Abstract
The red macroalga Gelidium latifolium is widely distributed in the coastal areas of Indonesia. However, current knowledge on its potential biological activities is still limited. In this study, we investigated the potential bioactive compounds in Gelidium latifolium ethanol extract (GLE), and its cytotoxic effects against the murine B16-F10 melanoma cell line. GLE shows high total phenolic content (107.06 ± 17.42 mg GAE/g) and total flavonoid content (151.77 ± 3.45 mg QE/g), which potentially contribute to its potential antioxidant activity (DPPH = 650.42 ± 2.01 µg/mL; ABTS = 557.01 ± 1.94 µg/mL). ESI-HR-TOF-MS analysis revealed large absorption in the [M-H]- of 327.2339 m/z, corresponding to the monoisotopic molecular mass of brassicolene. The presence of this compound potentially contributes to GLE’s cytotoxic activity (IC50 = 84.29 ± 1.93 µg/mL). Furthermore, GLE significantly increased the number of apoptotic cells (66.83 ± 3.06%) compared to controls (18.83 ± 3.76%). Apoptosis was also confirmed by changes in the expression levels of apoptosis-related genes (i.e., p53, Bax, Bak, and Bcl2). Downregulated expression of Bcl2 indicates an intrinsic apoptotic pathway. Current results suggest that components of Gelidium latifolium should be further investigated as possible sources of novel antitumor drugs.
1. Introduction
Malignant melanoma is the most aggressive form of skin cancer, and currently accounts for approximately 3% of all cases of malignant tumors [1]. The most common carcinogen responsible for the development of melanoma is ultraviolet radiation (UVR) [2]. The global incidence of melanoma is increasing due to the increase in UVR reaching the Earth’s surface [3]. Currently, melanoma, as with other types of cancer, is mainly treated with radiation or chemotherapy. The main problems with these treatments include severe adverse effects and the development of multidrug resistance [4]. Hence, various phytochemicals obtained from natural resources have been extensively investigated for their anticancer activities due to their safety and low-to-moderate toxicity compared to conventional cancer chemotherapeutic agents [5].
Marine macroalgae, also referred to as seaweeds, are well known as a rich source of structurally diverse bioactive compounds with various biological activities, as well as for their importance as a source of novel bioactive substances [6,7,8]. Several studies have revealed that seaweeds have biological activity against cancer, cardiovascular-related diseases, diabetes, inflammation, thrombosis, and obesity [9,10,11,12,13,14,15,16]. In particular, red macroalgae are a critical source of numerous bioactive compounds—unlike other groups of green and brown macroalgae. Bioactive compounds that are commonly present in red macroalgae—such as sulfated galactans, carrageenans, agars, phytoalexins, unsaturated fatty acids, terpenoids, and amino acids—have demonstrated anticancer activity in some cancer cell models [9,17,18,19,20,21,22].
Several other Gelidiaceae family members have been reported to have a variety of biological activities, including the prevention of cancer and cardiovascular disease, lowering blood pressure, and lowering blood glucose levels [23,24,25,26,27]. The red macroalgae genus Gelidium is mainly known as the source of agars that are used in the food industry. Approximately 20% of the largest agar industrial sources come from Gelidium spp. The agar content of Gelidium amansii has demonstrated anti-obesity, antioxidant, and anticarcinogenic activity. However, information regarding other phytochemical constituents in Gelidium species remains largely unknown. The species Gelidium latifolium is one of the most abundant red macroalgae in coastal areas of West Lombok, Indonesia; however, there remains limited information regarding its potential biological activities. In this study, we evaluated the phytochemical constituents of Gelidium latifolium ethanol extract (GLE) and its antiproliferative effect against melanoma cells using murine B16-F10 cells as a melanoma cell model.
2. Materials and Methods
2.1. Macroalgal Material
The red macroalga Gelidium latifolium was collected from the Batu Layar coast, West Lombok, Indonesia (8°24’11.7396” S, 116°4’1.9056” E). The macroalgae samples were identified based on an electronic algae database [28]. Corresponding material was deposited in the herbarium of Pusat Unggulan Biosains dan Bioteknologi (PUBB), University of Mataram. Fresh samples were washed with distilled water to remove adhering debris. Cleaned samples were then sprayed with 1% fungicide (Rely+On, Virkon) to prevent fungal contamination. The macroalgae samples were then dried at room temperature (24 °C) with air conditioning. After approximately 5 days, the macroalgae samples were incubated in an oven (Binder ED 56) at 40 °C until reaching a constant weight [29]. Dried samples were then ground with a blade grain miller (food-grade stainless steel) and kept in powder form at 4 °C until further use [30].
2.2. Chemicals and Reagents
For extraction, ACS-grade 96% ethanol (Merck, Germany) was used. The MTT (3-(3,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium) reagent was purchased from BioVision, Inc. (Milpitas, CA, USA). Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS) (South American origin), and penicillin were purchased from Gibco, Life Technologies (Paisley, UK). The fluorescence probes calcein-AM, propidium iodide (PI), and Hoechst33342 were purchased from Dojindo, Japan.
2.3. Preparation of Gelidium Latifolium Ethanol Extracts
Fifty grams (50 g) of dried algal powder was mixed with absolute ethanol solvent at a volume of five times the volume of the sample weight (w/v). The algal sample was extracted by maceration and incubated at room temperature for 3 × 24 h with constant stirring. After every 24 h, the mixture was filtered with Whatman No. 1 filter paper. All of the filtrates were collected and evaporated with a rotary evaporator (40 °C, 50 rpm) to evaporate the ethanol solvent. The resulting pasty extracts were collected and stored at 4 °C until future use [31].
2.4. Determination of GLE’s Total Phenolic Content and Total Flavonoid Content
The total phenolic content (TPC) of GLE was determined via the Folin–Ciocalteu colorimetric method [32]. Gallic acid equivalents (GAE) were used as the standard. A stock solution of GAE was prepared by dissolving 10 mg with 10 mL of ethanol (1 mg/mL). A dilution series of GAE concentrations (10–500 µg/mL) was prepared for the generation of the standard curve. An exact amount of 100 µL of sample (1 mg/mL) was combined with 0.75 mL of the Folin–Ciocalteu reagent (diluted 10-fold in distilled water before use). The mixture was incubated at room temperature for 5 min. A volume of 750 µL of sodium carbonate (Na2CO3) was added to the mixture and mixed gently with pipetting. The reaction took ~90 min. Absorbance was measured at 725 nm with a UV–Vis spectrophotometer microplate reader (Multiskan GO, Thermo Fisher Scientific). The TPC value obtained was revealed as gallic acid equivalents per 100 g of the dry extract.
The total flavonoid content (TFC) was measured according to the method described by Aregan et al., with minor modifications [33]. A volume of 100 µL of sample was added, along with 4 mL of distilled water, followed by the addition of 300 µL of 5% sodium nitrite. After 5 min, 300 µL of 10% aluminum chloride was added. The mixture was incubated for an additional 6 min before the addition of 2 mL of 1 M sodium hydroxide. Immediately, the mixture was diluted by the addition of 3.3 mL of distilled water, and then vortexed. The absorbance was determined at 510 nm versus a blank. Quercetin was used as the standard for the calibration curve. The total flavonoid content of the sample was expressed as mg of quercetin equivalents per 100 g of the dry extract.
2.5. Antioxidant Activity of GLE (DPPH and ABTS Assays)
The determination of GLE antioxidant capacity was based on DPPH (2,2-diphenyl-2-picrylhydrazyl) radical scavenging activity [34]. A volume of 100 µL of GLE or ascorbic acid (AA) at various concentrations (10–4000 µg/mL) was mixed with 100 µL of 200 µM freshly prepared DPPH in methanol. The reaction was conducted in the dark at room temperature for 30 min, and then the absorbance was measured at 517 nm. The measurement was repeated three times, and free-radical-inhibiting activity was calculated by Equation (1):
The ABTS (2,2’-azinobis(3-ethylbenzothiazolin-6-sulfonic acid)) radical cation method was also used to determine the antioxidant activity of GLE. The ABTS reagent was prepared by mixing 5 mL of 7 mM ABTS with 88 µL of 140 mM potassium persulfate [35]. The mixture was incubated in the dark at room temperature for 24 h to allow the generation of free radicals. For measurement of the scavenging activity of macroalgae extracts based on ABTS assay, a volume of 100 µL of ABTS reagent was mixed with 100 µL of sample or ascorbic acid (standard) in a 96-well microplate and incubated at room temperature for 6 min. After incubation, the absorbance was measured at 734 nm using a UV–Vis spectrometer (Multiskan GO, Thermo Fisher Scientific). The experiment was repeated three times, and the ABTS scavenging activity was measured using Equation (2):
2.6. Electrospray Ionization High-Resolution Time-of-Flight Mass Spectrometry (ESI-HR-TOF-MS)
High-resolution mass spectra were provided by a Bruker Daltonics maXis 4G time-of-flight mass spectrometer with electrospray ionization (ESI-HR-TOF-MS). Approximately 5 µL of GLE was subjected to the reversed phase of a C18 column (Phenomenex® Luna Omega 1.6 µm Polar C18, 100 Å, 150 × 2.1 mm) with a flow rate of 0.75 mL/min to disperse the analytes. Mass spectra (MS) analysis was performed in negative mode (50 eV). MS-grade 90% water with 0.1% formic acid to 100% MS-grade acetonitrile with 0.06% formic acid was used.
2.7. B16-F10 Murine Melanoma Cell Culture
B16-F10 murine melanoma cells (ECACC 92101204) and NIH-3T3 normal murine fibroblasts (ECACC 93061524) were purchased from the European Collection of Authenticated Cell Cultures. The cells were routinely cultivated in Dulbecco’s modified Eagle medium (DMEM, Wako) supplemented with 10% FBS for B16-F10 and 5% for NIH-3T3. The cells were kept at 37 °C in a 5% CO2 humidified incubator (Forma Steri-Cycle, Thermo Fisher Scientific). For all experiments, cells were grown in T-25 cell culture flasks with a seeding density of 0.8 × 106 cells/mL. After reaching 80–90% confluence, cells were then seeded according to the experimental requirements.
2.8. Cytotoxicity Assay
Cytotoxicity was estimated via the MTT cytotoxic assay [36]. B16-F10 and NIH-3T3 cells were cultured in 96-well culture plates at a seeding density of 1 × 104 cells/well. After 24 h, the culture media were discarded and replaced with new media containing various concentrations of GLE or doxorubicin (1–1000 µg/mL). After 72 h of incubation, the wells were supplemented with 50 µL of MTT reagent in 50 µL of serum-free DMEM. The 96-well plates were then kept at 37 °C with 5% CO2 for 3 h. After 3 h of incubation, the solution was discarded and replaced with 150 µL of MTT solvent, and then rotated for 15 min. Color change depending on MTT activity was then measured by absorbance at 590 nm with a UV–Vis microplate reader (Multiskan GO, Thermo Fisher Scientific). Cytotoxicity was calculated by Equation (3). The absorbance of the control group at 590 nm (A590 control) refers to the absorbance of the wells with no GLE or doxorubicin. A590 treated cells refers to the absorbance of the wells treated with various concentrations of GLE or doxorubicin. All experiments were performed in triplicate. Cell morphology was observed at 20× magnification with a phase-inverted microscope (Zeiss primo vert, Zeiss, Germany).
2.9. Calcein-AM and Propidium Iodide Staining (Viability Analyses)
Calcein acetoxymethyl (calcein-AM) and propidium iodide (PI) viability staining were performed to determine the cells’ apoptotic features [9]. The cells were seeded at a density of 3 × 104 cells/well in a 35 mm dish at 37 °C and a 5% CO2 atmosphere in 2 mL media (DMEM supplemented with 10% FBS and 1% penicillin). After 24 h of incubation, the medium was replaced with medium containing GLE or doxorubicin at IC50 concentration. After 72 h, the cells were washed twice with PBS, followed by the addition of calcein-AM (5 µM) and PI (5 µM). The stained cells were incubated for 15 min at 37 °C before observation with a fluorescence-inverted microscope (Axio observer Z1, Zeiss, Germany). Cells that emitted green fluorescence were live cells, whereas the cells that emitted red fluorescence were dead cells [37]. Further cell analysis was conducted using ImageJ software.
2.10. GLE’s Effect on DNA Condensation (Hoechst33342 Nuclear Staining)
Staining cells with the fluorescence dye Hoechst33342 is one of the ideal assays to determine apoptotic events in cells [38]. The B16-F10 cells were grown in 35 mm tissue culture dishes at a seeding density of 0.3 × 106 cells/mL. After 24 h, the media were discarded and replaced with non-treated medium or media containing GLE IC50 or doxorubicin IC50. The cells were then incubated for another 72 h at 37 °C with 5% CO2. After 72 h, the media were discarded and replaced with culture medium containing 40 µM of Hoechst33342. The cells were then incubated for 20 min at 37 °C with 5% CO2. Finally, the cells were washed twice with PBS and observed under a fluorescence-inverted microscope with 350 nm excitation filters. The cell images were taken at 20× magnification and analyzed with ImageJ to determine the apoptotic nuclei percentage. The fluorescence intensity of condensed nuclear DNA was determined using the corrected total cell fluorescence (CTCF) equation [39]: CTCF = integrated density—(area of selected cell × mean fluorescence of background readings).
2.11. GLE’s Effect on DNA Fragmentation
One of the biochemical hallmarks of apoptosis is the condensation and fragmentation of genomic DNA [40]. This is an irreversible event that commits the cell to die, and occurs before changes in plasma membrane permeability. The cells were seeded in 24-well plates for 24 h at a seeding density of 5 × 104 cells/mL. The next day, the culture medium was replaced with an IC50 concentration of GLE or doxorubicin and incubated for a further 72 h. After 72 h of incubation, the total cellular DNA was extracted with the DNeasy Kit (Qiagen, USA). Retrieved genomic DNA was run in 1.5% agarose and subjected to electrophoresis (80 V, 40 min). A DNA size marker of 1 kb (GeneRuler 1 kb, Thermo Scientific) was used as the standard DNA ladder. The gels were documented with the GelDoc imaging system (Cambridge, UK).
2.12. RNA Isolation and Semi-Quantitative PCR Analysis
Total RNA was extracted from both untreated and treated B16-F10 cells using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The B16-F10 cells were seeded in 24-well plates at a cell density of 5 × 104 cells/mL. After 24 h of incubation, the medium was replaced with an IC50 concentration of GLE or doxorubicin and incubated for a further 72 h. The total RNA was isolated from B16-F10 cells according to the manufacturer’s instructions. The obtained RNA was then converted to cDNA with a PrimeScript 1st strand cDNA synthesis kit (Takara, Japan), and PCR was performed using a TopTaq Master Mix PCR kit (Qiagen, USA). The expression of the apoptosis-related genes p53, bak, bax, and Bcl2 was investigated using primers that corresponded to data in GenBank. The primers were ordered from Fasmac, Japan. The semi-quantitative analyses of PCR products were determined relative to the housekeeping gene (GAPDH) with Image Lab software (Bio-Rad, Hercules, CA, USA).
2.13. Statistical Analyses
One-way ANOVA followed by Tukey’s multiple comparison post hoc test was conducted for multiple comparisons between treatment groups and controls. Experiments were repeated at least three times, and the data were represented as the mean ± SD. All statistical analyses were performed using GraphPad Prism software ver.9.2.0 (San Diego, CA, USA). A p-value of less than 0.05 was considered to be statistically significant, whereas a p-value of less than 0.01 was considered to be highly significant.
3. Results
3.1. The Morphology and Phytochemical Analysis of GLE
The macroalga Gelidium latifolium has a dark-red-colored thallus with a relatively small morphological size ranging from 5 to 7 cm (Figure 1A). The extract yield obtained from extraction of 50 g of dried G. latifolium powder was 0.87 ± 0.15 g. Using the Folin–Ciocalteu method, the total phenolic content (TPC) of G. latifolium was investigated. In this study, the red alga G. latifolium was found to obtain TPC of 107.06 ± 17.42 mg GAE/g (Figure 1B). Previous research has found lower TPC levels in ethanolic extracts of Sargassum muticum (94.20 mg GAE/g), Turbinaria conoides (0.09 mg GAE/g), Laminaria ochroleuca (83 mg GAE/g), and Halopteris scoparia (2.92 mg GAE/g) [32,33]. Other findings in relation to Gelidium species also reported low TPC in Gelidium sp. (6.33 mg GAE/g) and Gelidium chilense (9.9 ± 1.3 mg GAE/g) [34,41]. These differences could be influenced by numerous factors, such as species, season, age, environmental conditions, and extraction methods [42]. This could be seen in the study by Alvarez-Gomez et al., where the red alga Gelidium pusillum was reported to contain a higher amount of phenolic compounds compared to the related species Gelidium corneum [43].
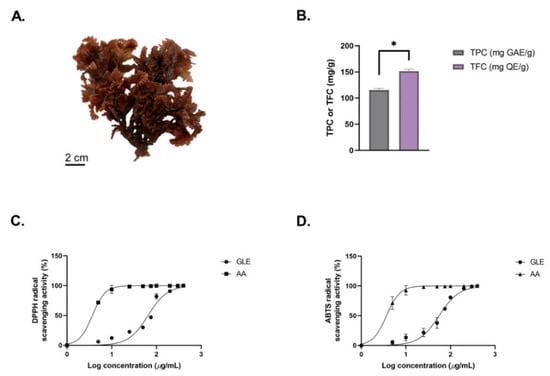
Figure 1.
(A) Morphological features of the red seaweed Gelidium latifolium. (B) Total phenolic content (TPC) and total flavonoid content (TFC) of GLE. (C) DPPH radical scavenging activity of GLE. (D) ABTS radical scavenging activity of GLE. Data represented are the mean ± SEM of 3 independent experiments; * indicates significant differences between groups (p < 0.05).
The TFC of GLE was also determined, using quercetin (QE) as standard (Figure 1B). The TFC value of GLE (151.77 ± 3.45 mg QE/g) was higher compared to Sargassum vulgar (187 ± 5 mg QE/g) and Gracilaria corticata (105 ± 10 mg QE/g) [44]. In addition, GLE showed significantly higher TFC compared to ethanolic extract of Gelidium pusillum (4.46 ± 0.004 mg QE/g) [45]. GLE’s potential antioxidant activity was determined based on the DPPH and ABTS assays (Figure 1C). The antioxidant IC50 values (DPPH = 650.42 ± 2.01 µg/mL; ABTS = 557.01 ± 1.94 µg/mL) obtained from the ethanolic crude extract of G. latifolium were lower compared to previous findings. A previous study by Chakraborty et al. reported that the methanolic extracts of H. musciformis, H. valentiae, and J. rubens had TFCs above 2 mg/mL [46]. The antioxidant activity IC50 value of GLE was also lower compared to crude extracts of other wild red macroalgae, such as Pterocladiella capillacea (7.90 ± 0.04 mg/mL), Hypnea spinella (11.59 ± 0.05 mg/mL), Dermocorynus dichotomus (4.69 ± 0.01 mg/mL), Halopithys incurva (2.62 ± 0.02 mg/mL), and Laurencia dendroidea (2.48 ± 0.01 mg/mL) [47]. Meanwhile, the IC50 value of 200–1000 µg/mL from crude extracts could be considered to have potential antioxidant activity [48]. Hence, this highlights the antioxidant potential of G. latifolium, which correlates with its high phenolic and flavonoid contents.
3.2. Brassicolene Detected in GLE by ESI-HR-TOF-MS
ESI-HR-TOF-MS was used to assess the phytochemical composition of GLE. GLE extract contains putative bioactive compounds including 2H-pyran; 5-methyl-2-octyl-3(2H)-furanone; trans, cis-2,6-nonadienal; 12-oxo-trans-10-dodecenoic acid; palmitoleic acid, (2E)-2-tetradecenal; and eicosenoic acid (Figure S1). Therefore, the main component was probably brassicolene, or a closely related molecule with [M-H]− of 327.2339 m/z—as shown in Figure 2A–C, which depicted (A) the extracted ion chromatogram of putative brassicolene, and (B,C) the MS1 and MS2 data, respectively. Brassicolene has the chemical formula C22H32O2, with a mass error of 0.2 ppm, and a round double bond (rdb) of 7, signifying seven degrees of unsaturation. Furthermore, UV absorption at 200 nm confirmed the presence of a conjugated diene (Figure 2D). The ESI-HR-TOF-MS and UV analyses of putative brassicolene found in GLE were consistent with the findings of Duh et al. [49].
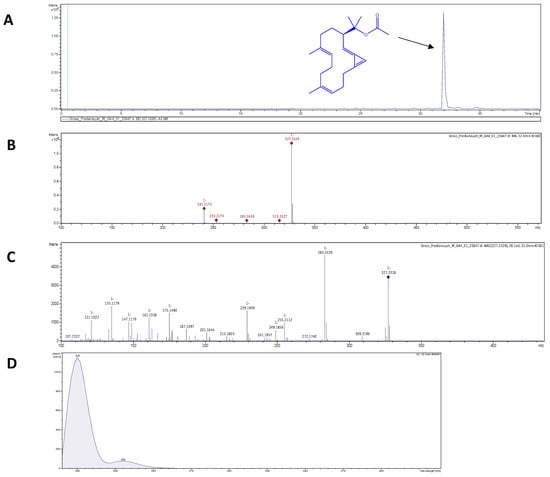
Figure 2.
ESI-HR-TOF-MS analysis of putative brassicolene in GLE: (A) Extracted ion chromatogram and chemical structure of brassicolene (a m/z of 327.2339 [M-H]-). (B) Negative mode MS1 of brassicolene. (C) Negative mode MS2 of brassicolene. (D) UV absorption in mass spectrometric analysis of brassicolene.
3.3. Cytotoxic Activity of GLE on B16-F10 Melanoma Cells
The cytotoxic effect of GLE on B16-F10 melanoma cells was evaluated with the MTT assay. GLE solution was diluted to various concentrations (1–1000 µg/mL), and effective doses were calculated from the dose–response curve in GraphPad Prism. After 72 h of administration, GLE shows moderate toxicity, with an IC50 value of 84.29 ± 1.93 µg/mL (Figure 3A). Based on US National Cancer Institute (NCI) guidelines, the value IC50 ≤ 20 µg/mL is considered highly cytotoxic, IC50 ranging between 21 and 200 µg/mL is considered moderately cytotoxic, IC50 ranging between 201 and 500 µg/mL is weakly cytotoxic, and IC50 above 500 µg/mL is considered to have no cytotoxic activity [50]. Hence, GLE could be considered moderately cytotoxic based on the presence of potential cytotoxic compounds. Furthermore, GLE treatment induces changes in the morphology of B16-F10 cells (Figure 3B). The cell death process is accompanied by changes in cell morphology, such as cell shrinkage and rounding. These features are observable in cells treated with GLE—especially at higher concentrations (50–200 µg/mL). In addition, our preliminary study showed the low cytotoxicity of GLE in NIH-3T3 normal fibroblast cells, with IC50 > 500 µg/mL (Figure S2). Previous cytotoxic analysis of extracts of the red macroalga Gelidium amansii showed growth-inhibitory effects at concentrations > 500 µg/mL after 7 days of treatment. No growth-inhibitory effect was seen at the concentration range of 500–7500 µg/mL after 3 days of treatment [51]. This actually indicates the low cytotoxic activity of Gelidium extracts in normal fibroblast cells. In addition, the low cytotoxic activity of macroalgae extracts and their phytochemical constituents in normal cells has been previously reported [52,53,54,55].
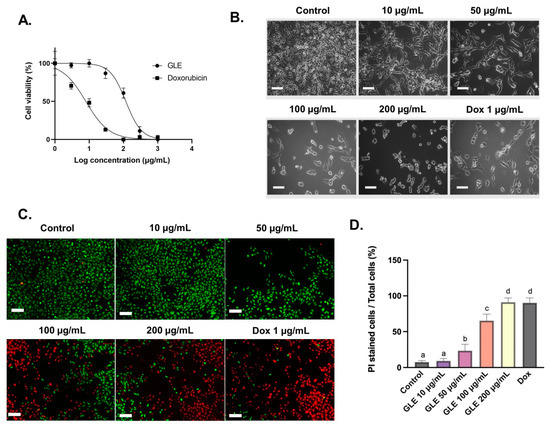
Figure 3.
Cytotoxic effects of GLE in B16-F10 cells treated for 72 h: (A) The dose–response curve of GLE and doxorubicin (1–1000 µg/mL) cytotoxic activity in B16-F10 melanoma cells. (B) Morphological observation of B16-F10 melanoma cancer cells treated with GLE or doxorubicin for 72 h, scale = 50 µm. (C) Viability staining with calcein-AM (green) and PI (red) in B16-F10 murine melanoma cells treated with GLE or doxorubicin for 72 h. (D) The percentage of PI-stained cells represent dead cells. Different letters denote significant differences between treatments. Scale = 100 µm.
3.4. Cell Viability Analyses with Fluorescence Double Staining (Calcein-AM/PI)
Cell viability can be measured using the fluorescent probes calcein-AM and PI, which can differentiate between living and dead cells [9]. In living cells, intracellular esterase can convert calcein-AM to calcein, which stays in the living cells and emits green fluorescence. Meanwhile, PI is cell-impermeable—it can only penetrate cells with impaired plasma membrane integrity to bind with DNA and emit red fluorescence. The number of dead cells in B16-F10 cells treated with GLE was increased in a concentration-dependent manner. The semi-quantitative analyses were conducted with ImageJ software based on the images obtained via fluorescence-inverted microscopy (Figure 3C). The B16-F10 cells treated with higher doses of GLE (100–200 µg/mL) resulted in a significant reduction in the number of living cells and an increase in the number of dead cells (Figure 3D).
3.5. Effect of GLE on B16-F10 Apoptosis
Chromatin condensation is one of the key features of cell apoptosis [56]. The fluorescence dye Hoechst33342 can be used to stain the condensed nuclei of apoptotic cells [57]. The B16-F10 cells treated with the IC50 concentration of GLE were seen to exhibit condensed nuclei (Figure 4A). To determine chromatin condensation between treatments, the fluorescence intensity of the treated cells could be calculated as corrected total cell fluorescence (CTCF) [39]. Based on the CTCF readout, there was a significant increase in fluorescence intensity in cells treated with GLE (CTCF = 13,676.31) compared to the control group (CTCF = 7390.22) (Figure 4B). Furthermore, the percentage of apoptotic cells treated with GLE (66.83 ± 3.06%) was higher than in the control group (18.83% ± 3.76%), but not significantly higher than in the doxorubicin-treated cells (77.50 ± 5.36%) (Figure 4C). In addition to nuclear DNA condensation, the fragmentation of DNA is also a key sign of cell apoptosis events, which can be observed via gel electrophoresis [40,58]. The formation of smaller DNA fragments can be observed in DNA samples of cells treated with IC50 concentrations of GLE and doxorubicin (Figure 4D).
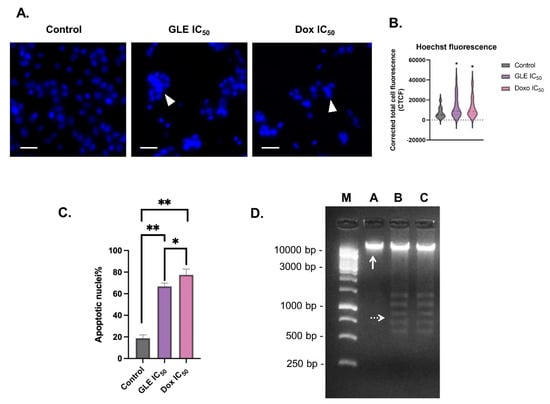
Figure 4.
Apoptosis-inducing effects of GLE in B16-F10 melanoma cells: (A) Staining with Hoechst33342 reveals chromatin condensation in apoptotic cells; arrowheads = apoptotic nuclei. (B) Corrected total cell fluorescence (CTCF) values of B16-F10 cells treated with GLE and doxorubicin at IC50 concentrations. (C) Calculation of apoptotic nuclei percentage in B16-F10 cells treated with IC50 concentrations of GLE and doxorubicin. (D) Fragmentation of B16-F10 genomic DNA treated with GLE; a 1 kb DNA ladder (250–10,000 bp) was used as a standard; M: DNA 1 kb marker; A: control; B: GLE IC50 concentration; C: doxorubicin IC50 concentration. * Indicates significant difference compared to control (p < 0.05); ** indicates highly significant difference compared to control (p < 0.01). Arrowheads = apoptotic nuclei. White arrows indicate genomic DNA. White dotted arrows show fragmented DNA. Scale = 25 µm.
3.6. Effects of GLE on Bak, Bax, and Bcl2 Expression
The effect of GLE on the expression of apoptosis-related genes via RT-PCR was analyzed using Image Lab software (Figure 5A). GLE altered the expression of apoptosis-related genes after 72 h of treatment in B16-F10 melanoma cells. Compared to the untreated cells, pro-apoptotic mRNA levels were markedly increased. However, Bcl2 expression was decreased by GLE treatment (Figure 5B). Several previous studies have shown that marine macroalgae extracts can alter the expression of apoptosis-related genes [59,60,61]. A previous study revealed the cytotoxic activity of various crude macroalgae extracts against MCF-10A [62]; among them are Gelidium spinosum and Gelidium pulchellum, which show very high cytotoxicity. However, to the best of our knowledge, no study has yet been conducted on the cytotoxic activity of Gelidium latifolium in melanoma cells.
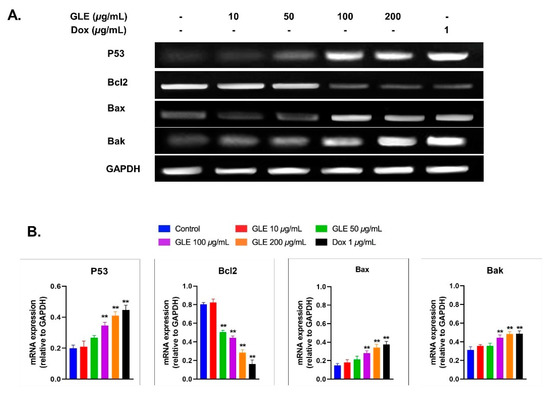
Figure 5.
(A) Amplification of the apoptosis-related genes p53, Bax, Bak, and Bcl2. (B) Analyses of gene expression based on mRNA expression relative to the housekeeping gene GAPDH. ** indicates highly significant difference compared to control (p < 0.01).
4. Discussion
Macroalgae have been extensively investigated as sources of new bioactive chemicals with a variety of biological properties [63,64,65]. Gelidium, a red macroalgae genus, has previously been proven to inhibit cell proliferation in cultured cells [51,66,67]. G. latifolium is a red macroalga that is commonly found in the coastal areas of the island of Lombok, Indonesia; however, there remains limited information regarding its potential biological activity. In this study, we investigated the phytochemical properties and cytotoxic activity of G. latifolium ethanol extract (GLE) in B16-F10 melanoma cells.
Algal secondary metabolites such as phenolic compounds, polysaccharides, and polyunsaturated acids have been shown to have a wide variety of biological functions [9,53,68]. Due to the agar content of the red macroalga Gelidium, it is one of the most important edible marine algae [69]. Gelidium agar has also been proven in previous studies to have anti-obesity, antioxidant, and anticarcinogenic properties [24,70,71]. Thus, agar has been shown to possess a variety of biological activities with potential pharmacological and therapeutic applications. However, data on additional possible bioactive chemicals in Gelidium are quite limited. The red macroalga G. latifolium exhibits a high level of antioxidant activity when compared to other crude macroalgal preparations [72]. Some investigations have concluded that there is no association between free radical scavenging and cytotoxicity [73]; however, there are also other reports that show positive correlation between antioxidant and cytotoxic activity [74,75].
In our study, ESI-HR-TOF-MS analysis revealed a significant amount of brassicolene in the ethanol extract of native Gelidium latifolium from Lombok. Brassicolene is a diterpenoid found in the soft coral Nephthea brassica Kükenthal that has been shown to have cytotoxic effects in a variety of tumor cells [49,76]. Numerous studies have noted the presence of diterpenoids from marine macroalgae, and their cytotoxicity in various cancer cells [65,77,78]. However, this is the first study to demonstrate the presence of the diterpenoid brassicolene in the red alga Gelidium latifolium. Our preliminary results also show that GLE also contains putative fatty acids such as trans, cis-2,6-nonadienal, 12-oxo-trans-10-dodecenoic acid, palmitoleic acid, (2E)-2-tetradecenal, and eicosenoic acid (Figure S1). In addition, 2H-pyran and 5-methyl-2-octyl-3(2h)-furanone were also detected. All of the above were present at small levels compared to brassicolene. However, none of these compounds have been reported to exhibit antiproliferative activity in tumor cells. Hence, we suggest that brassicolene potentially contributes to GLE’s cytotoxic activity.
The main purpose of anticancer treatments is to kill the cancer cells without damaging normal cells. However, chemotherapy and radiotherapy possess limited efficacy, and exert their actions on both tumor and normal cells. This results in adverse side effects on patients, such as anemia, delirium, and peripheral neuropathy [79]. Thus, the development of a more effective treatment that has anticancer activity with lower cytotoxicity and fewer side effects is still needed. Natural products such as crude extracts from marine macroalgae are well studied for their moderate-to-low cytotoxic activity against various cancer cells [65].
Based on our results, GLE shows stronger cytotoxic activity compared to crude macroalgae extracts in other studies [53,80,81]. However, the cytotoxic activity of GLE was quite similar to Laurencia papillosa—125.8 ± 2.1 µg/mL and 121. 64 µg/mL, respectively [18]. The IC50 values could differ in different tumor models [82]; hence, in order to establish GLE antitumor activity, further investigations are needed in different cell lines. In addition, GLE shows low cytotoxic activity against normal murine fibroblast cells (Figure S2). Similar studies also showed the minimal cytotoxic activity of macroalgae extracts against non-tumorigenic cell lines [52,53,54,55]. The macroalga Cystoseira tamariscifolia also showed selective cytotoxic activity against the normal cell line HUVEC [82]. Our previous study also showed that the bioactive compound sulfated polysaccharide from red algae exerts no significant cytotoxic effect against HUVEC cells [9]; a possible reason for this is that the active substances of macroalgae interact with specific cancer-associated receptors or proteins, thus triggering a certain mechanism that causes cancer cell death [83]; however, further experiments are still needed in order to elucidate the precise mechanism.
Evading apoptosis is one of the hallmarks of cancer treatment to restrain the survival of abnormal cells. Hence, anticancer therapies commonly target apoptosis for the prevention and treatment of cancer. In general, the apoptotic pathway consists of several biochemical events, including the activation of apoptotic genes such as p53, Bcl2, Bax, and Bak [84]. Based on mRNA levels, GLE increased the expression of pro-apoptotic genes (i.e., p53, Bax, and Bak). Hence, GLE may promote apoptosis in cancer cells via a mitochondria-dependent intrinsic pathway. In addition, other essential regulatory proteins for apoptotic pathways include Bcl2. The mRNA expression of the anti-apoptotic gene Bcl2 was decreased in B16-F10 cells treated with GLE. GLE concentrations above 100 µg/mL significantly increased the expression of Bax and Bak, while decreasing the expression of Bcl2 (Figure 5B). This indicates that GLE potentially induces apoptosis via the mitochondrial apoptotic pathway [85].
5. Conclusions
Our study’s results show the potential pharmaceutical and medicinal value of the red macroalga G. latifolium. The ethanol extract of G. latifolium (GLE) shows promising phytochemical properties and antioxidant activity. GLE had a weaker antiproliferative activity compared to doxorubicin. The present study is based on the main component that is found in the G. latifolium ethanol extract—which is putatively brassicolene or a closely related molecule. Other minor bioactive compounds may interact with brassicolene in a synergistic or antagonistic manner. Hence, future research on the isolation of brassicolene would be welcome in order to better understand how it affects cell viability. Nevertheless, GLE cytotoxicity induced apoptosis based on morphological observation and altered expression of apoptosis-related genes. The upregulation of the pro-apoptotic gene p53 and the downregulation of the anti-apoptotic gene Bcl2 suggest that the mechanism of apoptosis takes place through an intrinsic pathway. At this point, our results show the presence of a potential bioactive compound that could be useful in the discovery of novel macroalgae-based antiproliferative compounds.
Supplementary Materials
The following are available online: Figure S1: HPLC analysis of GLE determined that brassicolene was substantially more present as a prominent peak than the other compounds; Figure S2: Cytotoxic effects of GLE in NIH-3T3 normal fibroblast cells treated for 72 h.
Author Contributions
Conceptualization, E.S.P., A.F. and H.S.; methodology, E.S.P., A.F. and N.W.R.M.; software, E.S.P. and A.F.; validation, A.L.S. and S.W.; formal analysis, E.S.P. and A.F.; investigation, E.S.P., A.F., H.P., N.A. and N.W.R.M.; resources, A.N. and E.S.P.; data curation, E.S.P., N.W.R.M. and A.F.; writing—original draft preparation, E.S.P.; writing—review and editing, E.S.P. and A.F.; visualization, E.S.P. and A.F.; supervision, A.N. and S.W.; project administration, B.T.K.I.; funding acquisition, E.S.P., A.N. and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Indonesian Ministry of Education, Culture, Research and Technology grant number WCR 3939/UN.18.L1/PP/2021 and The APC was funded by The Indonesian Ministry of Education, Culture, Research and Technology.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding and first author.
Acknowledgments
All authors pay their condolences to one of the author H. Sunarpi, who passed away on 24 December 2020. He was our teacher, family, colleague and a great scientist.
Conflicts of Interest
No competing interest between authors.
Sample Availability
Samples of the compounds are available from the authors.
References
- Dimitriou, F.; Krattinger, R.; Ramelyte, E.; Barysch, M.J.; Micaletto, S.; Dummer, R.; Goldinger, S.M. The World of Melanoma: Epidemiologic, Genetic, and Anatomic Differences of Melanoma Across the Globe. Curr. Oncol. Rep. 2018, 20, 87. [Google Scholar] [CrossRef] [PubMed]
- Savoye, I.; Olsen, C.M.; Whiteman, D.C.; Bijon, A.; Wald, L.; Dartois, L.; Clavel-Chapelon, F.; Boutron-Ruault, M.-C.; Kvaskoff, M. Patterns of Ultraviolet Radiation Exposure and Skin Cancer Risk: The E3N-SunExp Study. J. Epidemiol. 2018, 28, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.; Holman, D.M.; Maguire-Eisen, M. Ultraviolet Radiation Exposure and Its Impact on Skin Cancer Risk. Semin. Oncol. Nurs. 2016, 32, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Ashraf, M.A. Phytochemicals as Potential Anticancer Drugs: Time to Ponder Nature’s Bounty. BioMed Res. Int. 2020, 2020, 8602879. [Google Scholar] [CrossRef]
- Admassu, H.; Gasmalla, M.A.A.; Yang, R.; Zhao, W. Identification of Bioactive Peptides with α-Amylase Inhibitory Potential from Enzymatic Protein Hydrolysates of Red Seaweed (Porphyra Spp). J. Agric. Food Chem. 2018, 66, 4872–4882. [Google Scholar] [CrossRef] [PubMed]
- Carson, M.A.; Clarke, S.A. Bioactive Compounds from Marine Organisms: Potential for Bone Growth and Healing. Mar. Drugs 2018, 16, 340. [Google Scholar] [CrossRef]
- Øverland, M.; Mydland, L.T.; Skrede, A. Marine Macroalgae as Sources of Protein and Bioactive Compounds in Feed for Monogastric Animals. J. Sci. Food Agric. 2018, 99, 13–24. [Google Scholar] [CrossRef]
- Prasedya, E.S.; Miyake, M.; Kobayashi, D.; Hazama, A. Carrageenan Delays Cell Cycle Progression in Human Cancer Cells in Vitro Demonstrated by FUCCI Imaging. BMC Complement. Altern. Med. 2016, 16, 270. [Google Scholar] [CrossRef]
- Prasedya, E.S.; Syafitri, S.M.; Geraldine, B.A.F.D.; Hamdin, C.D.; Frediansyah, A.; Miyake, M.; Kobayashi, D.; Hazama, A.; Sunarpi, H. UVA Photoprotective Activity of Brown Macroalgae Sargassum Cristafolium. Biomedicines 2019, 7, 77. [Google Scholar] [CrossRef]
- Prasedya, E.S.; Martyasari, N.W.R.; Abidin, A.S.; Pebriani, S.A.; Ilhami, B.T.K.; Frediansyah, A.; Sunarwidhi, A.L.; Widyastuti, S.; Sunarpi, H. Macroalgae Sargassum Cristaefolium Extract Inhibits Proinflammatory Cytokine Expression in BALB/C Mice. Scientifica 2020, 2020, 9769454. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, E.; Lukova, P.; Baldzhieva, A.; Katsarov, P.; Nikolova, M.; Iliev, I.; Peychev, L.; Trica, B.; Oancea, F.; Delattre, C.; et al. Immunomodulatory and Anti-Inflammatory Effects of Fucoidan: A Review. Polymers 2020, 12, 2338. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.-F.; Ge, W.-W.; Lin, M.-X.; Li, Q.-S. A Review of the Components of Seaweeds as Potential Candidates in Cancer Therapy. Anticancer Agents Med. Chem. 2018, 18, 354–366. [Google Scholar] [CrossRef]
- Ghannam, A.; Murad, H.; Jazzara, M.; Odeh, A.; Allaf, A.W. Isolation, Structural Characterization, and Antiproliferative Activity of Phycocolloids from the Red Seaweed Laurencia Papillosa on MCF-7 Human Breast Cancer Cells. Int. J. Biol. Macromol. 2018, 108, 916–926. [Google Scholar] [CrossRef]
- Desamero, M.J.; Kakuta, S.; Chambers, J.K.; Uchida, K.; Hachimura, S.; Takamoto, M.; Nakayama, J.; Nakayama, H.; Kyuwa, S. Orally Administered Brown Seaweed-Derived β-Glucan Effectively Restrained Development of Gastric Dysplasia in A4gnt KO Mice That Spontaneously Develop Gastric Adenocarcinoma. Int. Immunopharmacol. 2018, 60, 211–220. [Google Scholar] [CrossRef]
- Rocha, D.H.A.; Seca, A.M.L.; Pinto, D.C.G.A. Seaweed Secondary Metabolites In Vitro and In Vivo Anticancer Activity. Mar. Drugs 2018, 16, 410. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-C.; Hou, M.-F.; Huang, H.-W.; Chang, F.-R.; Yeh, C.-C.; Tang, J.-Y.; Chang, H.-W. Marine Algal Natural Products with Anti-Oxidative, Anti-Inflammatory, and Anti-Cancer Properties. Cancer Cell Int. 2013, 13, 55. [Google Scholar] [CrossRef]
- Tannoury, M.; Saab, A.; Elia, J.; Harb, N.; Makhlouf, H.; Diab, M. In Vitro Cytotoxic Activity of Laurencia Papillosa, Marine Red Algae from the Lebanese Coast. J. Appl. Pharm. Sci. 2017, 7, 175–179. [Google Scholar]
- Pereira, L. Macroalgae. Encyclopedia 2021, 1, 177–188. [Google Scholar] [CrossRef]
- Khotimchenko, M.; Tiasto, V.; Kalitnik, A.; Begun, M.; Khotimchenko, R.; Leonteva, E.; Bryukhovetskiy, I.; Khotimchenko, Y. Antitumor Potential of Carrageenans from Marine Red Algae. Carbohydr. Polym. 2020, 246, 116568. [Google Scholar] [CrossRef]
- Aziz, E.; Batool, R.; Khan, M.U.; Rauf, A.; Akhtar, W.; Heydari, M.; Rehman, S.; Shahzad, T.; Malik, A.; Mosavat, S.H.; et al. An overview on red algae bioactive compounds and their pharmaceutical applications. J. Complement. Integr. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yanshin, N.; Kushnareva, A.; Lemesheva, V.; Birkemeyer, C.; Tarakhovskaya, E. Chemical Composition and Potential Practical Application of 15 Red Algal Species from the White Sea Coast (the Arctic Ocean). Molecules 2021, 26, 2489. [Google Scholar] [CrossRef]
- Paiva, L.; Lima, E.; Neto, A.I.; Marcone, M.; Baptista, J. Health-Promoting Ingredients from Four Selected Azorean Macroalgae. Food Res. Int. 2016, 89, 432–438. [Google Scholar] [CrossRef]
- Yang, T.-H.; Yao, H.-T.; Chiang, M.-T. Red Algae (Gelidium amansii) Hot-Water Extract Ameliorates Lipid Metabolism in Hamsters Fed a High-Fat Diet. J. Food Drug Anal. 2017, 25, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Pugazhendhi, A.; Prabakar, D.; Jacob, J.M.; Karuppusamy, I.; Saratale, R.G. Synthesis and Characterization of Silver Nanoparticles Using Gelidium amansii and Its Antimicrobial Property against Various Pathogenic Bacteria. Microb. Pathog. 2018, 114, 41–45. [Google Scholar] [CrossRef]
- Choi, J.; Kim, K.-J.; Koh, E.-J.; Lee, B.-Y. Gelidium Elegans Extract Ameliorates Type 2 Diabetes via Regulation of MAPK and PI3K/Akt Signaling. Nutrients 2018, 10, 51. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.-J.; Lee, M. The Suppressive Effect of Gelidium Amansi-EtOH Extracts on the Adipogenesis with MAPK Signals in Adipocytes with or without Macrophages. Food Sci. Biotechnol. 2017, 26, 1715–1723. [Google Scholar] [CrossRef] [PubMed]
- Algaebase: Listing the World’s Algae. Available online: https://www.algaebase.org/ (accessed on 30 August 2021).
- Valentina, F.; Pinheiro, C.M. Physicochemical Changes of Air-Dried and Salt-Processed Ulva rigida over Storage Time. Molecules 2019, 24, 2955. [Google Scholar] [CrossRef]
- Hideki Kanda, W. Direct Extraction of Lutein from Wet Macroalgae by Liquefied Dimethyl Ether without Any Pretreatment. ACS Omega 2020, 5, 24005. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.; Schellenberg, I. Comparison of Different Extraction Methods for the Determination of Essential Oils and Related Compounds from Aromatic Plants and Optimization of Solid-Phase Microextraction/Gas Chromatography. Anal. Bioanal. Chem. 2007, 387, 2207–2217. [Google Scholar] [CrossRef]
- Boonchumi, W.; Peerapornpisal, Y.; Kanjanapothi, D.; Pekkoh, J.; Pumas, C.; Jamjai, U.; Amornlerdpison, D.; Noiraksar, T.; Vacharapiyasophon, P. Antioxidant Activity of Some Seaweed from the Gulf of Thailand. Int. J. Agric. Biol. 2011, 13, 1560–8530. [Google Scholar]
- Neto, R.T.; Marçal, C.; Queirós, A.S.; Abreu, H.; Silva, A.M.S.; Cardoso, S.M. Screening of Ulva Rigida, Gracilaria Sp., Fucus Vesiculosus and Saccharina Latissima as Functional Ingredients. Int. J. Mol. Sci. 2018, 19, 2987. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Viedma, J.; Aguilera, J.M.; Flores, M.; Lemus-Mondaca, R.; Larrazabal, M.J.; Miranda, J.M.; Aubourg, S.P. Protective Effect of Red Algae (Rhodophyta) Extracts on Essential Dietary Components of Heat-Treated Salmon. Antioxidants 2021, 10, 1108. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, J.; Fan, J.; Clark, J.; Shen, P.; Li, Y.; Zhang, C. Microwave Assisted Extraction of Phenolic Compounds from Four Economic Brown Macroalgae Species and Evaluation of Their Antioxidant Activities and Inhibitory Effects on α-Amylase, α-Glucosidase, Pancreatic Lipase and Tyrosinase. Food Res. Int. 2018, 113, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.-J.; Xu, M.-X.; Zhou, B.-H.; Du, Y.-S.; Yao, J.-H.; Zhou, L. Effects of 2-Aryl-1-Cyano-1,2,3,4-Tetrohydroisoquinolines on Apoptosis Induction Mechanism in NB4 and MKN-45 Cells. Toxicol. Vitro Int. 2018, 54, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Bratosin, D.; Mitrofan, L.; Palii, C.; Estaquier, J.; Montreuil, J. Novel fluorescence assay using calcein-AM for the determination of human erythrocyte viability and aging. Cytom. Part A J. Int. Soc. Anal. Cytol. 2005, 66, 78–84. [Google Scholar] [CrossRef]
- Arora, S.; Tandon, S. DNA Fragmentation and Cell Cycle Arrest: A Hallmark of Apoptosis Induced by Ruta Graveolens in Human Colon Cancer Cells. Homeopathy 2015, 104, 36–47. [Google Scholar] [CrossRef]
- Jakic, B.; Buszko, M.; Cappellano, G.; Wick, G. Elevated Sodium Leads to the Increased Expression of HSP60 and Induces Apoptosis in HUVECs. PLoS ONE 2017, 12, e0179383. [Google Scholar] [CrossRef]
- Kasibhatla, S.; Amarante-Mendes, G.P.; Finucane, D.; Brunner, T.; Bossy-Wetzel, E.; Green, D.R. Analysis of DNA Fragmentation Using Agarose Gel Electrophoresis. CSH Protoc. 2006, 2006, pdb.prot4429. [Google Scholar] [CrossRef]
- Zakaria, F.R.; Prangdimurti, E.; Adawiyah, D.R.; Priosoeryanto, B.P.; Huda, N. Chemical Evaluation of a Nori-like Product (Geluring) Made from the Mixture of Gelidium Sp. and Ulva Lactuca Seaweeds. Curr. Res. Nutr. Food Sci. J. 2018, 6, 664–671. [Google Scholar]
- Generalić Mekinić, I.; Skroza, D.; Šimat, V.; Hamed, I.; Čagalj, M.; Popović Perković, Z. Phenolic Content of Brown Algae (Pheophyceae) Species: Extraction, Identification, and Quantification. Biomolecules 2019, 9, 244. [Google Scholar] [CrossRef]
- Alvarez-Gomez, F. Analysis of Antioxidant Capacity and Bioactive Compounds in Marine Macroalgal and Lichenic Extracts Using Different Solvents and Evaluation Methods. Cienc. Mar. 2016, 42, 271–288. [Google Scholar] [CrossRef]
- Pirian, K.; Jeliani, Z.Z.; Sohrabipour, J.; Arman, M.; Faghihi, M.M.; Yousefzadi, M. Nutritional and Bioactivity Evaluation of Common Seaweed Species from the Persian Gulf. Iran. J. Sci. Technol. Trans. Sci. 2018, 42, 1795–1804. [Google Scholar] [CrossRef]
- Agarwal, P.; Kayala, P.; Chandrasekaran, N.; Mukherjee, A.; Shah, S.; Thomas, J. Antioxidant and Antibacterial Activity of Gelidium Pusillum (Stackhouse) against Aeromonas Caviae and Its Applications in Aquaculture. Aquac. Int. 2021, 29, 845–858. [Google Scholar] [CrossRef]
- Chakraborty, K.; Joseph, D.; Praveen, N.K. Antioxidant Activities and Phenolic Contents of Three Red Seaweeds (Division: Rhodophyta) Harvested from the Gulf of Mannar of Peninsular India. J. Food Sci. Technol. 2015, 52, 1924–1935. [Google Scholar] [CrossRef]
- Vega, J.; Álvarez-Gómez, F.; Güenaga, L.; Figueroa, F.L.; Gómez-Pinchetti, J.L. Antioxidant Activity of Extracts from Marine Macroalgae, Wild-Collected and Cultivated, in an Integrated Multi-Trophic Aquaculture System. Aquaculture 2020, 522, 735088. [Google Scholar] [CrossRef]
- Molyneux, P. The Use of the Stable Free Radical Diphenylpicrylhydrazyl (DPPH) for Estimating Antioxidant. Songklanakarin J. Sci. Technol. 2004, 26, 211–219. [Google Scholar]
- Duh, C.-Y.; Wang, S.-K.; Weng, Y.-L. Brassicolene, A Novel Cytotoxic Diterpenoid from the Formosan Soft Coral Nephthea Brassica. Tetrahedron Lett. 2000, 41, 1401–1403. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Ta, Q.T.H.; Pham, Q.T.; Luong, T.N.H.; Phung, V.T.; Duong, T.-H.; Vo, V.G. Anticancer Activity of Novel Plant Extracts and Compounds from Adenosma Bracteosum (Bonati) in Human Lung and Liver Cancer Cells. Molecules 2020, 25, 2912. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Tu, C.-J.; Wu, H.-T. Growth-Inhibitory Effects of the Red Alga Gelidium amansii on Cultured Cells. Biol. Pharm. Bull. 2004, 27, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Harada, H.; Noro, T.; Kamei, Y. Selective antitumor activity in vitro from marine algae from Japan coasts. Biol. Pharm. Bull. 1997, 20, 541–546. [Google Scholar] [CrossRef]
- Martins, R.M.; Nedel, F.; Guimarães, V.B.S.; da Silva, A.F.; Colepicolo, P.; de Pereira, C.M.P.; Lund, R.G. Macroalgae Extracts from Antarctica Have Antimicrobial and Anticancer Potential. Front. Microbiol. 2018, 9, 412. [Google Scholar] [CrossRef]
- Shalaby, E. Algae as Promising Organisms for Environment and Health. Plant Signal. Behav. 2011, 6, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Shiao, W.-C.; Kuo, C.-H.; Tsai, Y.-H.; Hsieh, S.-L.; Kuan, A.-W.; Hong, Y.-H.; Huang, C.-Y. In Vitro Evaluation of Anti-Colon Cancer Potential of Crude Extracts of Fucoidan Obtained from Sargassum Glaucescens Pretreated by Compressional-Puffing. Appl. Sci. 2020, 10, 3058. [Google Scholar] [CrossRef]
- Yuan, J.; Yankner, B.A. Apoptosis in the Nervous System. Nature 2000, 407, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Pelicci, S.; Diaspro, A.; Lanzanò, L. Chromatin Nanoscale Compaction in Live Cells Visualized by Acceptor-to-Donor Ratio Corrected Förster Resonance Energy Transfer between DNA Dyes. J. Biophotonics 2019, 12, e201900164. [Google Scholar] [CrossRef]
- Walker, P.R.; LeBlanc, J.; Sikorska, M. Evidence That DNA Fragmentation in Apoptosis Is Initiated and Propagated by Single-Strand Breaks. Cell Death Differ. 1997, 4, 506–515. [Google Scholar] [CrossRef]
- Bernardini, G.; Minetti, M.; Polizzotto, G.; Biazzo, M.; Santucci, A. Pro-Apoptotic Activity of French Polynesian Padina Pavonica Extract on Human Osteosarcoma Cells. Mar. Drugs 2018, 16, 504. [Google Scholar] [CrossRef]
- Güner, A.; Nalbantsoy, A.; Sukatar, A.; Karabay Yavaşoğlu, N.Ü. Apoptosis-Inducing Activities of Halopteris Scoparia L. Sauvageau (Brown Algae) on Cancer Cells and Its Biosafety and Antioxidant Properties. Cytotechnology 2019, 71, 687–704. [Google Scholar] [CrossRef]
- Al Monla, R.; Dassouki, Z.; Kouzayha, A.; Salma, Y.; Gali-Muhtasib, H.; Mawlawi, H. The Cytotoxic and Apoptotic Effects of the Brown Algae Colpomenia Sinuosa Are Mediated by the Generation of Reactive Oxygen Species. Molecules 2020, 25, 1993. [Google Scholar] [CrossRef]
- Çelenk, F.G.; Özkaya, A.B.; Sukatar, A. Macroalgae of Izmir Gulf: Dictyotaceae Exhibit High in Vitro Anti-Cancer Activity Independent from Their Antioxidant Capabilities. Cytotechnology 2016, 68, 2667–2676. [Google Scholar] [CrossRef]
- Olasehinde, T.A.; Olaniran, A.O.; Okoh, A.I. Macroalgae as a Valuable Source of Naturally Occurring Bioactive Compounds for the Treatment of Alzheimer’s Disease. Mar. Drugs 2019, 17, 609. [Google Scholar] [CrossRef]
- Haq, S.H.; Al-Ruwaished, G.; Al-Mutlaq, M.A.; Naji, S.A.; Al-Mogren, M.; Al-Rashed, S.; Ain, Q.T.; Al-Amro, A.A.; Al-Mussallam, A. Antioxidant, Anticancer Activity and Phytochemical Analysis of Green Algae, Chaetomorpha Collected from the Arabian Gulf. Sci. Rep. 2019, 9, 18906. [Google Scholar] [CrossRef]
- Ismail, M.M.; Alotaibi, B.S.; EL-Sheekh, M.M. Therapeutic Uses of Red Macroalgae. Molecules 2020, 25, 4411. [Google Scholar] [CrossRef]
- Alghazeer, R.; Howell, N.K.; El-Naili, M.B.; Awayn, N. Anticancer and Antioxidant Activities of Some Algae from Western Libyan Coast. Nat. Sci. 2018, 10, 232. [Google Scholar] [CrossRef][Green Version]
- Abdala Díaz, R.T.; Casas Arrojo, V.; Arrojo Agudo, M.A.; Cárdenas, C.; Dobretsov, S.; Figueroa, F.L. Immunomodulatory and Antioxidant Activities of Sulfated Polysaccharides from Laminaria Ochroleuca, Porphyra Umbilicalis, and Gelidium Corneum. Mar. Biotechnol. 2019, 21, 577–587. [Google Scholar] [CrossRef]
- Sithranga Boopathy, N.; Kathiresan, K. Anticancer Drugs from Marine Flora: An Overview. J. Oncol. 2010, 2010, 214186. [Google Scholar] [CrossRef] [PubMed]
- Häder, D.-P. Chapter 9—Phycocolloids from macroalgae. In Natural Bioactive Compounds; Sinha, R.P., Häder, D.-P., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 187–201. ISBN 978-0-12-820655-3. [Google Scholar]
- Kim, M.S.; Kim, J.Y.; Choi, W.H.; Lee, S.S. Effects of Seaweed Supplementation on Blood Glucose Concentration, Lipid Profile, and Antioxidant Enzyme Activities in Patients with Type 2 Diabetes Mellitus. Nutr. Res. Pract. 2008, 2, 62–67. [Google Scholar] [CrossRef]
- Kang, M.-C.; Kang, N.; Kim, S.-Y.; Lima, I.S.; Ko, S.-C.; Kim, Y.-T.; Kim, Y.-B.; Jeung, H.-D.; Choi, K.-S.; Jeon, Y.-J. Popular Edible Seaweed, Gelidium amansii Prevents against Diet-Induced Obesity. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2016, 90, 181–187. [Google Scholar] [CrossRef]
- Nursid, M.; Khatulistiani, T.S.; Noviendri, D.; Hapsari, F.; Hardiyati, T. Total Phenolic Content, Antioxidant Activity and Tyrosinase Inhibitor from Marine Red Algae Extract Collected from Kupang, East Nusa Tenggara. IOP Conf. Ser. Earth Environ. Sci. 2020, 493, 012013. [Google Scholar] [CrossRef]
- Sammar, M.; Abu-Farich, B.; Rayan, I.; Falah, M.; Rayan, A. Correlation between Cytotoxicity in Cancer Cells and Free Radical-Scavenging Activity: In Vitro Evaluation of 57 Medicinal and Edible Plant Extracts. Oncol. Lett. 2019, 18, 6563–6571. [Google Scholar] [CrossRef] [PubMed]
- Hassan, L.E.A.; Ahamed, M.B.K.; Majid, A.S.A.; Baharetha, H.M.; Muslim, N.S.; Nassar, Z.D.; Majid, A.M.A. Correlation of Antiangiogenic, Antioxidant and Cytotoxic Activities of Some Sudanese Medicinal Plants with Phenolic and Flavonoid Contents. BMC Complement. Altern. Med. 2014, 14, 406. [Google Scholar] [CrossRef]
- ZiemLewska, A.; Zagórska-Dziok, M.; Nizioł-Łukaszewska, Z. Assessment of Cytotoxicity and Antioxidant Properties of Berry Leaves as By-Products with Potential Application in Cosmetic and Pharmaceutical Products. Sci. Rep. 2021, 11, 3240. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Gustafson, K.R. Marine Pharmacology in 2000: Antitumor and Cytotoxic Compounds. Int. J. Cancer 2003, 105, 291–299. [Google Scholar] [CrossRef] [PubMed]
- El Gamal, A.A. Biological Importance of Marine Algae. Saudi Pharm. J. 2010, 18, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Murray, J.; Delgoda, R.; Gallimore, W. Two New Oxodolastane Diterpenes from the Jamaican Macroalga Canistrocarpus Cervicornis. Mar. Drugs 2017, 15, 150. [Google Scholar] [CrossRef]
- Altun, İ.; Sonkaya, A. The Most Common Side Effects Experienced by Patients Were Receiving First Cycle of Chemotherapy. Iran. J. Public Health 2018, 47, 1218–1219. [Google Scholar]
- Gambato, G.; Baroni, É.G.; Garcia, C.S.C.; Frassini, R.; Frozza, C.O.S.; Moura, S.; Pereira, C.M.P.; Fujii, M.T.; Colepicolo, P.; Lambert, A.P.F.; et al. Brown Algae Himantothallus Grandifolius (Desmarestiales, Phaeophyceae) Suppresses Proliferation and Promotes Apoptosis-Mediated Cell Death in Tumor Cells. Adv. Biol. Chem. 2014, 04, 98–108. [Google Scholar] [CrossRef]
- Canoy, J.L.; Bitacura, J.G. Cytotoxicity and Antiangiogenic Activity of Turbinaria Ornata Agardh and Padina Australis Hauck Ethanolic Extracts. Anal. Cell. Pathol. 2018, 2018, e3709491. [Google Scholar] [CrossRef]
- Hazekawa, M.; Nishinakagawa, T.; Kawakubo-Yasukochi, T.; Nakashima, M. Evaluation of IC50 Levels Immediately after Treatment with Anticancer Reagents Using a Real-time Cell Monitoring Device. Exp. Ther. Med. 2019, 18, 3197–3205. [Google Scholar] [CrossRef]
- Namvar, F.; Baharara, J.; Mahdi, A.A. Antioxidant and Anticancer Activities of Selected Persian Gulf Algae. Indian J. Clin. Biochem. 2014, 29, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Boise, L.H.; Shanmugam, M. Cancer Metabolism and the Evasion of Apoptotic Cell Death. Cancers 2019, 11, 1144. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Kalimuthu, K.; Park, Y.S.; Luo, X.; Choudry, M.H.A.; Bartlett, D.L.; Lee, Y.J. BAX-Dependent Mitochondrial Pathway Mediates the Crosstalk between Ferroptosis and Apoptosis. Apoptosis 2020, 25, 625–631. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).