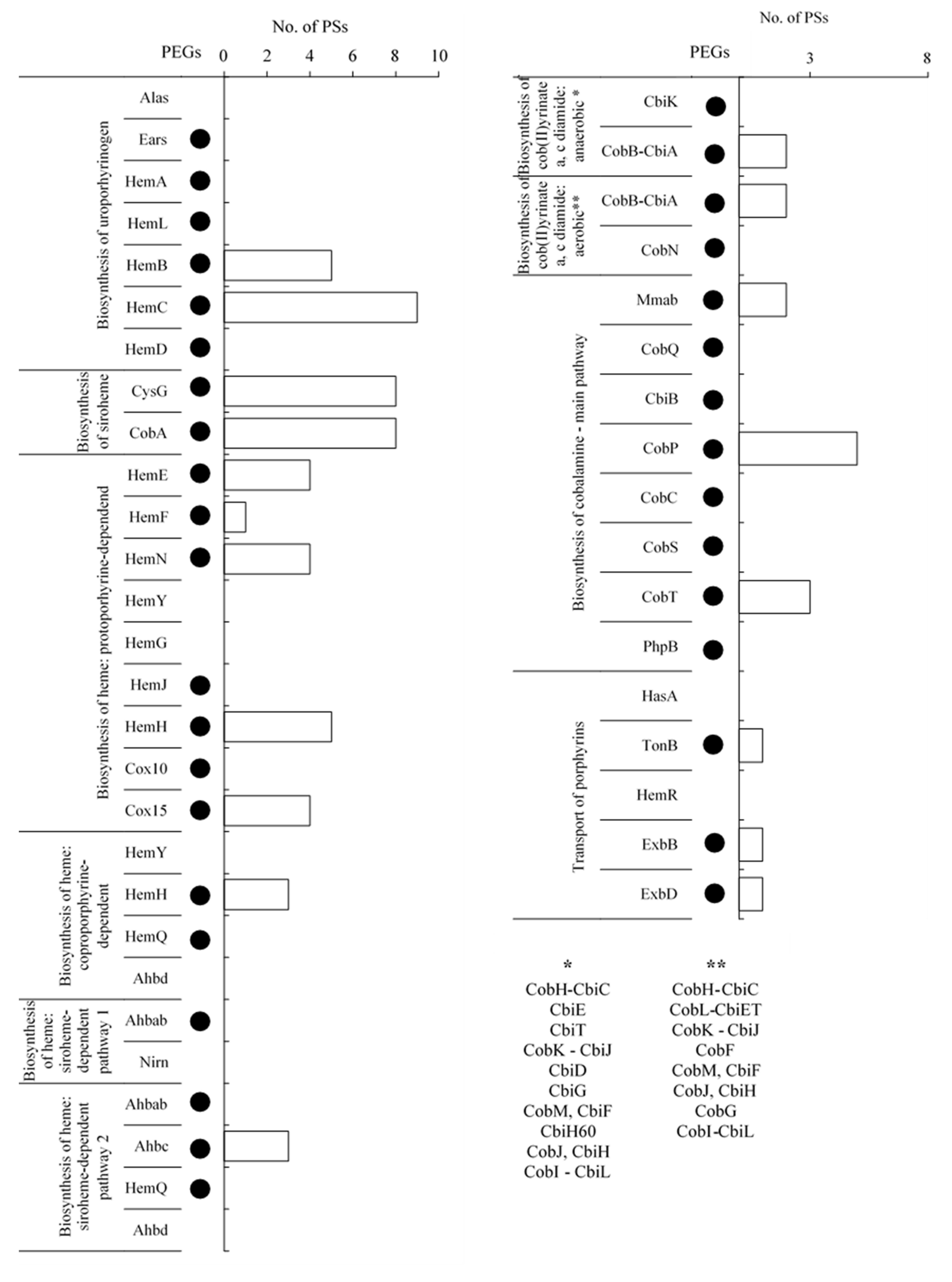
2.3.1. Uroporphyrinogen III Biosynthesis
2.3.2. Siroheme Biosynthesis
2.3.3. Heme Biosynthesis
Protoporphyrin-Dependent Heme Biosynthetic Pathway
Coproporphyrin-Dependent Heme Biosynthetic Pathway
Siroheme-Dependent Heme Biosynthetic Pathway
2.3.4. Cobalamin Biosynthesis
Biosynthesis of Anaerobic and Aerobic Cob(II)yrinate a,c-Diamide
Biosynthesis of Cobalamin from Cob(II)yrinate a,c-Diamide
2.3.5. Transport of Exogenous Porphyrins
2.3.6. Tetrapyrrole Cofactor-Containing Enzymes Produced by BC
3. Discussion
4. Materials and Methods
4.1. Site and Sample Description
4.2. Isolation of DNA
4.3. Sequencing and Analysis of DNA
4.4. Isolation of Proteins
4.5. Identification of Proteins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Barton, L.L.; Fardeau, M.; Fauque, G.D. Hydrogen sulfide: A toxic gas produced by dissimilatory sulfate and sulfur reduction and consumed by microbial oxidation. In The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment; Springer: New York, NY, USA, 2014; pp. 237–277. [Google Scholar]
- Roth, J.R.; Lawrence, J.G.; Bobik, T.A. Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol. 1996, 50, 137–181. [Google Scholar] [CrossRef]
- Romine, M.F.; Rodionov, D.A.; Maezato, Y.; Anderson, L.N.; Nandhikonda, P.; Rodionova, I.A.; Wright, A.T. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc. Natl. Acad. Sci. USA 2017, 114, E1205–E1214. [Google Scholar] [CrossRef]
- Banerjee, R. Chemistry and Biochemistry of B12; John Wiley & Sons: Hoboken, NJ, USA, 1999. [Google Scholar]
- Janssen, D.B.; Oppentocht, J.E.; Poelarends, G.J. Microbial dehalogenation. Curr. Opin. Biotechnol. 2001, 12, 254–258. [Google Scholar] [CrossRef]
- Rodionov, D.A.; Vitreschak, A.G.; Mironov, A.A.; Gelfand, M.S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003, 278, 41148–41159. [Google Scholar] [CrossRef]
- Stephen, W.; Ragdale, G. Chapter 6. Biochemistry of Methyl-Coenzyme M Reductase: The Nickel Metalloenzyme that Catalyzes the Final Step in Synthesis and the First Step in Anaerobic Oxidation of the Greenhouse Gas Methane. Met. Ions Life Sci. 2014, 14, 125–145. [Google Scholar]
- Eckardt, C.B.; Wolf, M.; Maxwell, J.R. Iron porphyrins in the Permian Kupferschiefer of the lower Rhine Basin, NW Germany. Org. Geochem. 1989, 14, 659–666. [Google Scholar] [CrossRef]
- Wolf, M.; David, P.; Eckardt, C.B.; Hagemann, H.W.; Püttmann, W. Facies and rank of the Permian Kupferschiefer from the Lower Rhine Basin and NW Germany. Int. J. Coal Geol. 1989, 14, 119–136. [Google Scholar] [CrossRef]
- Czechowski, F. Metalloporphyrin composition and a model for the early diagenetic mineralization of the Permian Kupferschiefer, SW Poland. In Organic Matter and Mineralisation: Thermal Alteration, Hydrocarbon Generation and Role in Metallogenesis; Springer: Dordrecht, The Netherlands, 2000; pp. 243–259. [Google Scholar]
- Szubert, A.; Sadowski, Z.; Gros, C.P.; Barbe, J.M.; Guilard, R. Identification of metalloporphyrins extracted from the copper bearing black shale of Fore Sudetic Monocline (Poland). Miner. Eng. 2006, 19, 1212–1215. [Google Scholar] [CrossRef]
- Stasiuk, R.; Matlakowska, R. Postdiagenetic bacterial transformation of nickel and vanadyl sedimentary porphyrins of organic-rich shale rock (Fore-Sudetic Monocline, Poland). Front. Microbiol. 2021. [Google Scholar] [CrossRef]
- Grice, K.; Schaeffer, P.; Schwark, L.; Maxwell, J.R. Changes in palaeoenvironmental conditions during deposition of the Permian Kupferschiefer (Lower Rhine Basin, northwest Germany) inferred from molecular and isotopic compositions of biomarker components. Org. Geochem. 1997, 26, 677–690. [Google Scholar] [CrossRef]
- Tait, G.H. Aminolaevulinate synthetase of Micrococcus denitrificans. Purification and properties of the enzyme, and the effects of growth conditions on the enzyme activity in cells. Biochemistry 1973, 131, 389–403. [Google Scholar] [CrossRef][Green Version]
- Battersby, A.R. Tetrapyrroles: The pigments of life. Nat. Prod. Rep. 2000, 17, 507–526. [Google Scholar] [CrossRef]
- Schauer, S.; Chaturvedi, S.; Randau, L.; Moser, J.; Kitabatake, M.; Lorenz, S.; Jahn, D. Escherichia coli glutamyl-tRNA reductase: Trapping the thioester intermediate. J. Biol. Chem. 2002, 277, 48657–48663. [Google Scholar] [CrossRef]
- Schubert, H.L.; Raux, E.; Brindley, A.A.; Leech, H.K.; Wilson, K.S.; Hill, C.P.; Warren, M.J. The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase. EMBO Rep. 2002, 21, 2068–2075. [Google Scholar] [CrossRef]
- Lobo, S.A.; Brindley, A.; Warren, M.J.; Saraiva, L.M. Functional characterization of the early steps of tetrapyrrole biosynthesis and modification in Desulfovibrio vulgaris Hildenborough. Biochemistry 2009, 420, 317–326. [Google Scholar] [CrossRef]
- Johansson, P.; Hederstedt, L. Organization of genes for tetrapyrrole biosynthesis in gram-positive bacteria. Microbiology 1999, 145, 529–538. [Google Scholar] [CrossRef]
- Layer, G.; Verfu, K.; Mahlitz, E.; Jahn, D. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J. Biol. Chem 2002, 277, 34136–34142. [Google Scholar] [CrossRef]
- Dailey, H.A.; Dailey, T.A.; Gerdes, S.; Jahn, D.; Jahn, M.; O’Brian, M.R.; Warren, M.J. Prokaryotic heme biosynthesis: Multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 2017, 81, e00048-16. [Google Scholar] [CrossRef]
- Dailey, H.A. Enzymes of heme biosynthesis. J. Biol. Inorg. Chem. 1997, 2, 411–417. [Google Scholar] [CrossRef]
- Qin, X.; Sun, L.; Wen, X.; Yang, X.; Tan, Y.; Jin, H.; Shen, Y. Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis. J. Struct. Biol. 2010, 170, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Bali, S.; Lawrence, A.D.; Lobo, S.A.; Saraiva, L.M.; Golding, B.T.; Palmer, D.J.; Warren, M.J. Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc. Natl. Acad. Sci. USA 2011, 108, 18260–18265. [Google Scholar] [CrossRef] [PubMed]
- Adamczack, J.; Hoffmann, M.; Papke, U.; Haufschildt, K.; Nicke, T.; Bröring, M.; Layer, G. NirN protein from Pseudomonas aeruginosa is a novel electron-bifurcating dehydrogenase catalyzing the last step of heme d1 biosynthesis. J. Biol. Chem. 2014, 289, 30753–30762. [Google Scholar] [CrossRef] [PubMed]
- Kühner, M.; Haufschildt, K.; Neumann, A.; Storbeck, S.; Streif, J.; Layer, G. The alternative route to heme in the methanogenic archaeon Methanosarcina barkeri. Archaea 2014, 327–340. [Google Scholar]
- Celis, A.I.; Gauss, G.H.; Streit, B.R.; Shisler, K.; Moraski, G.C.; Rodgers, K.R.; DuBois, J.L. Structure-based mechanism for oxidative decarboxylation reactions mediated by amino acids and heme propionates in coproheme decarboxylase (HemQ). J. Am. Chem. Soc. 2017, 139, 1900–1911. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.J.; Raux, E.; Schubert, H.L.; Escalante-Semerena, J.C. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 2002, 19, 390–412. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, Y.; Santander, P.J.; Roessner, C.A.; Pérez, L.M.; Scott, A.I. Genetically Engineered Synthesis and Structural Characterization of Cobalt− Precorrin 5A and− 5B, Two New Intermediates on the Anaerobic Pathway to Vitamin B12: Definition of the Roles of the CbiF and CbiG Enzymes. J. Am. Chem. Soc. 2006, 128, 9971–9978. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.J.; Biedendieck, R.; Lawrence, A.D.; Deery, E.; Howard, M.J.; Rigby, S.E.; Warren, M.J. Characterization of the enzyme CbiH60 involved in anaerobic ring contraction of the cobalamin (vitamin B12) biosynthetic pathway. J. Biol. Chem. 2013, 288, 297–305. [Google Scholar] [CrossRef]
- Lobo, S.A.; Videira, M.A.; Pacheco, I.; Wass, M.N.; Warren, M.J.; Teixeira, M.; Saraiva, L.M. Desulfovibrio vulgaris CbiKP cobaltochelatase: Evolution of a haem binding protein orchestrated by the incorporation of two histidine residues. Environ. Microbiol. 2017, 19, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Mera, P.E.; Escalante-Semerena, J.C. Dihydroflavin-driven adenosylation of 4-coordinate Co (II) corrinoids: Are cobalamin reductases enzymes or electron transfer proteins? J. Biol. Chem. 2010, 285, 2911–2917. [Google Scholar] [CrossRef]
- Spencer, J.B.; Stolowich, N.J.; Roessner, C.A.; Scott, A.I. The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett. 1993, 335, 57–60. [Google Scholar] [CrossRef]
- Hogle, S.L.; Barbeau, K.A.; Gledhill, M. Heme in the marine environment: From cells to the iron cycle. Metallomics 2014, 6, 1107–1120. [Google Scholar] [CrossRef]
- Pan, J.; Zhou, Z.; Béjà, O.; Cai, M.; Yang, Y.; Liu, Y.; Li, M. Genomic and transcriptomic evidence of light-sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome 2020, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 2018, 18, 1–13. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, A.P. Metagenomic insights into the microbial diversity in manganese-contaminated mine tailings and their role in biogeochemical cycling of manganese. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Doxey, A.C.; Kurtz, D.A.; Lynch, M.D.; Sauder, L.A.; Neufeld, J.D. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015, 9, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Panek, H.; O’Brian, M.R. A whole genome view of prokaryotic haem biosynthesis. Microbiology 2002, 148, 2273–2282. [Google Scholar] [CrossRef]
- Glanville, D.G.; Mullineaux-Sanders, C.; Corcoran, C.J.; Burger, B.T.; Imam, S.; Donohue, T.J.; Ulijasz, A.T. A High-Throughput Method for Identifying Novel Genes That Influence Metabolic Pathways Reveals New Iron and Heme Regulation in Pseudomonas aeruginosa. Msystems 2021, 6, e00933-20. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Heal, K.R.; Ingalls, A.E.; Doxey, A.C.; Neufeld, J.D. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 2020, 14, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Balabanova, L.; Averianova, L.; Marchenok, M.; Son, O.; Tekutyeva, L. Microbial and genetic resources for cobalamin (vitamin B12) biosynthesis: From ecosystems to industrial biotechnology. Int. J. Mol. Sci. 2021, 22, 4522. [Google Scholar] [CrossRef]
- Boynton, T.O.; Gerdes, S.; Craven, S.H.; Neidle, E.L.; Phillips, J.D.; Dailey, H.A. Discovery of a gene involved in a third bacterial protoporphyrinogen oxidase activity through comparative genomic analysis and functional complementation. Appl. Environ. Microbiol. 2011, 77, 4795–4801. [Google Scholar] [CrossRef]
- Cavallaro, G.; Decaria, L.; Rosato, A. Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J. Proteome Res. 2008, 7, 4946–4954. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson, B.M.; Roe, K.L.; Barbeau, K.A. Heme uptake by Microscilla marina and evidence for heme uptake systems in the genomes of diverse marine bacteria. Appl. Environ. Microbiol. 2008, 74, 6263–6270. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dent, A.T.; Wilks, A. Contributions of the heme coordinating ligands of the Pseudomonas aeruginosa outer membrane receptor HasR to extracellular heme sensing and transport. J. Biol. Chem. 2020, 295, 10456–10467. [Google Scholar] [CrossRef]
- Datta, R.; Kelkar, A.; Baraniya, D.; Molaei, A.; Moulick, A.; Meena, R.S.; Formanek, P. Enzymatic degradation of lignin in soil: A review. Sustainability 2017, 9, 1163. [Google Scholar] [CrossRef]
- Wang, M.; Nie, Y.; Wu, X.L. Extracellular heme recycling and sharing across species by novel mycomembrane vesicles of a Gram-positive bacterium. ISME J. 2020, 15, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, D.R.; White, D.C.; Bryant, M.P.; Doetsch, R.N. Specificity of the heme requirement for growth of Bacteroides ruminicola. J. Bacteriol. Res. 1965, 90, 1645–1654. [Google Scholar] [CrossRef]
- Duwat, P.; Sourice, S.; Cesselin, B.; Lamberet, G.; Vido, K.; Gaudu, P. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. Res. 2001, 183, 4509–4516. [Google Scholar] [CrossRef] [PubMed]
- White, D.C.; Granick, S. Hemin biosynthesis in Haemophilus. J. Bacteriol. Res. 1963, 85, 842–850. [Google Scholar] [CrossRef]
- Fournier, C.; Smith, A.; Delepelaire, P. Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol. Microbiol. 2011, 80, 133–148. [Google Scholar] [CrossRef]
- Jacobs, N.J.; Jacobs, J.M.; Brent, P. Formation of protoporphyrin from coproporphyrinogen in extracts of various bacteria. J. Bacteriol. Res. 1970, 102, 398–403. [Google Scholar] [CrossRef]
- Lawrance, A.D.; Nemoto-Smith, E.; Deery, E.; Baker, J.A.; Schroeder, S.; Brown, D.G.; Warren, M.J. Construction of fluorescent analogs to follow the uptake and distribution of cobalamin (vitamin B12) in bacteria, worms, and plants. Cell Chem. Biol. 2018, 25, 941–951.e6. [Google Scholar] [CrossRef]
- Bernal, V.; Castaño-Cerezo, S.; Gallego-Jara, J.; Écija-Conesa, A.; de Diego, T.; Iborra, J.L.; Cánovas, M. Regulation of bacterial physiology by lysine acetylation of proteins. New Biotechnol. 2014, 31, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Cerezo, S.; Bernal, V.; Post, H.; Fuhrer, T.; Cappadona, S.; Sánchez-Díaz, N.C.; Cánovas, M. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 2014, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Hentchel, K.L.; Escalante-Semerena, J.C. Acylation of biomolecules in prokaryotes: A widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 2015, 79, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Demopoulos, B.J.; Anderson, H.J.; Loader, C.E.; Faber, K. Pyrrole chemistry. XXVI. A synthesis of porphobilinogen from pyrrole. Can. J. Chem. 1983, 61, 2415–2422. [Google Scholar] [CrossRef]
- Robinson, R. Structural Relations of Natural Products; Clarendon Press: Oxford, UK, 1955. [Google Scholar]
- Heal, K.R.; Qin, W.; Ribalet, F.; Bertagnolli, A.D.; Coyote-Maestas, W.; Hmelo, L.R.; Ingalls, A.E. Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc. Natl. Acad. Sci. USA 2017, 114, 364–369. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. J. Bioinform. 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Agarwala, R.; Rotmistrovsky, K. BMTagger: Best Match Tagger for Removing Human Reads from Metagenomics Datasets. 2011. Available online: Ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/ (accessed on 21 February 2014).
- Wood, D.E.; Lu, J.; Langmead, B. mproved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. J. Bioinform. 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. J. Bioinform. 2020, 36, 2251–2252. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab: Berkeley, CA, USA, 2014. [Google Scholar]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Lakin, S.M.; Dean, C.; Noyes, N.R.; Dettenwanger, A.; Ross, A.S.; Doster, E.; Boucher, C. MEGARes: An antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017, 45, D574–D580. [Google Scholar] [CrossRef] [PubMed]
- Ram, R.J.; VerBerkmoes, N.C.; Thelen, M.P.; Tyson, G.W.; Baker, B.J.; Blake, R.C., II; Shah, M.; Hettich, R.L.; Banfield, J.F. Community proteomics of a natural microbial biofilm. Science 2005, 308, 1915–1920. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Ravel, J.M.; Wang, S.F.; Heinemeyer, C.; Shive, W. Glutamyl and glutaminyl ribonucleic acid synthetases of Escherichia coli W: Separation, properties, and stimulation of adenosine triphosphate-pyrophosphate exchange by acceptor ribonucleic acid. J. Biol. Chem. 1965, 240, 432–438. [Google Scholar] [CrossRef]
- Beale, S.I. Biosynthesis of 5-aminolevulinic acid. In Chlorophylls and Bacteriochlorophylls; Springer: Dordrecht, The Netherlands, 2006; pp. 147–158. [Google Scholar]
- Friedman, J.; Lad, L.; Li, H.; Wilks, A.; Poulos, T.L. Structural basis for novel δ-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry. 2004, 43, 5239–5245. [Google Scholar] [CrossRef]
- Tian, B.X.; Erdtman, E.; Eriksson, L.A. Catalytic mechanism of porphobilinogen synthase: The chemical step revisited by QM/MM calculations. J. Phys. Chem. B 2012, 116, 12105–12112. [Google Scholar] [CrossRef]
- Battersby, A.R.; Fookes, C.J.; Matcham, G.W.; McDonald, E. Biosynthesis of the pigments of life: Formation of the macrocycle. Nature 1980, 285, 17–21. [Google Scholar] [CrossRef]
- Kohno, H.; Furukawa, T.; Yoshinaga, T.; Tokunaga, R.; Taketani, S. Coproporphyrinogen oxidase. Purification, molecular cloning, and induction of mRNA during erythroid differentiation. J. Biol. Chem. 1993, 268, 21359–21363. [Google Scholar] [CrossRef]
- Boynton, T.O.; Daugherty, L.E.; Dailey, T.A.; Dailey, H.A. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 2009, 48, 6705–6711. [Google Scholar] [CrossRef] [PubMed]
- Möbius, K.; Arias-Cartin, R.; Breckau, D.; Hännig, A.L.; Riedmann, K.; Biedendieck, R.; Schröder, S.; Becher, D.; Magalon, A.; Moser, J.; et al. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc. Natl. Acad. Sci. USA 2010, 107, 10436–10441. [Google Scholar] [CrossRef] [PubMed]
- Skotnicová, P.; Sobotka, R.; Shepherd, M.; Hájek, J.; Hrouzek, P.; Tichý, M. The cyanobacterial protoporphyrinogen oxidase HemJ is a new b-type heme protein functionally coupled with coproporphyrinogen III oxidase. J. Biol. Chem. 2018, 293, 12394–12404. [Google Scholar] [CrossRef]
- Al-Karadaghi, S.; Hansson, M.; Nikonov, S.; Jönsson, B.; Hederstedt, L. Crystal structure of ferrochelatase: The terminal enzyme in heme biosynthesis. Structure 1997, 5, 1501–1510. [Google Scholar] [CrossRef]
- Mogi, T. Over-expression and characterization of Bacillus subtilis heme O synthase. J. Biochem. 2009, 145, 669–675. [Google Scholar] [CrossRef]
- Niwa, S.; Takeda, K.; Kosugi, M.; Tsutsumi, E.; Mogi, T.; Miki, K. Crystal structure of heme A synthase from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2018, 115, 11953–11957. [Google Scholar] [CrossRef]
- Spencer, P.; Stolowich, N.J.; Sumner, L.W.; Scott, A.I. Definition of the redox states of cobalt-precorrinoids: Investigation of the substrate and redox specificity of CbiL from Salmonella typhimurium. Biochemistry 1998, 37, 14917–14927. [Google Scholar] [CrossRef]
- Debussche, L.; Thibaut, D.; Cameron, B.; Crouzet, J.; Blanche, F. Biosynthesis of the corrin macrocycle of coenzyme B12 in Pseudomonas denitrificans. J. Bacteriol 1993, 175, 7430–7440. [Google Scholar] [CrossRef]
- Moore, S.J.; Lawrence, A.D.; Biedendieck, R.; Deery, E.; Frank, S.; Howard, M.J.; Rigby, S.E.; Warren, M.J. Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc. Natl. Acad. Sci. USA 2013, 110, 14906–14911. [Google Scholar] [CrossRef]
- Fresquet, V.; Williams, L.; Raushel, F.M. Mechanism of cobyrinic acid a, c-diamide synthetase from Salmonella typhimurium LT2. Biochemistry 2004, 43, 10619–10627. [Google Scholar] [CrossRef]
- Roessner, C.A.; Warren, M.J.; Santander, P.J.; Atshaves, B.P.; Ozaki, S.I.; Stolowich, N.J.; Iida, K.; Scott, A.I. Expression of 9 Salmonella typhimurium enzymes for cobinamide synthesis Identification of the 11-methyl and 20-methyl transferases of corrin biosynthesis. FEBS Lett. 1992, 301, 73–78. [Google Scholar] [CrossRef]
- Deery, E.; Schroeder, S.; Lawrence, A.D.; Taylor, S.L.; Seyedarabi, A.; Waterman, J.; Wilson, K.S.; Brown, D.; Geeves, M.A.; Howard, M.J.; et al. An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis. Nat. Chem. Biol. 2012, 8, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.R.; Lawrence, J.G.; Rubenfield, M.; Kieffer-Higgins, S.; Church, G.M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. Res. 1993, 175, 3303–3316. [Google Scholar] [CrossRef] [PubMed]
- Cheong, C.G.; Bauer, C.B.; Brushaber, K.R.; Escalante-Semerena, J.C.; Rayment, I. Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica. Biochemistry 2002, 41, 4798–4808. [Google Scholar] [CrossRef] [PubMed]
- Watkins, H.A.; Baker, E.N. Structural and functional characterization of an RNase HI domain from the bifunctional protein Rv2228c from Mycobacterium tuberculosis. J. Bacteriol. Res. 2010, 192, 2878–2886. [Google Scholar] [CrossRef]
| Organic Compound | Name | Reference |
|---|---|---|
| Primary geoporphyrins | Iron aetioporphyrins (octaethyl porphyrins), | [8] |
| Iron cycloalkanoporphyrins | ||
| Iron di-cycloalkanoporphyrins | ||
| Iron benz-cycloalkanoporphyrins | ||
| Vanadyl-cycloalkano-porphyrins | [9] | |
| Etio and DPEP iron porphyrins | [10] | |
| Etio,DPEP benzo-etio and benzo-DPEP vanadyl porphyrins | ||
| Vanadyl porphyrins of series etio/DPEP | [11] | |
| Octaethyl nickel porphyrin | [12] | |
| Octaethyl vanadyl porphyrin | ||
| Meso-tetraphenyl vanadyl porphyrin | ||
| Meso-tetraphenyl nickel porphyrin | ||
| Protoporphyrin IX cobalt | ||
| Modified geoporphyrins | Diphenyl vanadyl porphyrin | [12] |
| Tetraethyl vanadyl porphyrin | ||
| Vanadyl porphyrin | ||
| Tetraethyl nickel porphyrin | ||
| Nickel porphyrin | ||
| Organic compounds containing 3 pyrrole rings | 3-[2-[[3-(2-Carboxyethyl)-5-[(3.4-dimethyl-5-oxopyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl]methylidene]-4-methyl-5-oxopyrrol-3-yl]propanoic acid | [12] |
| 3-[(5Z)-5-[[4-Ethenyl-5-[(Z)-(4-ethenyl-3-methyl-5-oxopyrrol-2-ylidene)methyl]-3-methyl-1H-pyrrol-2-yl]methylidene]-4-methyl-2-oxopyrrol-3-yl]propanoate | ||
| Organic compounds containing 2 pyrrole rings | 3,3′-Bipyrrole | |
| 2,2′-Bipyrrole | ||
| 1,1′-Bipyrrole-2,2′,5,5′-tetraone | ||
| 3,3′,4,4′-Tetramethyl-1H,1′H-2,2′-bipyrrole-5,5′-dicarboxylic acid | ||
| Organic compounds containing 1 pyrrole ring | 1H-Pyrrole | |
| 1H-Pyrrole-2-carboxylic acid | ||
| 2H-Pyrrol-2-one, 5-[[2-[(4-aminophenyl)methylene]-3,4-dimethyll]methylene]-3-ethyl-1,5-dihydro-4-methyl | ||
| Indole acetic acid | ||
| 1H-Indole-2-carboxylic acid | ||
| Indole carbaldehyde | ||
| 1H-pyrrole-2,5-diones | [13] |
| Enzyme | Accession | Score | Seq(sig) | emPAI | Genus/Species |
|---|---|---|---|---|---|
| Heme-Containing Cytochromes | |||||
| cbb3-type cytochrome c oxidase subunit ii | gi|653251020 | 62 | 1 | 0.19 | Dechloromonas agitata |
| cb-type cytochrome c oxidase ccoo subunit | gi|4519209 | 55 | 1 | 0.12 | Magnetospirillum magnetotacticum |
| cytochrome b6 | gi|499245897 | 128 | 2 | 0.13 | Geobacter sulfurreducens |
| cytochrome b6 | gi|493924522 | 117 | 2 | 0.13 | Legionella drancourtii |
| cytochrome bd-type quinol oxidase, subunit 1 | gi|493975388 | 67 | 1 | 0.07 | Desulfovibrio magneticus |
| cytochrome c | gi|504001995 | 71 | 2 | 0.1 | Azospira oryzae |
| cytochrome c oxidase cbb3-type, subunit iii | gi|330949201 | 56 | 1 | 0.77 | Pseudomonas syringa. 1704B |
| cytochrome c1 | gi|268584477 | 58 | 1 | 0.11 | Neisseria gonorrhoeae PID18 |
| cytochrome cbb3 | gi|499630345 | 55 | 1 | 0.27 | Thiobacillus denitrificans |
| cytochrome d ubiquinol oxidase | gi|498185039 | 55 | 1 | 0.06 | Lactobacillus acidipiscis |
| cytochrome o ubiquinol oxidase | gi|657198419 | 40 | 1 | 0.12 | Aeromonas caviae |
| cytochrome p450 | gi|503190589 | 20 | 1 | 0.1 | Frankia sp. EuI1c |
| cytochrome p450 | gi|664433299 | 44 | 1 | 0.07 | Streptomyces sp. NRRL F-5140 |
| cytochrome soxa | gi|499630342 | 44 | 1 | 0.11 | Thiobacillus denitrificans |
| cytochrome soxa | gi|517333069 | 74 | 1 | 0.11 | Thiobacillus thioparu |
| multispecies: cytochrome c | gi|494962102 | 68 | 1 | 0.25 | Sphingobium sp. |
| flavocytochrome c sulfide dehydrogenase | gi|519012058 | 39 | 1 | 0.07 | Methylotenera sp. |
| succinate dehydrogenase cytochrome b-556 subunit | gi|254672872 | 66 | 1 | 0.39 | Neisseria meningitidis alpha275 |
| thiosulfate reductase cytochrome b subunit | gi|488713780 | 61 | 1 | 0.15 | Myxococcus sp. |
| Heme-Containing Catalases | |||||
| catalase | gi|491324047 | 515 | 11 | 0.84 | Acinetobacter sp.CIP 53.82 |
| catalase | gi|3927890 | 86 | 1 | 0.06 | Desulfovibrio vulgaris |
| catalase | gi|520401 | 60 | 1 | 0.06 | Haemophilus influenzae |
| catalase | gi|500251659 | 103 | 2 | 0.13 | Pseudomonas stutzeri |
| catalase | gi|504938131 | 67 | 1 | 0.09 | Synechococcus sp. PCC 6312 |
| Heme-Containing Peroxidases | |||||
| hydroperoxidase | gi|647531474 | 46 | 1 | 0.04 | Shewanella marina |
| hydroperoxidase II | gi|489375670 | 84 | 2 | 0.09 | Pseudomonas stutzeri |
| hydroperoxidase II | gi|515815228 | 105 | 2 | 0.09 | Pseudomonas stutzeri |
| peroxidase | gi|491142120 | 58 | 1 | 0.14 | Nitrococcus mobilis |
| Sirroheme-Containing Enzymes | |||||
| nitrite reductase (NAD(P)H) large subunit | gi|159882975 | 62 | 1 | 0.04 | Hydrogenivirga sp. 128-5-R1-1 |
| nitrite reductase | gi|491129994 | 58 | 1 | 0.08 | Streptomyces ghanaensis |
| nitrite reductase | gi|23392987 | 65 | 1 | 0.14 | uncultured bacterium |
| sulfite reductase | gi|521065095 | 689 | 6 | 0.47 | Thiothrix disciformis |
| sulfite reductase | gi|655041650 | 53 | 1 | 0.08 | Thiothrix lacustris |
| sulfite reductase | gi|488797739 | 2076 | 7 | 1.18 | Thiothrix nivea |
| sulfite reductase | gi|488797740 | 251 | 3 | 0.27 | Thiothrix nivea |
| dissimilatory sulfite reductase alpha subunit | gi|30525497 | 52 | 1 | 0.08 | uncultured sulfate-reducing bacterium |
| reverse-type dissimilatory siroheme sulfite reductase subunit A | gi|162072844 | 2187 | 7 | 1.3 | Thiothrix nivea DSM 5205 |
| reverse-type dissimilatory sulfite reductase (rDSR), alpha subunit (DsrA) | gi|385763698 | 48 | 1 | 0.07 | uncultured bacterium 172H5 |
| Cobalamin-Containing Enzymes | |||||
| methylcrotonoyl-CoA carboxylase | gi|648618195 | 36 | 1 | 0.06 | Niabella aurantiaca |
| methylmalonyl-CoA carboxyltransferase | gi|587641191 | 40 | 1 | 0.06 | Skermanella stibiiresistens SB22 |
| methionyl-tRNA synthetase | gi|516410008 | 68 | 1 | 0.04 | Erwinia toletana |
| methionyl-tRNA synthetase | gi|588476233 | 35 | 1 | 0.04 | Lactobacillus composti JCM 14202 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stasiuk, R.; Krucoń, T.; Matlakowska, R. Biosynthesis of Tetrapyrrole Cofactors by Bacterial Community Inhabiting Porphyrine-Containing Shale Rock (Fore-Sudetic Monocline). Molecules 2021, 26, 6746. https://doi.org/10.3390/molecules26216746
Stasiuk R, Krucoń T, Matlakowska R. Biosynthesis of Tetrapyrrole Cofactors by Bacterial Community Inhabiting Porphyrine-Containing Shale Rock (Fore-Sudetic Monocline). Molecules. 2021; 26(21):6746. https://doi.org/10.3390/molecules26216746
Chicago/Turabian StyleStasiuk, Robert, Tomasz Krucoń, and Renata Matlakowska. 2021. "Biosynthesis of Tetrapyrrole Cofactors by Bacterial Community Inhabiting Porphyrine-Containing Shale Rock (Fore-Sudetic Monocline)" Molecules 26, no. 21: 6746. https://doi.org/10.3390/molecules26216746
APA StyleStasiuk, R., Krucoń, T., & Matlakowska, R. (2021). Biosynthesis of Tetrapyrrole Cofactors by Bacterial Community Inhabiting Porphyrine-Containing Shale Rock (Fore-Sudetic Monocline). Molecules, 26(21), 6746. https://doi.org/10.3390/molecules26216746






