A Molecular Probe with Both Chromogenic and Fluorescent Units for Detecting Serine Proteases
Abstract
1. Introduction
2. Results and Discussion
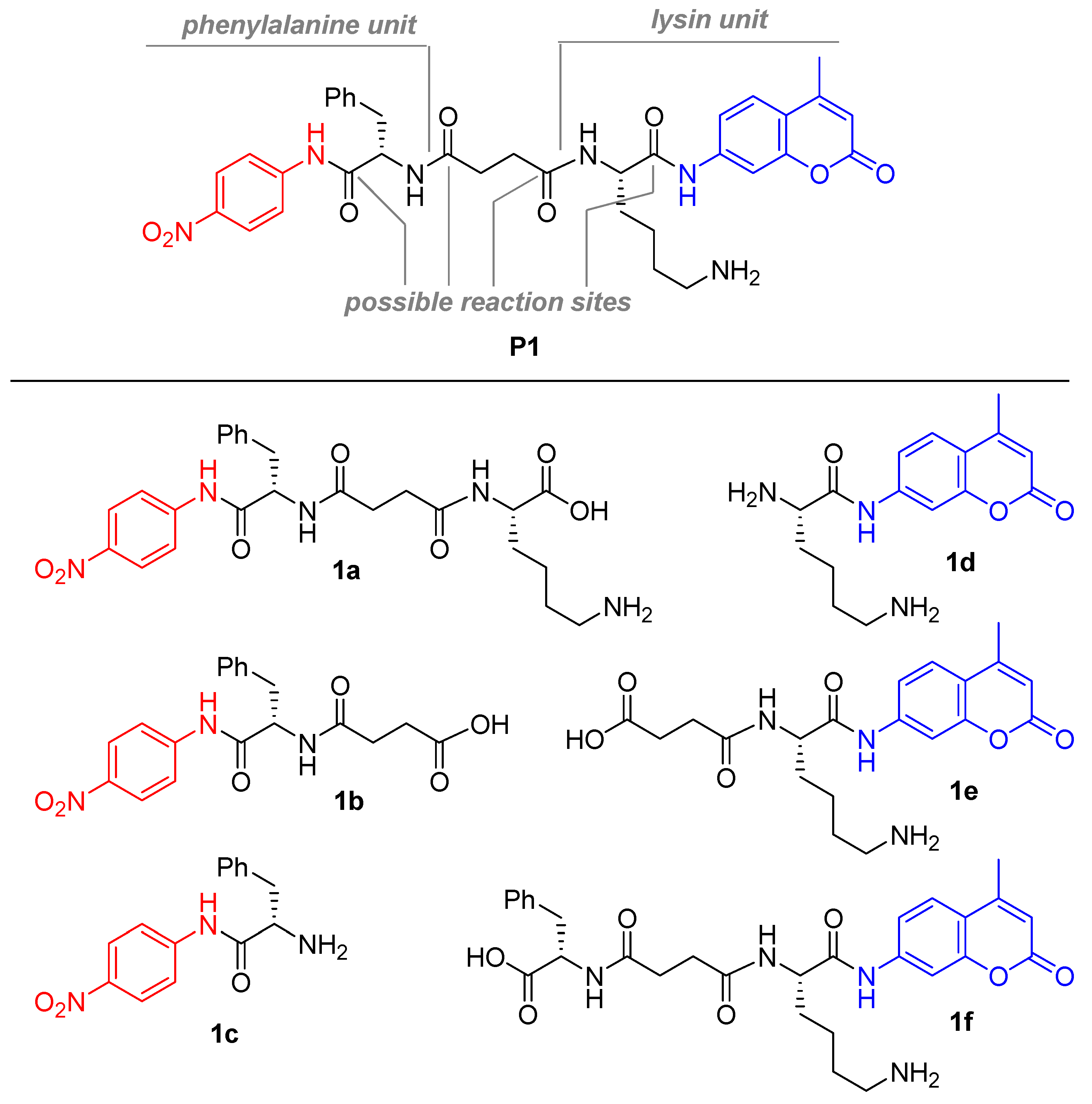
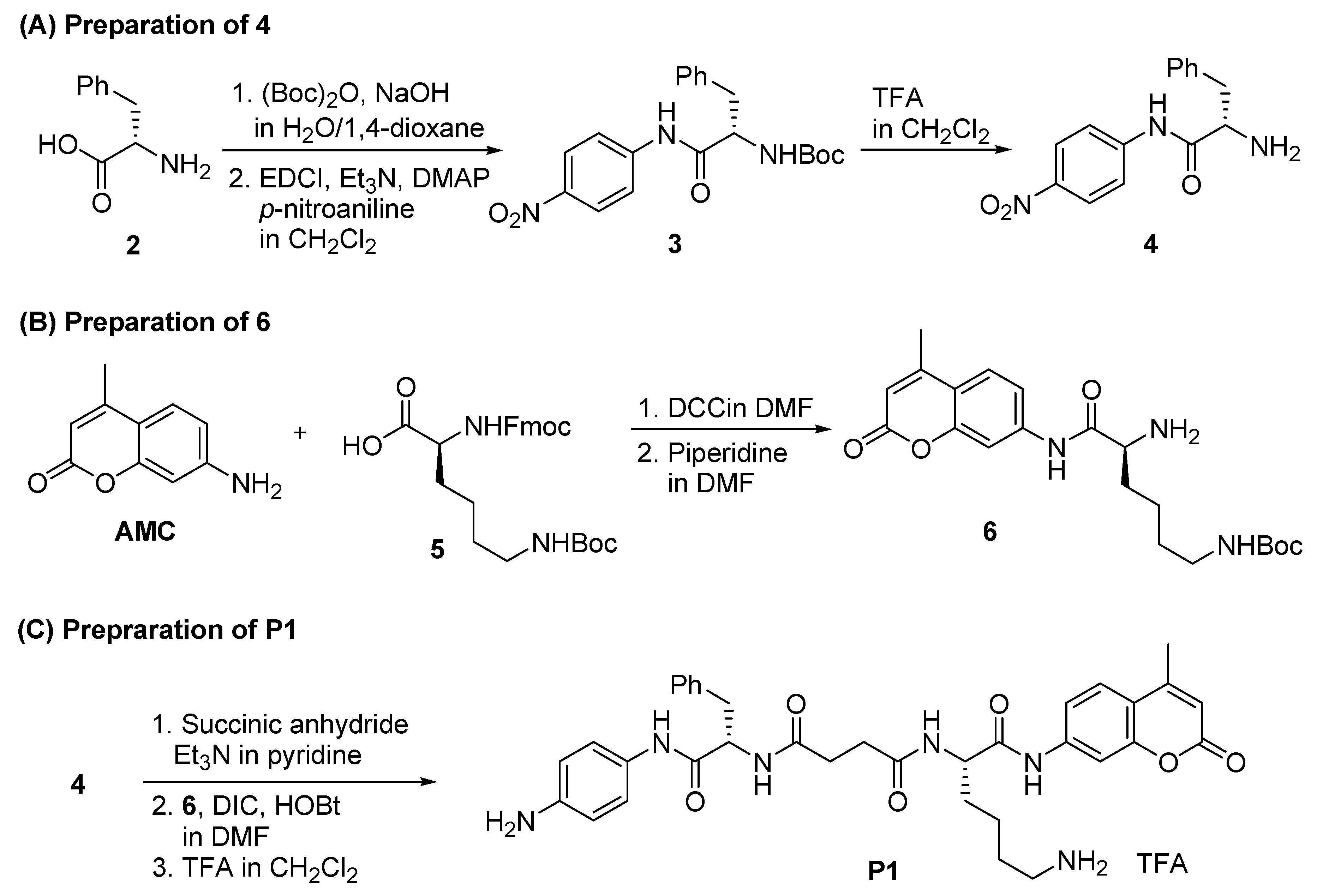
2.1. Synthesis of P1
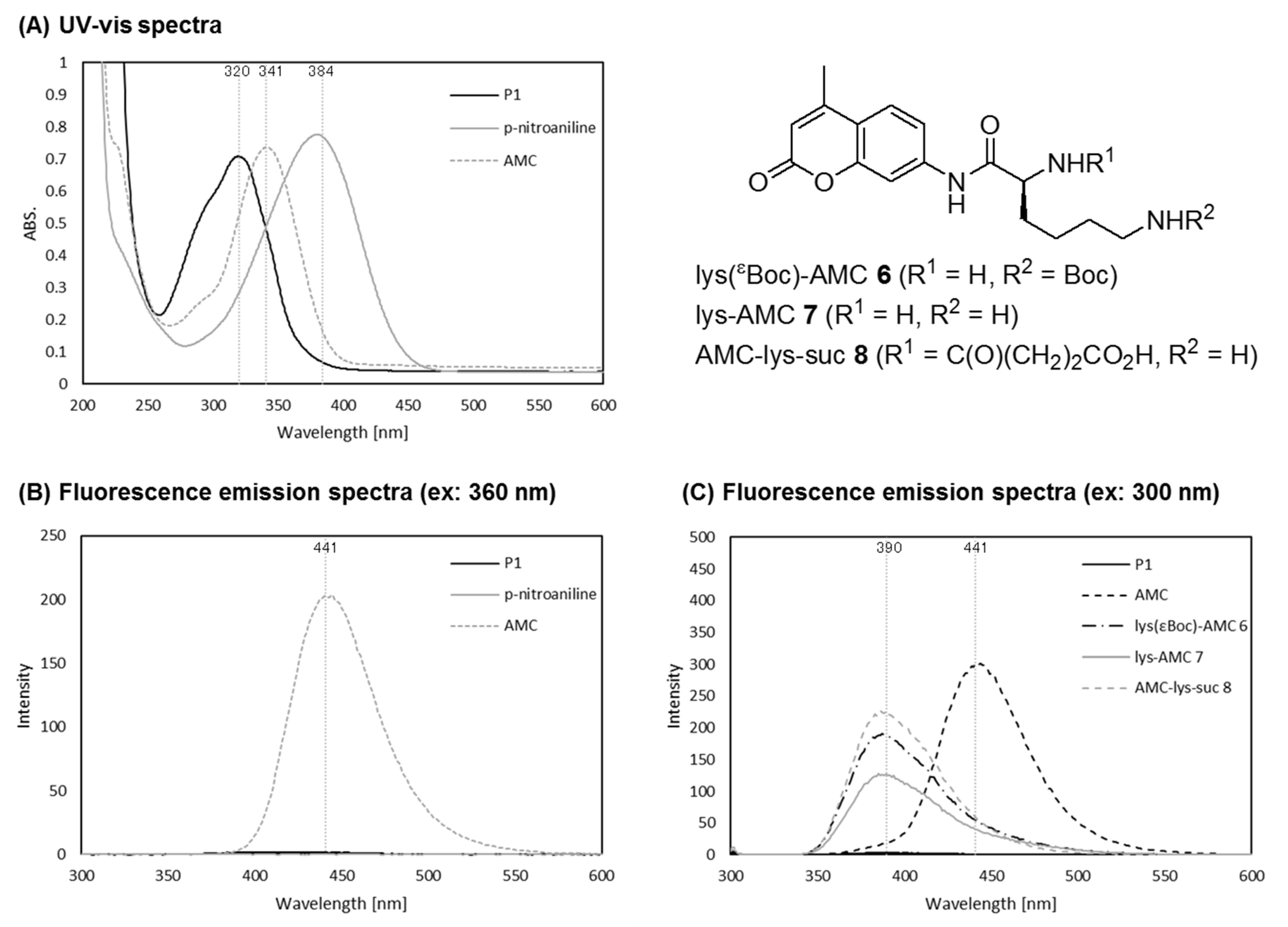
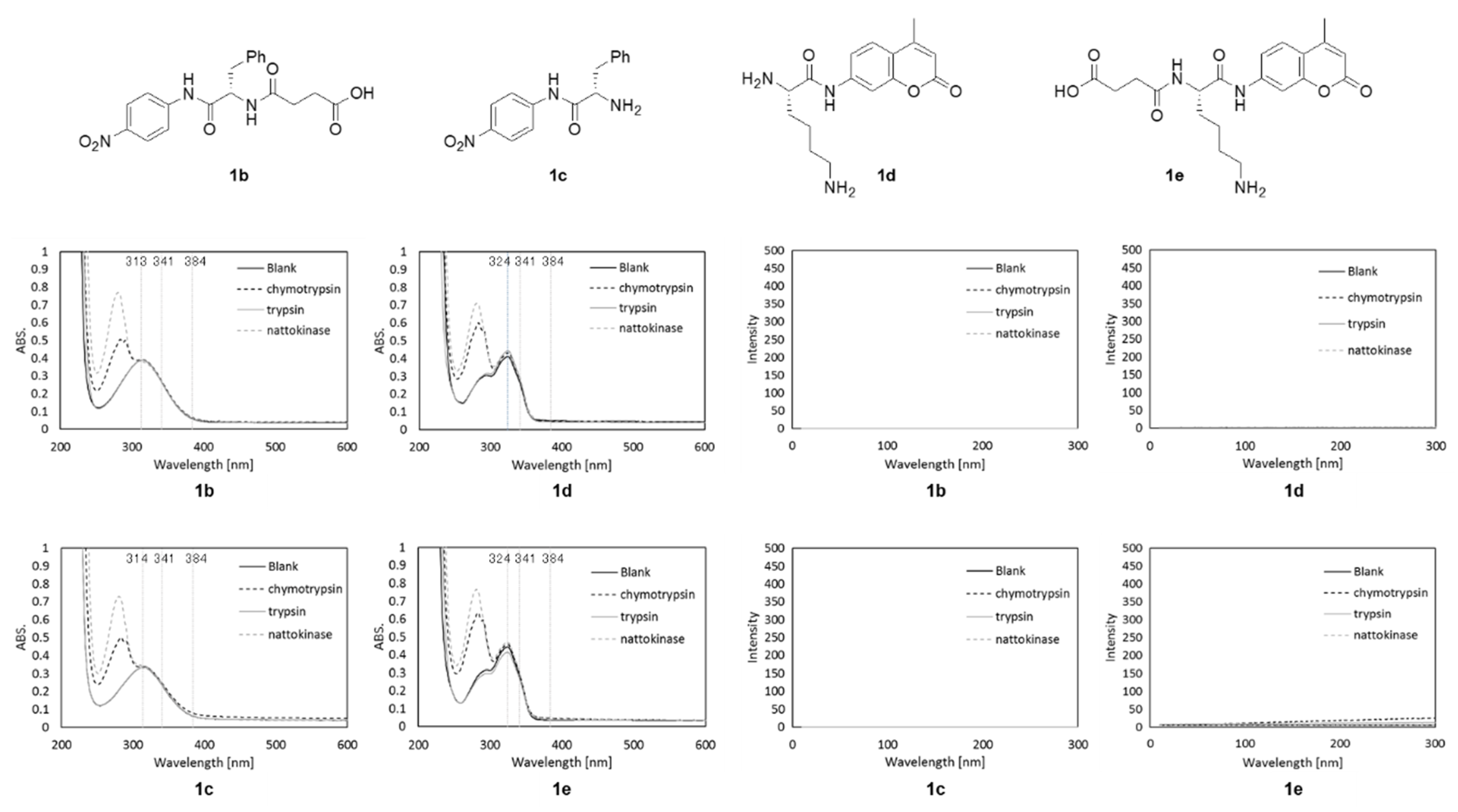
2.2. UV and Fluorescence Spectra of P1
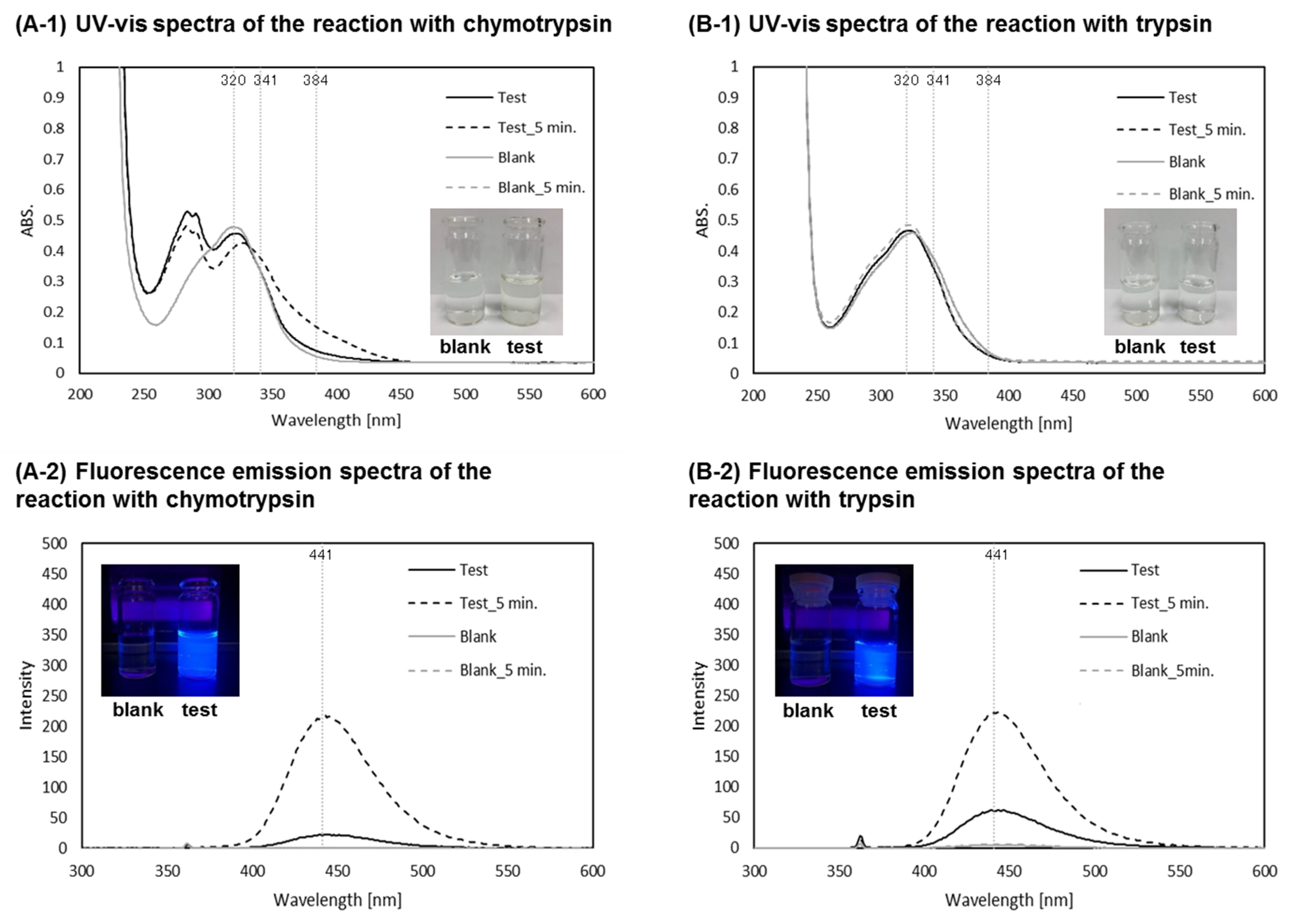
2.3. Reaction of P1 with Proteases
3. Materials and Methods
3.1. General Methods
3.2. Synthesis of Substrates
3.2.1. Synthesis of N-(Boc)-l-phenylalanine p-nitroanilide (3)
3.2.2. Synthesis of l-phenylalanine p-nitroanilide (4)
3.2.3. Synthesis of α-Fmoc-ε-Boc-l-lysin 4-methylcoumaryl-7-amide
3.2.4. Synthesis of ε-Boc-l-lysin 4-methylcoumaryl-7-amide (6)
3.2.5. Synthesis of N-Succinyl-l-phenylalanine p-nitroanilide
3.2.6. Synthesis of Boc-Protected Probe Boc-P1
3.2.7. Synthesis of P1
3.2.8. Synthesis of l-lysin 4-methylcoumaryl-7-amide (1d)
3.2.9. Synthesis of AMC-lys(Boc)-suc
3.2.10. Synthesis of AMC-lys-suc (Fragment Probe 1e)
3.3. Preparation of Solutions
3.3.1. pH 8.0 Tris-HCl Buffer Solution
3.3.2. pH 7.4 PBS Buffer Solution
3.3.3. pH 2.2 Glycine-HCl Buffer Solution
3.3.4. 0.59 mM P1 Solution
3.3.5. 0.59 mM p-Nitroaniline and AMC Solution
3.3.6. 0.59 mM Solution of Fragment Probe 1b–e
3.3.7. 36 µM P1 in PBS Buffer Solution
3.3.8. 36 µM Solution of Fragment Probe 1b and 1e in PBS
3.3.9. Solution of α-Chymotrypsin, Pepsin, Elastase, Proteinase K, and Papain in HCl
3.3.10. Solution of Trypsin in PBS Buffer
3.4. Reaction of Molecular Probes with Proteases
3.4.1. Reaction of P1 with α-Chymotrypsin
3.4.2. Reaction of P1 with Trypsin
3.4.3. Reaction of P1 with Proteases
Preparation of Test Solutions
Determination of p-Nitroaniline Concentration after 5 min
Time Course of p-Nitroaniline Concentration
Determination of AMC Concentration
Time Course of AMC Concentration
3.4.4. Reaction of P1 Fragment Probes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug. Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef]
- Vasiljeva, O.; Menendez, E.; Nguyen, M.; Craik, C.S.; Kavanaugh, M.W. Monitoring protease activity in biological tissues using antibody prodrugs as sensing probes. Sci. Rep. 2020, 10, 5894. [Google Scholar] [CrossRef] [PubMed]
- Koblinski, J.E.; Ahram, M.; Sloane, B.F. Unraveling the role of proteases in cancer. Clin. Chim. Acta 2000, 291, 113–135. [Google Scholar] [CrossRef]
- Hao Ong, I.L.; Yang, K.-L. Recent developments in protease activity assays and sensors. Analyst 2017, 142, 1867–1881. [Google Scholar] [CrossRef]
- Welser, K.; Adsley, R.; Moore, B.M.; Chan, W.C.; Aylott, J.W. Protease sensing with nanoparticle based platforms. Analyst 2011, 136, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Hosta-Rigau, L.; Chung, S.F.; Postma, A.; Chandrawati, R.; Städler, B.; Caruso, F. Capsosomes with “Free-Floating” Liposomal Subcompartments. Adv. Mater. 2011, 23, 4082–4087. [Google Scholar] [CrossRef] [PubMed]
- Sadat Ebrahimi, M.M.; Schönherr, H. Enzyme-Sensing Chitosan Hydrogels. Langmuir 2014, 30, 7842–7850. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Takemoto, Y.; Nsiama, T.K.; Kato, T.; Nishino, N.; Ito, A.; Yoshida, M. Design and synthesis of peptide-MCA substrates for a novel assay of histone methyltransferases and their inhibitors. Bioorg. Med. Chem. 2014, 22, 1268–1275. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Parta, A.; Mitra, R.K. Modulation of anionic reverse micellar interface with non-ionic surfactants can regulate enzyme activity within the micellar waterpool. Colloid Polym. Sci. 2016, 294, 715–726. [Google Scholar] [CrossRef]
- Lesner, A.; Brzozowski, K.; Kupryszewski, G.; Rolka, K. Design, Chemical Synthesis and Kinetic Studies of Trypsin Chromogenic Substrates Based on the Proteinase Binding Loop of Cucurbita maxima Trypsin Inhibitor (CMTI-III). Biochem. Biophys. Res. Commun. 2000, 269, 81–84. [Google Scholar] [CrossRef]
- Hong, R.; Emrick, T.; Rotello, V.M. Monolayer-Controlled Substrate Selectivity Using Noncovalent Enzyme−Nanoparticle Conjugates. J. Am. Chem. Soc. 2004, 126, 13572–13573. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, S.; Matsumoto, T.; Yamashita, S.; Wang, Y.; Lu, Z.-R. Conjugation of poorly absorptive drugs with mucoadhesive polymers for the improvement of oral absorption of drugs. J. Control. Release 2007, 123, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Tomar, R.; Kumar, R.; Jagannadham, M.V. A Stable Serine Protease, Wrightin, from the Latex of the Plant Wrightia tinctoria (Roxb.) R. Br.: Purification and Biochemical Properties. J. Agric. Food Chem. 2008, 56, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.T.; Lee, B.; Zhang, M.; Jun, S.-H.; Shim, J.; Lee, J.; Kim, J.; Gu, M.B. Immobilization and stabilization of subtilisin Carlsberg in magnetically-separable mesoporous silica for transesterification in an organic solvent. Green Chem. 2012, 14, 1884–1887. [Google Scholar] [CrossRef]
- Moyano, F.; Setien, E.; Silber, J.J.; Correa, N.M. Enzymatic Hydrolysis of N-Benzoyl-l-Tyrosine p-Nitroanilide by α-Chymotrypsin in DMSO-Water/AOT/n-Heptane Reverse Micelles. A Unique Interfacial Effect on the Enzymatic Activity. Langmuir 2013, 29, 8245–8254. [Google Scholar] [CrossRef]
- Ono, S.; Murai, J.; Nakai, T.; Kuroda, H.; Horino, Y.; Yoshimura, T.; Oyama, H.; Umezaki, M. Site-selective Chemical Modification of α-Chymotrypsin Using a Peptidyl Diphenyl 1-Amino-2-phenylethylphosphonate Derivative. Chem. Lett. 2013, 42, 860–862. [Google Scholar] [CrossRef]
- Semashko, T.A.; Vorotnikova, E.A.; Sharikova, V.F.; Vinokurov, K.S.; Smirnova, Y.A.; Dunaevsky, Y.E.; Belozersky, M.A.; Oppert, B.; Elpidina, E.N.; Filippova, I.Y. Selective chromogenic and fluorogenic peptide substrates for the assay of cysteine peptidases in complex mixtures. Anal. Biochem. 2014, 449, 179–187. [Google Scholar] [CrossRef]
- Sato, D.; Kato, T. Novel fluorescent substrates for detection of trypsin activity and inhibitor screening by self-quenching. Bioorg. Med. Chem. Lett. 2016, 26, 5736–5740. [Google Scholar] [CrossRef]
- Ast, S.; Rutledge, P.J.; Todd, M.H. Reversing the Triazole Topology in a Cyclam-Triazole-Dye Ligand Gives a 10-Fold Brighter Signal Response to Zn2+ in Aqueous Solution. Eur. J. Inorg. Chem. 2012, 2012, 5611–5615. [Google Scholar] [CrossRef]
- De Martins, R.S.; Pereira, M.P.; de Castro, P.P.; Bombonato, F.I. Design and preparation of a novel prolinamide-based organocatalyst for the solvent-free asymmetric aldol reaction. Tetrahedron 2020, 76, 130855. [Google Scholar] [CrossRef]
- Dong, J.; Wang, Y.; Xiang, Q.; Lv, X.; Weng, W.; Zeng, Q. Synthesis of Chiral Amino Acid Anilides by Ligand-Free Copper-Catalyzed Selective N-Arylation of Amino Acid Amides. Adv. Synth. Catal. 2013, 355, 692–696. [Google Scholar] [CrossRef]
- Madsen, A.S.; Olsen, C.A. Substrates for Efficient Fluorometric Screening Employing the NAD-Dependent Sirtuin 5 Lysine Deacylase (KDAC) Enzyme. J. Med. Chem. 2012, 55, 5582–5590. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Tsuda, Y.; Teno, N.; Nadamatsu, Y.; Okamoto, U. Amino Acids and Peptides. XXII. Synthesis of Substrates and Inhibitors of Human Leukocyte Cathepsin G. Chem. Pharm. Bull. 1988, 36, 4794–4801. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishida, K.; Nakamura, Y.; Ohta, T.; Oe, Y. A Molecular Probe with Both Chromogenic and Fluorescent Units for Detecting Serine Proteases. Molecules 2021, 26, 482. https://doi.org/10.3390/molecules26020482
Ishida K, Nakamura Y, Ohta T, Oe Y. A Molecular Probe with Both Chromogenic and Fluorescent Units for Detecting Serine Proteases. Molecules. 2021; 26(2):482. https://doi.org/10.3390/molecules26020482
Chicago/Turabian StyleIshida, Kirara, Yushi Nakamura, Tetsuo Ohta, and Yohei Oe. 2021. "A Molecular Probe with Both Chromogenic and Fluorescent Units for Detecting Serine Proteases" Molecules 26, no. 2: 482. https://doi.org/10.3390/molecules26020482
APA StyleIshida, K., Nakamura, Y., Ohta, T., & Oe, Y. (2021). A Molecular Probe with Both Chromogenic and Fluorescent Units for Detecting Serine Proteases. Molecules, 26(2), 482. https://doi.org/10.3390/molecules26020482




