Revealing DNA Structure at Liquid/Solid Interfaces by AFM-Based High-Resolution Imaging and Molecular Spectroscopy
Abstract
1. Introduction
2. Preparation of DNA Samples for High-Resolution AFM Imaging
3. Basic Technical Developments
3.1. Dynamic AFM Modes
3.1.1. AM-AFM Imaging
3.1.2. PeakForce Tapping
3.1.3. FM-AFM Imaging
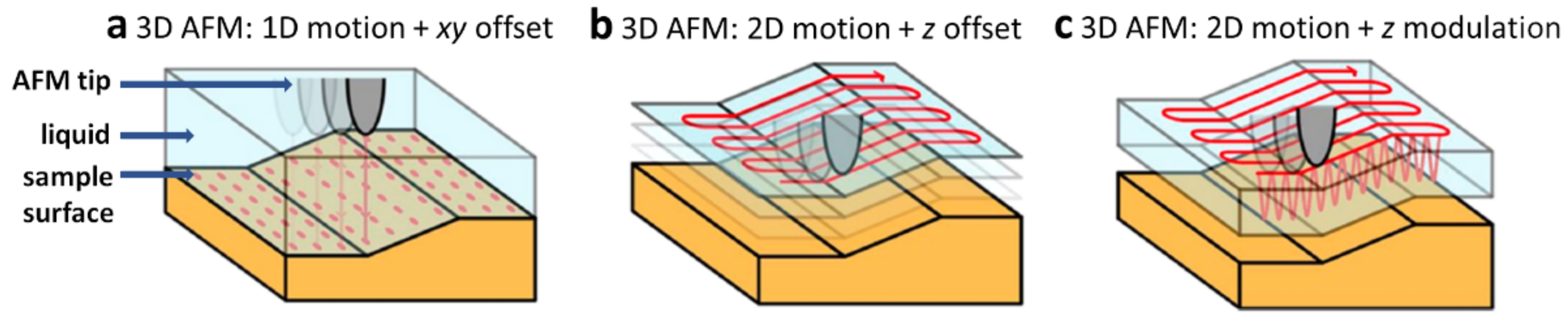
3.1.4. 3D AFM at Solid-Liquid Interfaces
4. AFM Profiling of DNA
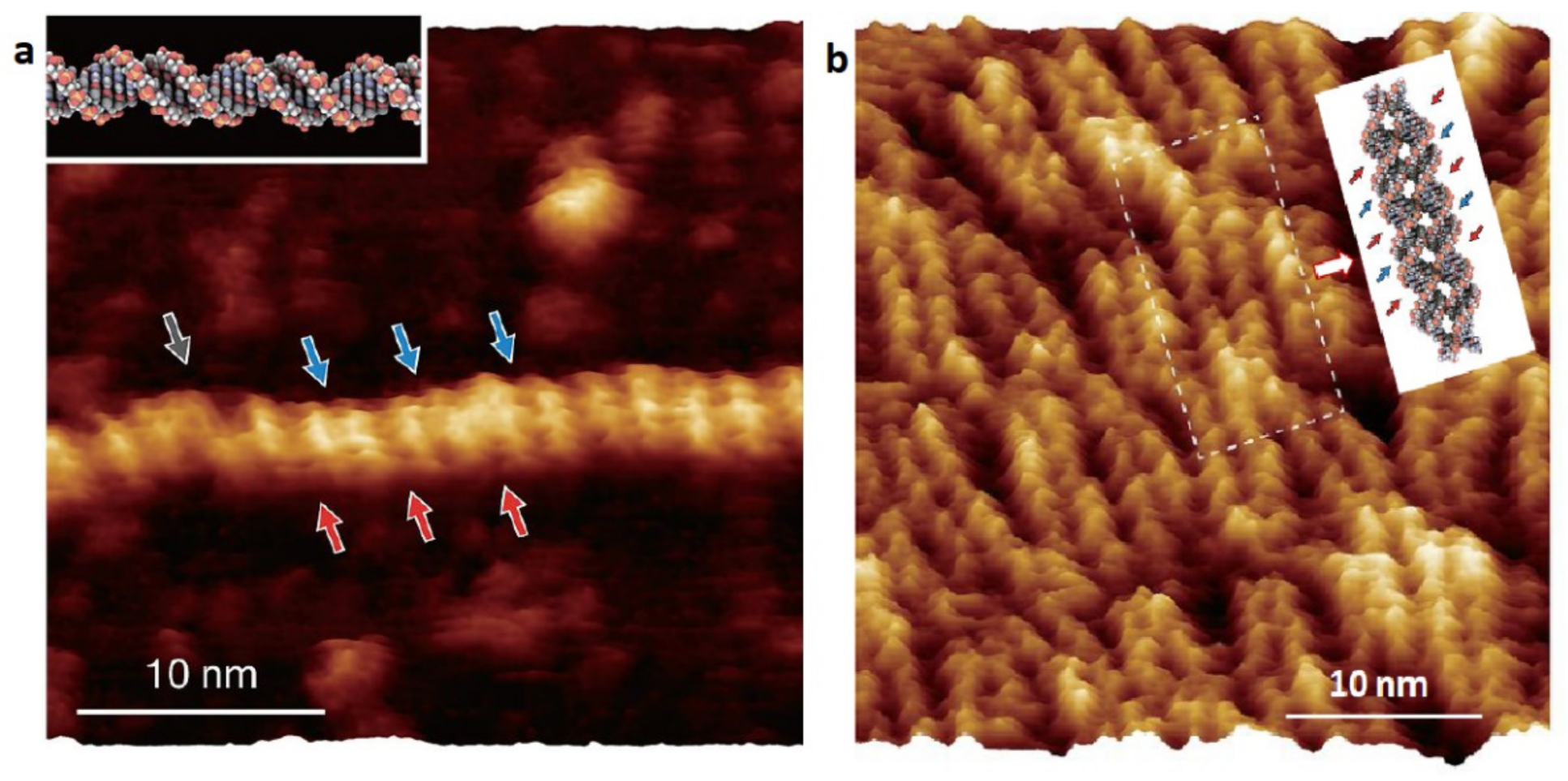
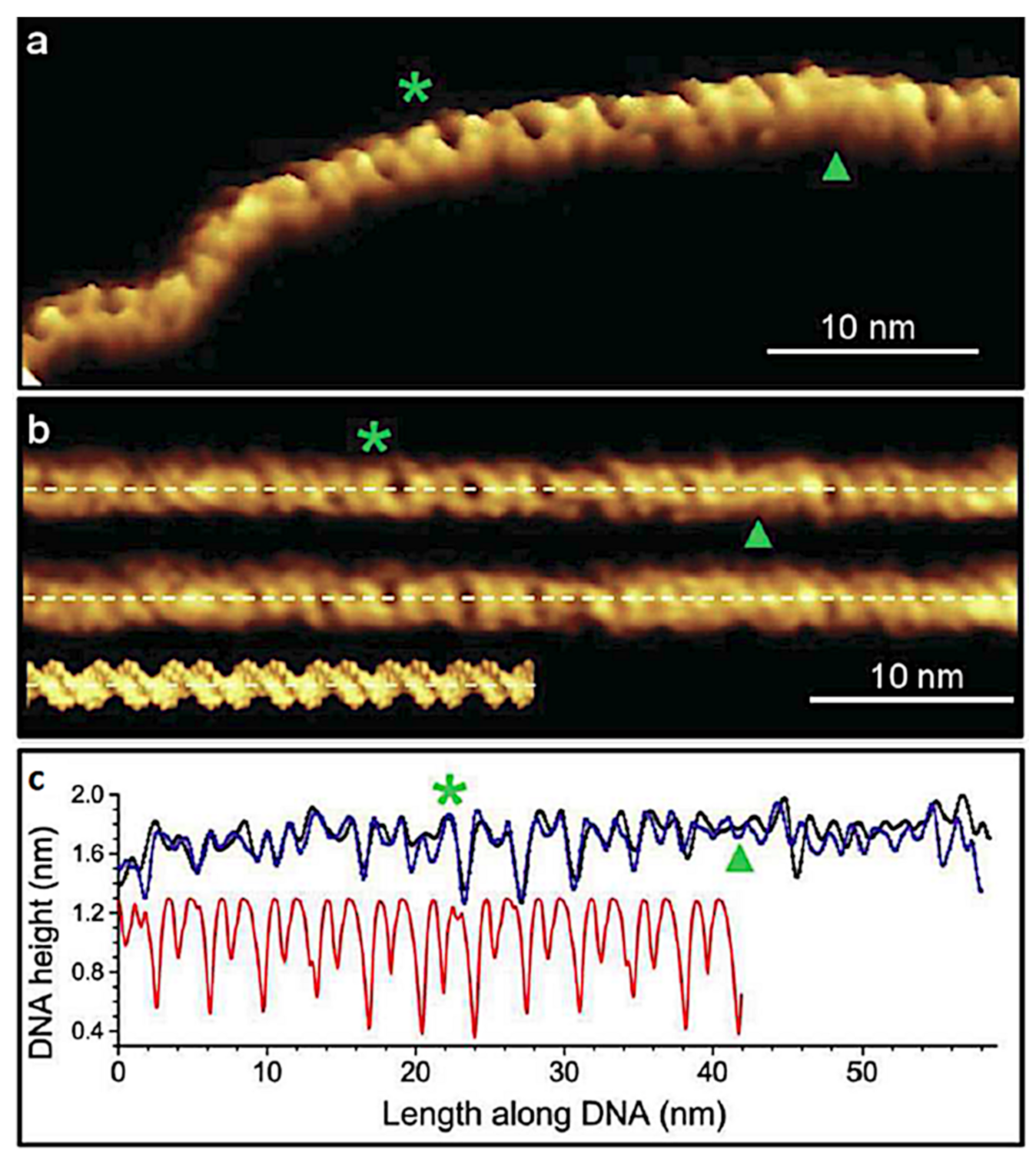
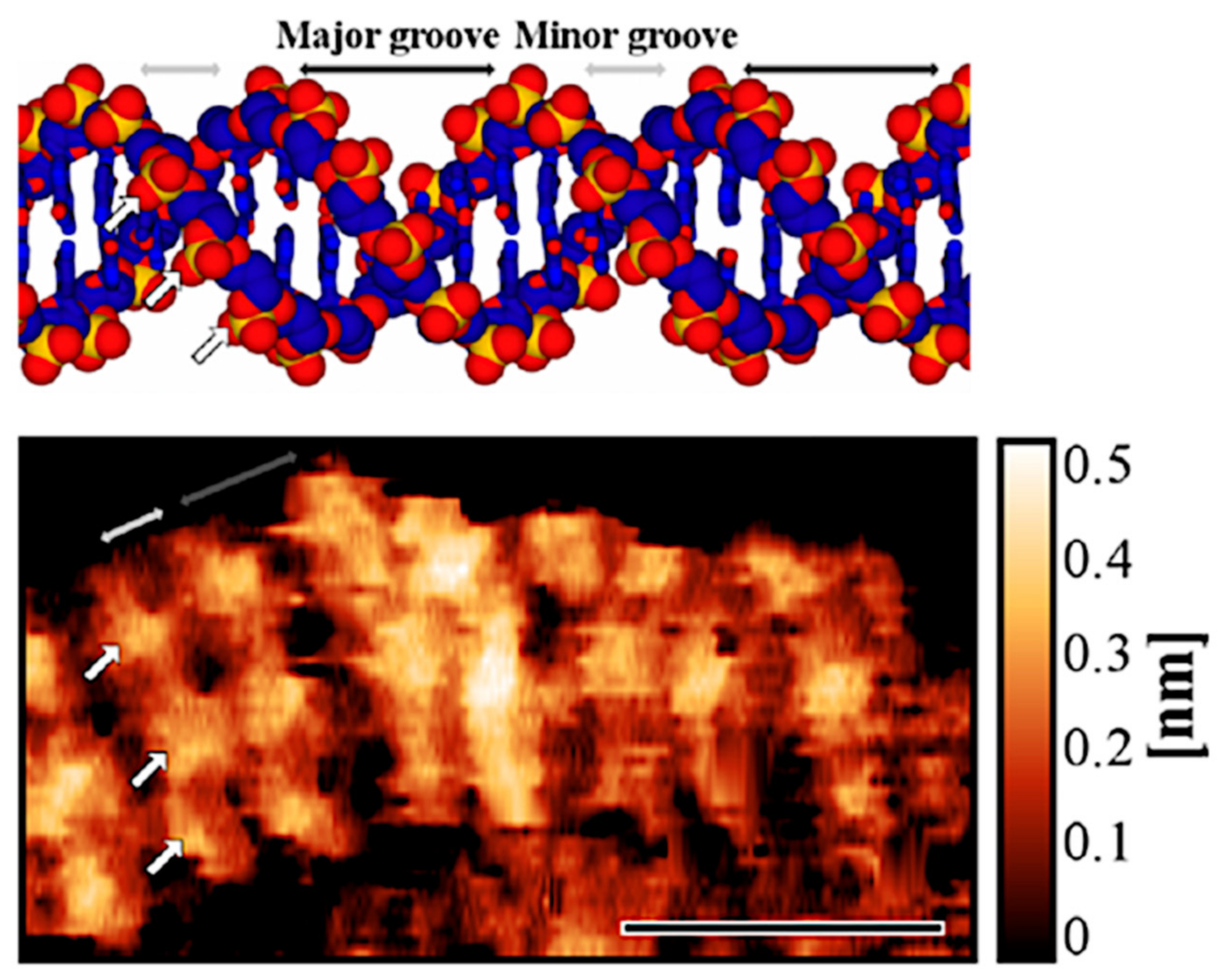
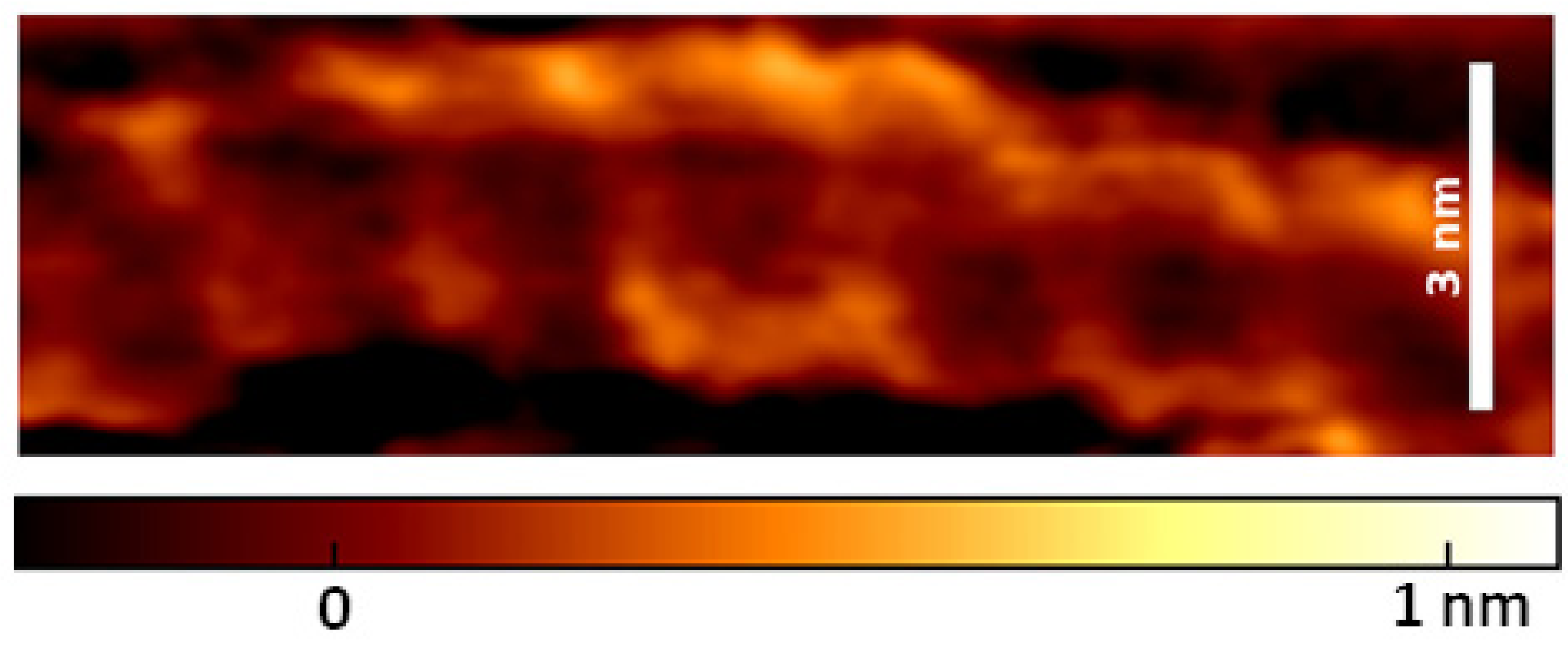
4.1. Revealing Submolecular Structure of Individual DNA Molecules
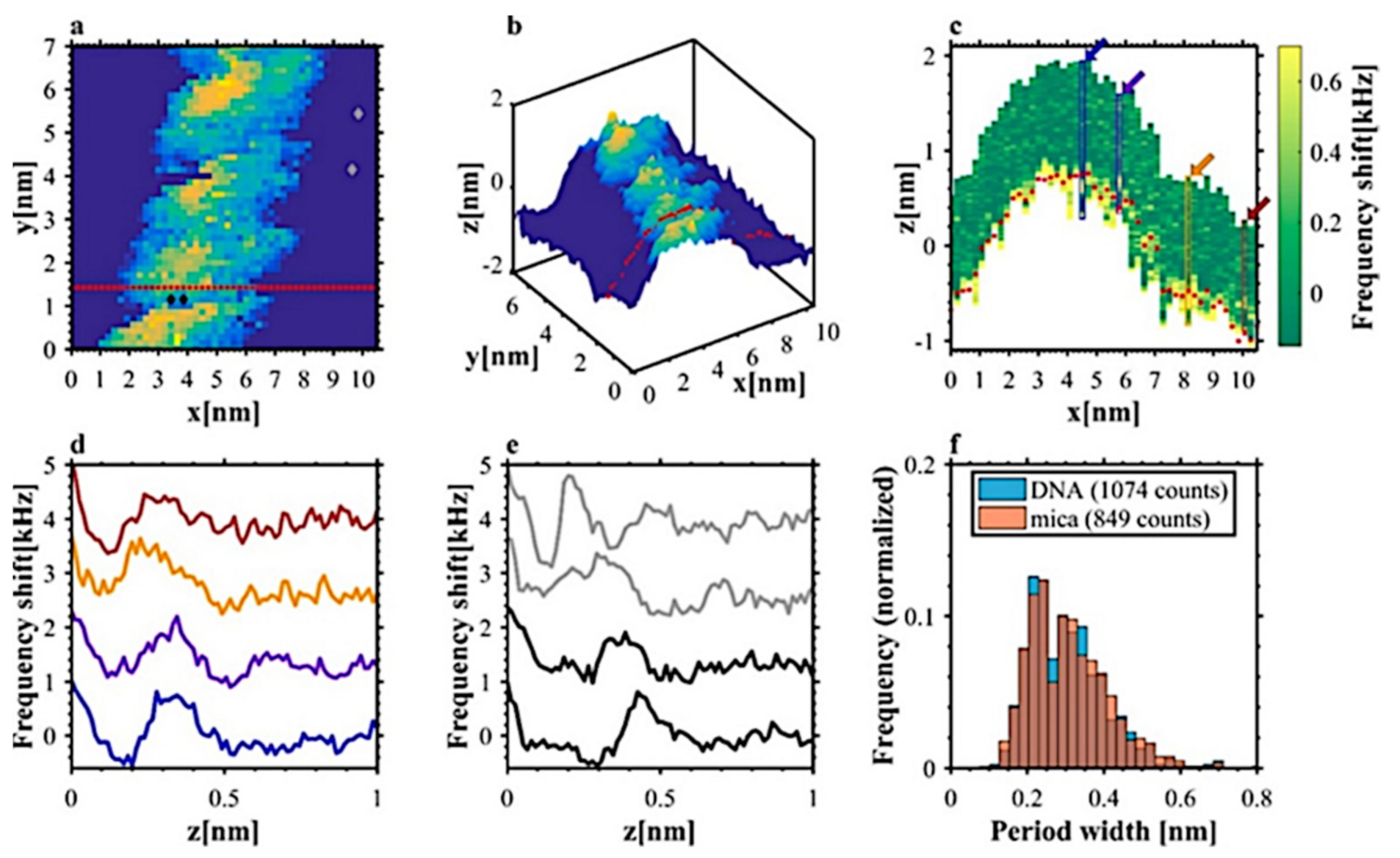
4.2. 3D AFM Imaging of DNA Associated Hydration Structures
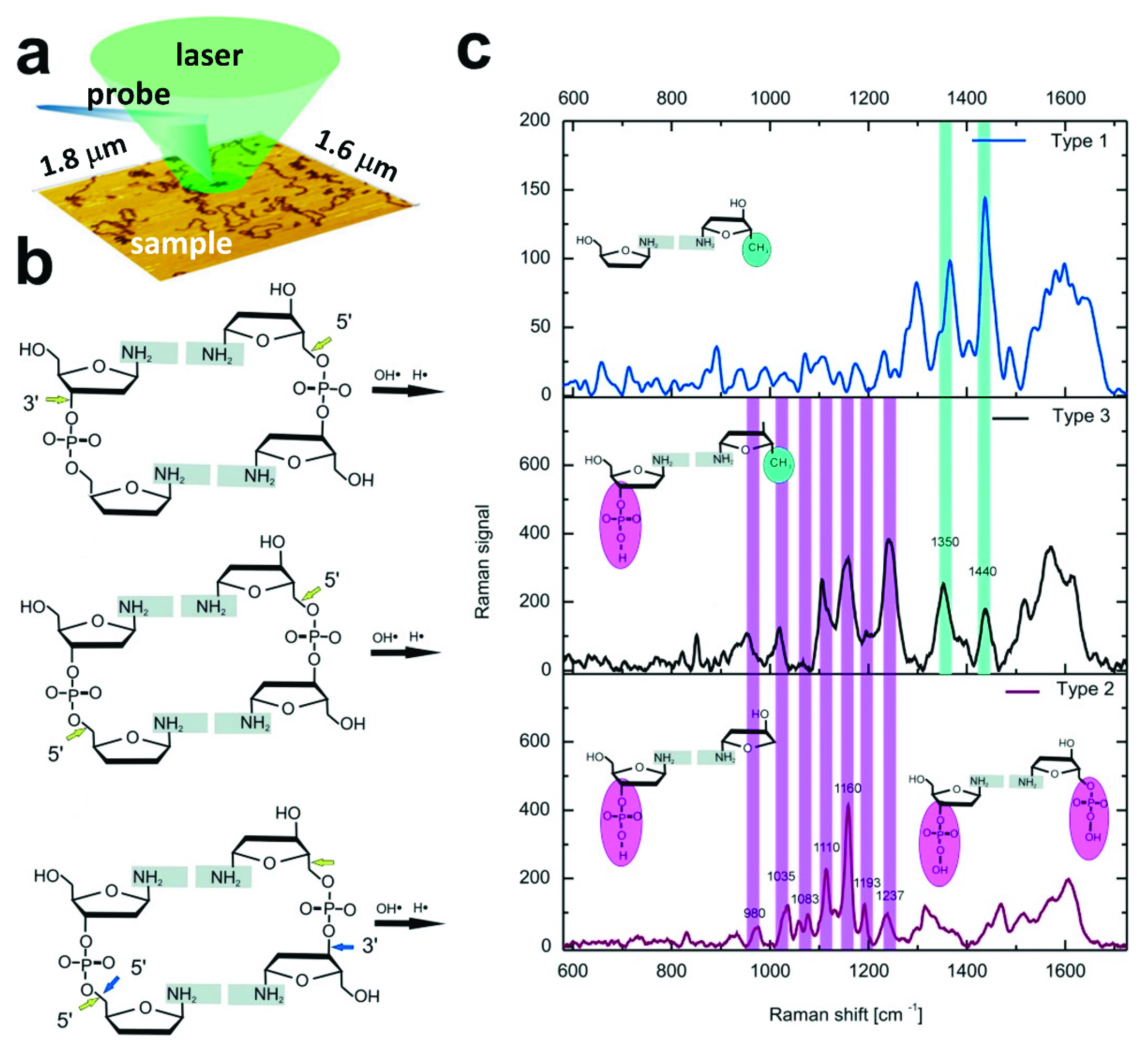
5. SERS and TERS Research on DNA Structure
6. Protective Role of Solvents
7. Concluding Remarks and Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.E.; Gosling, R.G. Molecular configuration in sodium thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Maurstad, G.; Prass, M.; Serpell, L.C.; Sikorski, P. Dehydration stability of amyloid fibrils studied by AFM. Eur. Biophys. J. 2009, 38, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef]
- Mou, J.; Czajkowsky, D.M.; Zhang, Y.; Shao, Z. High-resolution atomic-force microscopy of DNA: The pitch of the double helix. FEBS Lett. 1995, 371, 279–282. [Google Scholar] [CrossRef]
- Hegner, M.; Wagner, P.; Semenza, G. Immobilizing DNA on gold via thiol modification for Atomic Force Microscopy imaging in buffer solutions. FEBS Lett. 1993, 336, 452–456. [Google Scholar] [CrossRef]
- Medalia, O.; Englander, J.; Guckenberger, R.; Sperling, J. AFM imaging in solution of protein-DNA complexes formed on DNA anchored to a gold surface. Ultramicroscopy 2002, 90, 103–112. [Google Scholar] [CrossRef]
- Oliveira Brett, A.M.; Chiorcea, A.M. Atomic Force Microscopy of DNA immobilized onto a highly oriented pyrolytic graphite electrode surface. Langmuir 2003, 19, 3830–3839. [Google Scholar] [CrossRef]
- Dubrovin, E.V.; Dadinova, L.A.; Petoukhov, M.V.; Soshinskaya, E.Y.; Mozhaev, A.A.; Klinov, D.V.; Schäffer, T.E.; Shtykova, E.V.; Batishchev, O.V. Spatial organization of Dps and DNA–Dps complexes. J. Mol. Biol. 2021, 433, 166930. [Google Scholar] [CrossRef]
- Ricardo, K.B.; Xu, A.; Salim, M.; Zhou, F.; Liu, H. Deposition of DNA Nanostructures on Highly Oriented Pyrolytic Graphite. Langmuir 2017, 33, 3991–3997. [Google Scholar] [CrossRef]
- Shankla, M.; Aksimentiev, A. Step-defect guided delivery of DNA to a graphene nanopore. Nat. Nanotechnol. 2019, 14, 858–865. [Google Scholar] [CrossRef]
- Adamcik, J.; Klinov, D.V.; Witz, G.; Sekatskii, S.K.; Dietler, G. Observation of single-stranded DNA on mica and highly oriented pyrolytic graphite by Atomic Force Microscopy. FEBS Lett. 2006, 580, 5671–5675. [Google Scholar] [CrossRef]
- Rechendorff, K.; Witz, G.; Adamcik, J.; Dietler, G. Persistence length and scaling properties of single-stranded DNA adsorbed on modified graphite. J. Chem. Phys. 2009, 131, 095103. [Google Scholar] [CrossRef]
- Adamcik, J.; Tobenas, S.; Di Santo, G.; Klinov, D.; Dietler, G. Temperature-controlled assembly of high ordered/disordered dodecylamine layers on HOPG: Consequences for DNA patterning. Langmuir 2009, 25, 3159–3162. [Google Scholar] [CrossRef]
- Tobenas, S.; Bystrenova, E.; Radenovic, A.; Di Santo, G.; Dietler, G. Study of DNA in “glasslike state” by Atomic Force Microscopy: Importance of substrates. Jpn. J. Appl. Phys. 2006, 45, 2345–2348. [Google Scholar] [CrossRef]
- Hansma, H.G.; Vesenka, J.; Siegerist, C.; Kelderman, G.; Morrett, H.; Sinsheimer, R.L.; Elings, V.; Bustamante, C.; Hansma, P.K. Reproducible imaging and dissection of plasmid DNA under liquid with the atomic force microscope. Science 1992, 256, 1180–1184. [Google Scholar] [CrossRef]
- Japaridze, A.; Vobornik, D.; Lipiec, E.; Cerreta, A.; Szczerbinski, J.; Zenobi, R.; Dietler, G. Toward an Effective Control of DNA’s Submolecular Conformation on a Surface. Macromolecules 2016, 49, 643–652. [Google Scholar] [CrossRef]
- Heenan, P.R.; Perkins, T.T. Imaging DNA Equilibrated onto Mica in Liquid Using Biochemically Relevant Deposition Conditions. ACS Nano 2019, 13, 4220–4229. [Google Scholar] [CrossRef]
- Kuchuk, K.; Sivan, U. Hydration Structure of a Single DNA Molecule Revealed by Frequency-Modulation Atomic Force Microscopy. Nano Lett. 2018, 18, 2733–2737. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. Visualization of supercoiled DNA with Atomic Force Microscopy in situ. Proc. Natl. Acad. Sci. USA 1997, 94, 496–501. [Google Scholar] [CrossRef]
- Pope, L.H.; Davies, M.C.; Laughton, C.A.; Roberts, C.J.; Tendler, S.J.B.; Williams, P.M. Atomic Force Microscopy studies of intercalation-induced changes in plasmid DNA tertiary structure. J. Microsc. 2000, 199, 68–78. [Google Scholar] [CrossRef]
- Fang, Y.; Hoh, J.H. Early intermediates in spermidine-induced DNA condensation on the surface of mica. J. Am. Chem. Soc. 1998, 120, 8903–8909. [Google Scholar] [CrossRef]
- Hansma, H.G.; Golan, R.; Hsieh, W.; Lollo, C.P.; Mullen-Ley, P.; Kwoh, D. DNA condensation for gene therapy as monitored by Atomic Force Microscopy. Nucleic Acids Res. 1998, 26, 2481–2487. [Google Scholar] [CrossRef]
- Allison, D.P.; Kerper, P.S.; Doktycz, M.J.; Spain, J.A.; Modrich, P.; Larimer, F.W.; Thundat, T.; Warmack, R.J. Direct atomic force microscope imaging of EcoRI endonuclease site specifically bound to plasmid DNA molecules. Proc. Natl. Acad. Sci. USA 1996, 93, 8826–8829. [Google Scholar] [CrossRef]
- Moreno-Herrero, F.; Herrero, P.; Colchero, J.; Baró, A.M.; Moreno, F. Imaging and mapping protein-binding sites on DNA regulatory regions with Atomic Force Microscopy. Biochem. Biophys. Res. Commun. 2001, 280, 151–157. [Google Scholar] [CrossRef]
- Kasas, S.; Thomson, N.H.; Smith, B.L.; Hansma, H.G.; Zhu, X.; Guthold, M.; Bustamante, C.; Kool, E.T.; Kashlev, M.; Hansma, P.K. Escherichia coli RNA polymerase activity observed using Atomic Force Microscopy. Biochemistry 1997, 36, 461–468. [Google Scholar] [CrossRef]
- Hansma, H.G.; Laney, D.E. DNA binding to mica correlates with cationic radius: Assay by Atomic Force Microscopy. Biophys. J. 1996, 70, 1933–1939. [Google Scholar] [CrossRef]
- Zheng, J.; Li, Z.; Wu, A.; Zhou, H. AFM studies of DNA structures on mica in the presence of alkaline earth metal ions. Biophys. Chem. 2003, 104, 37–43. [Google Scholar] [CrossRef]
- Thomson, N.H.; Kasas, S.; Smith, B.; Hansma, H.G.; Hansma, P.K. Reversible binding of DNA to mica for AFM imaging. Langmuir 1996, 12, 5905–5906. [Google Scholar] [CrossRef]
- Kan, Y.; Tan, Q.; Wu, G.; Si, W.; Chen, Y. Study of DNA adsorption on mica surfaces using a surface force apparatus. Sci. Rep. 2015, 5, 8442. [Google Scholar] [CrossRef]
- Pastré, D.; Hamon, L.; Landousy, F.; Sorel, I.; David, M.O.; Zozime, A.; Le Cam, E.; Piétrement, O. Anionic polyelectrolyte adsorption on mica mediated by multivalent cations: A solution to DNA imaging by Atomic Force Microscopy under high ionic strengths. Langmuir 2006, 22, 6651–6660. [Google Scholar] [CrossRef] [PubMed]
- Lyubchenko, Y.; Shlyakhtenko, L.; Harrington, R.; Oden, P.; Lindsay, S. Atomic Force Microscopy of long DNA: Imaging in air and under water. Proc. Natl. Acad. Sci. USA 1993, 90, 2137–2140. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Pérez, R. Dynamic Atomic Force Microscopy methods. Surf. Sci. Rep. 2002, 47, 197–301. [Google Scholar] [CrossRef]
- Adams, J.D.; Erickson, B.W.; Grossenbacher, J.; Brugger, J.; Nievergelt, A.; Fantner, G.E. Harnessing the damping properties of materials for high-speed Atomic Force Microscopy. Nat. Nanotechnol. 2016, 11, 147–151. [Google Scholar] [CrossRef]
- Schäffer, T.E.; Cleveland, J.P.; Ohnesorge, F.; Walters, D.A.; Hansma, P.K. Studies of vibrating atomic force microscope cantilevers in liquid. J. Appl. Phys. 1998, 80, 3622. [Google Scholar] [CrossRef]
- Pittenger, B.; Erina, N.; Su, C. Mechanical property mapping at the nanoscale using PeakForce QNM scanning probe technique. Solid Mech. Appl. 2014, 203, 31–51. [Google Scholar] [CrossRef]
- Schillers, H.; Medalsy, I.; Hu, S.; Slade, A.L.; Shaw, J.E. PeakForce Tapping resolves individual microvilli on living cells. J. Mol. Recognit. 2016, 29, 95–101. [Google Scholar] [CrossRef]
- Main, K.H.S.; Provan, J.I.; Haynes, P.J.; Wells, G.; Hartley, J.A.; Pyne, A.L.B. Atomic Force Microscopy—A tool for structural and translational DNA research. APL Bioeng. 2021, 5, 031504. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Martínez-Martín, D.; Medalsy, I.; Alsteens, D.; Müller, D.J. Multiparametric imaging of biological systems by force-distance curve-based AFM. Nat. Methods 2013, 10, 847–854. [Google Scholar] [CrossRef]
- Albrecht, T.R.; Grütter, P.; Horne, D.; Rugar, D. Frequency modulation detection using high-Q cantilevers for enhanced force microscope sensitivity. J. Appl. Phys. 1991, 69, 668–673. [Google Scholar] [CrossRef]
- Giessibl, F.J. Advances in Atomic Force Microscopy. Rev. Mod. Phys. 2003, 75, 949. [Google Scholar] [CrossRef]
- Morita, S.; Gießibl, F.; Meyer, E.; Wiesendanger, R. Noncontact Atomic Force Microscopy; Springer International Publishing: Berlin, Germany, 2015; Volume 3, ISBN 978-3-319-15588-3. [Google Scholar]
- Fukuma, T.; Ueda, Y.; Yoshioka, S.; Asakawa, H. Atomic-Scale distribution of water molecules at the mica-Water interface visualized by three-Dimensional scanning force microscopy. Phys. Rev. Lett. 2010, 104, 016101. [Google Scholar] [CrossRef]
- Fukuma, T. Water distribution at solid/liquid interfaces visualized by frequency modulation Atomic Force Microscopy. Sci. Technol. Adv. Mater. 2010, 11, 033003. [Google Scholar] [CrossRef]
- Fukuma, T.; Garcia, R. Atomic- and Molecular-Resolution Mapping of Solid-Liquid Interfaces by 3D Atomic Force Microscopy. ACS Nano 2018, 12, 11785–11797. [Google Scholar] [CrossRef]
- Martin-Jimenez, D.; Chacon, E.; Tarazona, P.; Garcia, R. Atomically resolved three-dimensional structures of electrolyte aqueous solutions near a solid surface. Nat. Commun. 2016, 7, 12164. [Google Scholar] [CrossRef]
- Kocun, M.; Labuda, A.; Meinhold, W.; Revenko, I.; Proksch, R. Fast, High Resolution, and Wide Modulus Range Nanomechanical Mapping with Bimodal Tapping Mode. ACS Nano 2017, 11, 10097–10105. [Google Scholar] [CrossRef]
- Radmacher, M.; Fritz, M.; Hansma, H.G.; Hansma, P.K. Direct observation of enzyme activity with the atomic force microscope. Science 1994, 265, 1577–1579. [Google Scholar] [CrossRef]
- Argaman, M.; Golan, R.; Thomson, N.H.; Hansma, H.G. Phase imaging of moving DNA molecules and DNA molecules replicated in the atomic force microscope. Nucleic Acids Res. 1997, 25, 4379–4384. [Google Scholar] [CrossRef][Green Version]
- Dunlap, D.D.; Maggi, A.; Soria, M.R.; Monaco, L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997, 25, 3095–3101. [Google Scholar] [CrossRef]
- Ono, M.Y.; Spain, E.M. Dynamics of DNA condensates at the solid-liquid interface by Atomic Force Microscopy. J. Am. Chem. Soc. 1999, 121, 7330–7334. [Google Scholar] [CrossRef]
- Hansma, H.G.; Laney, D.E.; Bezanilla, M.; Sinsheimer, R.L.; Hansma, P.K. Applications for Atomic Force Microscopy of DNA. Biophys. J. 1995, 68, 1672–1677. [Google Scholar] [CrossRef]
- Ohnesorge, F.; Binnig, G. True atomic resolution by Atomic Force Microscopy through repulsive and attractive forces. Science 1993, 260, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.W.; Hwang, I.S.; Chen, Y.F.; Chang, C.S.; Tsai, D.P. Imaging of soft matter with tapping-mode Atomic Force Microscopy and non-contact-mode Atomic Force Microscopy. Nanotechnology 2007, 18, 084009. [Google Scholar] [CrossRef]
- Ares, P.; Fuentes-Perez, M.E.; Herrero-Galán, E.; Valpuesta, J.M.; Gil, A.; Gomez-Herrero, J.; Moreno-Herrero, F. High resolution Atomic Force Microscopy of double-stranded RNA. Nanoscale 2016, 8, 11818–11826. [Google Scholar] [CrossRef]
- Ido, S.; Kimura, K.; Oyabu, N.; Kobayashi, K.; Tsukada, M.; Matsushige, K.; Yamada, H. Beyond the helix pitch: Direct visualization of native DNA in aqueous solution. ACS Nano 2013, 7, 1817–1822. [Google Scholar] [CrossRef]
- Kominami, H.; Kobayashi, K.; Yamada, H. Molecular-scale visualization and surface charge density measurement of Z-DNA in aqueous solution. Sci. Rep. 2019, 9, 6851. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Zhuo, R. A ‘tile’ tale: Hierarchical self-assembly of DNA lattices. Appl. Mater. Today 2016, 2, 7–16. [Google Scholar] [CrossRef]
- Cerreta, A.; Vobornik, D.; Dietler, G. Fine DNA structure revealed by constant height frequency modulation AFM imaging. Eur. Polym. J. 2013, 49, 1916–1922. [Google Scholar] [CrossRef]
- Pyne, A.; Thompson, R.; Leung, C.; Roy, D.; Hoogenboom, B.W. Single-molecule reconstruction of oligonucleotide secondary structure by Atomic Force Microscopy. Small 2014, 10, 3257–3261. [Google Scholar] [CrossRef]
- Pyne, A.L.B.; Noy, A.; Main, K.H.S.; Velasco-Berrelleza, V.; Piperakis, M.M.; Mitchenall, L.A.; Cugliandolo, F.M.; Beton, J.G.; Stevenson, C.E.M.; Hoogenboom, B.W.; et al. Base-pair resolution analysis of the effect of supercoiling on DNA flexibility and major groove recognition by triplex-forming oligonucleotides. Nat. Commun. 2021, 12, 1053. [Google Scholar] [CrossRef]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef]
- Umeda, K.; Kobayashi, K.; Oyabu, N.; Matsushige, K.; Yamada, H. Molecular-scale quantitative charge density measurement of biological molecule by frequency modulation Atomic Force Microscopy in aqueous solutions. Nanotechnology 2015, 26, 285103. [Google Scholar] [CrossRef]
- Rhodes, D.; Klug, A. Helical periodicity of DNA determined by enzyme digestion. Nature 1980, 286, 573–578. [Google Scholar] [CrossRef]
- Bose, K.; Lech, C.J.; Heddi, B.; Phan, A.T. High-resolution AFM structure of DNA G-wires in aqueous solution. Nat. Commun. 2018, 9, 1959. [Google Scholar] [CrossRef]
- Calzolari, A.; Di Felice, R.; Molinari, E.; Garbesi, A. G-quartet biomolecular nanowires. Appl. Phys. Lett. 2002, 80, 3331–3333. [Google Scholar] [CrossRef]
- Sader, J.E.; Jarvis, S.P. Accurate formulas for interaction force and energy in frequency modulation force spectroscopy. Appl. Phys. Lett. 2004, 84, 1801–1803. [Google Scholar] [CrossRef]
- Baumann, C.G.; Smith, S.B.; Bloomfield, V.A.; Bustamante, C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA 1997, 94, 6185–6190. [Google Scholar] [CrossRef]
- Duboué-Dijon, E.; Fogarty, A.C.; Hynes, J.T.; Laage, D. Dynamical Disorder in the DNA Hydration Shell. J. Am. Chem. Soc. 2016, 138, 7610–7620. [Google Scholar] [CrossRef]
- Saenger, W.; Hunter, W.N.; Kennard, O. DNA conformation is determined by economics in the hydration of phosphate groups. Nature 1986, 324, 385–388. [Google Scholar] [CrossRef]
- Berman, H.M. Hydration of DNA. Curr. Opin. Struct. Biol. 1991, 1, 423–427. [Google Scholar] [CrossRef]
- Feig, M.; Montgomery Pettitt, B. A molecular simulation picture of DNA hydration around A- And B-DNA. Biopolymers 1998, 48, 199–209. [Google Scholar] [CrossRef]
- Mertens, J.; Rogero, C.; Calleja, M.; Ramos, D.; Martín-Gago, J.A.; Briones, C.; Tamayo, J. Label-free detection of DNA hybridization based on hydration-induced tension in nucleic acid films. Nat. Nanotechnol. 2008, 3, 301–307. [Google Scholar] [CrossRef]
- Denisov, V.P.; Carlström, G.; Venu, K.; Halle, B. Kinetics of DNA hydration. J. Mol. Biol. 1997, 268, 118–136. [Google Scholar] [CrossRef]
- Privé, G.G.; Heinemann, U.; Chandrasegaran, S.; Kan, L.S.; Kopka, M.L.; Dickerson, R.E. Helix geometry, hydration, and G·A mismatch in a B-DNA decamer. Science 1987, 238, 498–504. [Google Scholar] [CrossRef]
- Laage, D.; Elsaesser, T.; Hynes, J.T. Water Dynamics in the Hydration Shells of Biomolecules. Chem. Rev. 2017, 117, 10694–10725. [Google Scholar] [CrossRef]
- Santos, S.; Stefancich, M.; Hernandez, H.; Chiesa, M.; Thomson, N.H. Hydrophilicity of a single DNA molecule. J. Phys. Chem. C 2012, 116, 2807–2818. [Google Scholar] [CrossRef]
- Bonhommeau, S.; Lecomte, S. Tip-Enhanced Raman Spectroscopy: A Tool for Nanoscale Chemical and Structural Characterization of Biomolecules. ChemPhysChem 2018, 19, 8–18. [Google Scholar] [CrossRef]
- Sofińska, K.; Wilkosz, N.; Szymoński, M.; Lipiec, E. Molecular spectroscopic markers of DNA damage. Molecules 2020, 25, 561. [Google Scholar] [CrossRef]
- Chen, C.; Liu, W.; Tian, S.; Hong, T. Novel Surface-Enhanced Raman Spectroscopy techniques for DNA, protein and drug detection. Sensors 2019, 19, 1712. [Google Scholar] [CrossRef]
- Long, D.A. The Raman Effect: A Unified Treatment of the Theory of Raman Scattering by Molecules; John Wiley & Sons, Ltd.: Chichester, UK, 2002; Volume 8, ISBN 0471490288. [Google Scholar]
- Panikkanvalappil, S.R.; Mahmoud, M.A.; MacKey, M.A.; El-Sayed, M.A. Surface-Enhanced Raman Spectroscopy for real-time monitoring of reactive oxygen species-induced DNA damage and its prevention by platinum nanoparticles. ACS Nano 2013, 7, 7524–7533. [Google Scholar] [CrossRef]
- Yue, J.; Shen, Y.; Liang, L.; Guan, X.; Zhang, X.; Xu, S.; Liang, C.; Shi, W.; Xu, W. Tracing the molecular dynamics of living mitochondria under phototherapy via surface-enhanced Raman scattering spectroscopy. Analyst 2019, 144, 5521–5527. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, A.; Zhang, D.; Tam, F.; Halas, N.J. Surface-Enhanced Raman Spectroscopy of DNA. J. Am. Chem. Soc. 2008, 130, 5523–5529. [Google Scholar] [CrossRef] [PubMed]
- Crul, M.; Schellens, J.H.M.; Beijnen, J.H.; Maliepaard, M. Cisplatin resistance and DNA repair. Cancer Treat. Rev. 1997, 23, 341–366. [Google Scholar] [CrossRef]
- Jordan, P.; Carmo-Fonseca, M. Molecular mechanisms involved in cisplatin cytotoxicity. Cell. Mol. Life Sci. 2000, 57, 1229–1235. [Google Scholar] [CrossRef]
- Corda, Y.; Job, C.; Anin, M.F.; Leng, M.; Job, D. Transcription by Eucaryotic and Procaryotic RNA Polymerases of DNA Modified at a d(GG) or a d(AG) Site by the Antitumor Drug cis-Diamminedichloroplatinum(II). Biochemistry 1991, 30, 222–230. [Google Scholar] [CrossRef]
- Corda, Y.; Job, D.; Anin, M.F.; Leng, M. RNA Polymerases React Differently at D(Apg) and D(Gpg) Adducts in DNA Modified by Cis-Diamminedichloroplatinum(II). Biochemistry 1992, 31, 1904–1908. [Google Scholar] [CrossRef]
- Mymryk, J.S.; Zaniewski, E.; Archer, T.K. Cisplatin inhibits chromatin remodeling, transcription factor binding, and transcription from the mouse mammary tumor virus promoter in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 2076–2080. [Google Scholar] [CrossRef]
- Masetti, M.; Xie, H.N.; Krpetić, Ž.; Recanatini, M.; Alvarez-Puebla, R.A.; Guerrini, L. Revealing DNA interactions with exogenous agents by surface-enhanced raman scattering. J. Am. Chem. Soc. 2015, 137, 469–476. [Google Scholar] [CrossRef]
- Morjani, H.; Sharonov, S.; Manfait, M.; Sokolov, K.; Nabiev, I. SERS and micro-sers analysis of doxorubicin interaction in vitro and in living human cancer cells. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Paris, France, 29 October–1 November 1992; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 1992; Volume 1, pp. 330–331. [Google Scholar]
- Beljebbar, A.; Sockalingum, G.D.; Angiboust, J.F.; Manfait, M. Comparative FT SERS, resonance Raman and SERRS studies of doxorubicin and its complex with DNA. Spectrochim. Acta Part A Mol. Spectrosc. 1995, 51, 2083–2090. [Google Scholar] [CrossRef]
- Spadavecchia, J.; Perumal, R.; Casale, S.; Krafft, J.M.; Methivier, C.; Pradier, C.M. Polyethylene glycol gold-nanoparticles: Facile nanostructuration of doxorubicin and its complex with DNA molecules for SERS detection. Chem. Phys. Lett. 2016, 648, 182–188. [Google Scholar] [CrossRef]
- Kang, B.; Afifi, M.M.; Austin, L.A.; El-Sayed, M.A. Exploiting the nanoparticle plasmon effect: Observing drug delivery dynamics in single cells via Raman/fluorescence imaging spectroscopy. ACS Nano 2013, 7, 7420–7427. [Google Scholar] [CrossRef]
- Lin, D.; Wu, Q.; Qiu, S.; Chen, G.; Feng, S.; Chen, R.; Zeng, H. Label-free liquid biopsy based on blood circulating DNA detection using SERS-based nanotechnology for nasopharyngeal cancer screening. Nanomed. Nanotechnol. Biol. Med. 2019, 22, 102100. [Google Scholar] [CrossRef]
- Zeng, Y.; Ren, J.Q.; Shen, A.G.; Hu, J.M. Splicing Nanoparticles-Based “click” SERS Could Aid Multiplex Liquid Biopsy and Accurate Cellular Imaging. J. Am. Chem. Soc. 2018, 140, 10649–10652. [Google Scholar] [CrossRef]
- Pashaee, F.; Tabatabaei, M.; Caetano, F.A.; Ferguson, S.S.G.; Lagugné-Labarthet, F. Tip-Enhanced Raman Spectroscopy: Plasmid-free: Vs. plasmid-embedded DNA. Analyst 2016, 141, 3251–3258. [Google Scholar] [CrossRef]
- Domke, K.F.; Zhang, D.; Pettinger, B. Enhanced Raman spectroscopy: Single molecules or carbon? J. Phys. Chem. C 2007, 111, 8611–8616. [Google Scholar] [CrossRef]
- Lipiec, E.; Sekine, R.; Bielecki, J.; Kwiatek, W.M.; Wood, B.R. Molecular characterization of DNA double strand breaks with tip-enhanced Raman scattering. Angew. Chem. Int. Ed. 2014, 53, 169–172. [Google Scholar] [CrossRef]
- Najjar, S.; Talaga, D.; Schué, L.; Coffinier, Y.; Szunerits, S.; Boukherroub, R.; Servant, L.; Rodriguez, V.; Bonhommeau, S. Tip-Enhanced Raman Spectroscopy of combed double-stranded DNA bundles. J. Phys. Chem. C 2014, 118, 1174–1181. [Google Scholar] [CrossRef]
- He, Z.; Han, Z.; Kizer, M.; Linhardt, R.J.; Wang, X.; Sinyukov, A.M.; Wang, J.; Deckert, V.; Sokolov, A.V.; Hu, J.; et al. Tip-Enhanced Raman Imaging of Single-Stranded DNA with Single Base Resolution. J. Am. Chem. Soc. 2019, 141, 753–757. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, X.; Wang, H.; Zhang, Y.; Jiang, S.; Hu, C.; Zhang, Y.; Luo, Y.; Dong, Z. Distinguishing Individual DNA Bases in a Network by Non-Resonant Tip-Enhanced Raman Scattering. Angew. Chem.—Int. Ed. 2017, 56, 5561–5564. [Google Scholar] [CrossRef]
- He, Z.; Qiu, W.; Kizer, M.E.; Wang, J.; Chen, W.; Sokolov, A.V.; Wang, X.; Hu, J.; Scully, M.O. Resolving the Sequence of RNA Strands by Tip-Enhanced Raman Spectroscopy. ACS Photonics 2021, 8, 424–430. [Google Scholar] [CrossRef]
- Bailo, E.; Deckert, V. Tip-Enhanced Raman Spectroscopy of single RNA strands: Towards a novel direct-sequencing method. Angew. Chem. Int. Ed. 2008, 47, 1658–1661. [Google Scholar] [CrossRef]
- Treffer, R.; Lin, X.; Bailo, E.; Deckert-Gaudig, T.; Deckert, V. Distinction of nucleobases—A tip-enhanced Raman approach. Beilstein J. Nanotechnol. 2011, 2, 628–637. [Google Scholar] [CrossRef]
- Lipiec, E.; Japaridze, A.; Szczerbiński, J.; Dietler, G.; Zenobi, R. Preparation of Well-Defined DNA Samples for Reproducible Nanospectroscopic Measurements. Small 2016, 12, 4821–4829. [Google Scholar] [CrossRef]
- Lipiec, E.; Kaderli, J.; Kobierski, J.; Riek, R.; Skirlińska-Nosek, K.; Sofińska, K.; Szymoński, M.; Zenobi, R. Nanoscale Hyperspectral Imaging of Amyloid Secondary Structures in Liquid. Angew. Chem. 2021, 133, 4595–4600. [Google Scholar] [CrossRef]
- Richard-Lacroix, M.; Deckert, V. Direct molecular-level near-field plasmon and temperature assessment in a single plasmonic hotspot. Light Sci. Appl. 2020, 9, 35. [Google Scholar] [CrossRef]
- Zeng, Z.C.; Wang, H.; Johns, P.; Hartland, G.V.; Schultz, Z.D. Photothermal Microscopy of Coupled Nanostructures and the Impact of Nanoscale Heating in Surface-Enhanced Raman Spectroscopy. J. Phys. Chem. C 2017, 121, 11623–11631. [Google Scholar] [CrossRef]
- Caldarola, M.; Albella, P.; Cortés, E.; Rahmani, M.; Roschuk, T.; Grinblat, G.; Oulton, R.F.; Bragas, A.V.; Maier, S.A. Non-plasmonic nanoantennas for surface enhanced spectroscopies with ultra-low heat conversion. Nat. Commun. 2015, 6, 7915. [Google Scholar] [CrossRef]
- Lindquist, N.C.; de Albuquerque, C.D.L.; Sobral-Filho, R.G.; Paci, I.; Brolo, A.G. High-speed imaging of surface-enhanced Raman scattering fluctuations from individual nanoparticles. Nat. Nanotechnol. 2019, 14, 981–987. [Google Scholar] [CrossRef]
- Brongersma, M.L.; Halas, N.J.; Nordlander, P. Plasmon-induced hot carrier science and technology. Nat. Nanotechnol. 2015, 10, 25–34. [Google Scholar] [CrossRef]
- Clavero, C. Plasmon-induced hot-electron generation at nanoparticle/metal-oxide interfaces for photovoltaic and photocatalytic devices. Nat. Photonics 2014, 8, 95–103. [Google Scholar] [CrossRef]
- Szczerbiński, J.; Metternich, J.B.; Goubert, G.; Zenobi, R. How Peptides Dissociate in Plasmonic Hot Spots. Small 2020, 16, 1905197. [Google Scholar] [CrossRef] [PubMed]
- Szczerbiński, J.; Gyr, L.; Kaeslin, J.; Zenobi, R. Plasmon-Driven Photocatalysis Leads to Products Known from E-beam and X-ray-Induced Surface Chemistry. Nano Lett. 2018, 18, 6740–6749. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, J.; Rigor, J.; Large, N.; El-Khoury, P.Z.; Rogachev, A.Y.; Kurouski, D. Direct Experimental Evidence of Hot Carrier-Driven Chemical Processes in Tip-Enhanced Raman Spectroscopy (TERS). J. Phys. Chem. C 2020, 124, 2238–2244. [Google Scholar] [CrossRef]
- Linic, S.; Aslam, U.; Boerigter, C.; Morabito, M. Photochemical transformations on plasmonic metal nanoparticles. Nat. Mater. 2015, 14, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.; Wang, X.; Zhao, Q.Q.; Zhu, J.F.; Li, C.W.; He, Y.H.; Hu, S.; Sartin, M.M.; Yan, S.; Ren, B. Probing nanoscale spatial distribution of plasmonically excited hot carriers. Nat. Commun. 2020, 11, 4211. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lipiec, E.; Sofińska, K.; Seweryn, S.; Wilkosz, N.; Szymonski, M. Revealing DNA Structure at Liquid/Solid Interfaces by AFM-Based High-Resolution Imaging and Molecular Spectroscopy. Molecules 2021, 26, 6476. https://doi.org/10.3390/molecules26216476
Lipiec E, Sofińska K, Seweryn S, Wilkosz N, Szymonski M. Revealing DNA Structure at Liquid/Solid Interfaces by AFM-Based High-Resolution Imaging and Molecular Spectroscopy. Molecules. 2021; 26(21):6476. https://doi.org/10.3390/molecules26216476
Chicago/Turabian StyleLipiec, Ewelina, Kamila Sofińska, Sara Seweryn, Natalia Wilkosz, and Marek Szymonski. 2021. "Revealing DNA Structure at Liquid/Solid Interfaces by AFM-Based High-Resolution Imaging and Molecular Spectroscopy" Molecules 26, no. 21: 6476. https://doi.org/10.3390/molecules26216476
APA StyleLipiec, E., Sofińska, K., Seweryn, S., Wilkosz, N., & Szymonski, M. (2021). Revealing DNA Structure at Liquid/Solid Interfaces by AFM-Based High-Resolution Imaging and Molecular Spectroscopy. Molecules, 26(21), 6476. https://doi.org/10.3390/molecules26216476





