Identification of the New In Vivo Metabolites of Ilaprazole in Rat Plasma after Oral Administration by LC-MS: In Silico Prediction of the H+/K+-ATPase Inhibitor
Abstract
1. Introduction
2. Results and Discussion
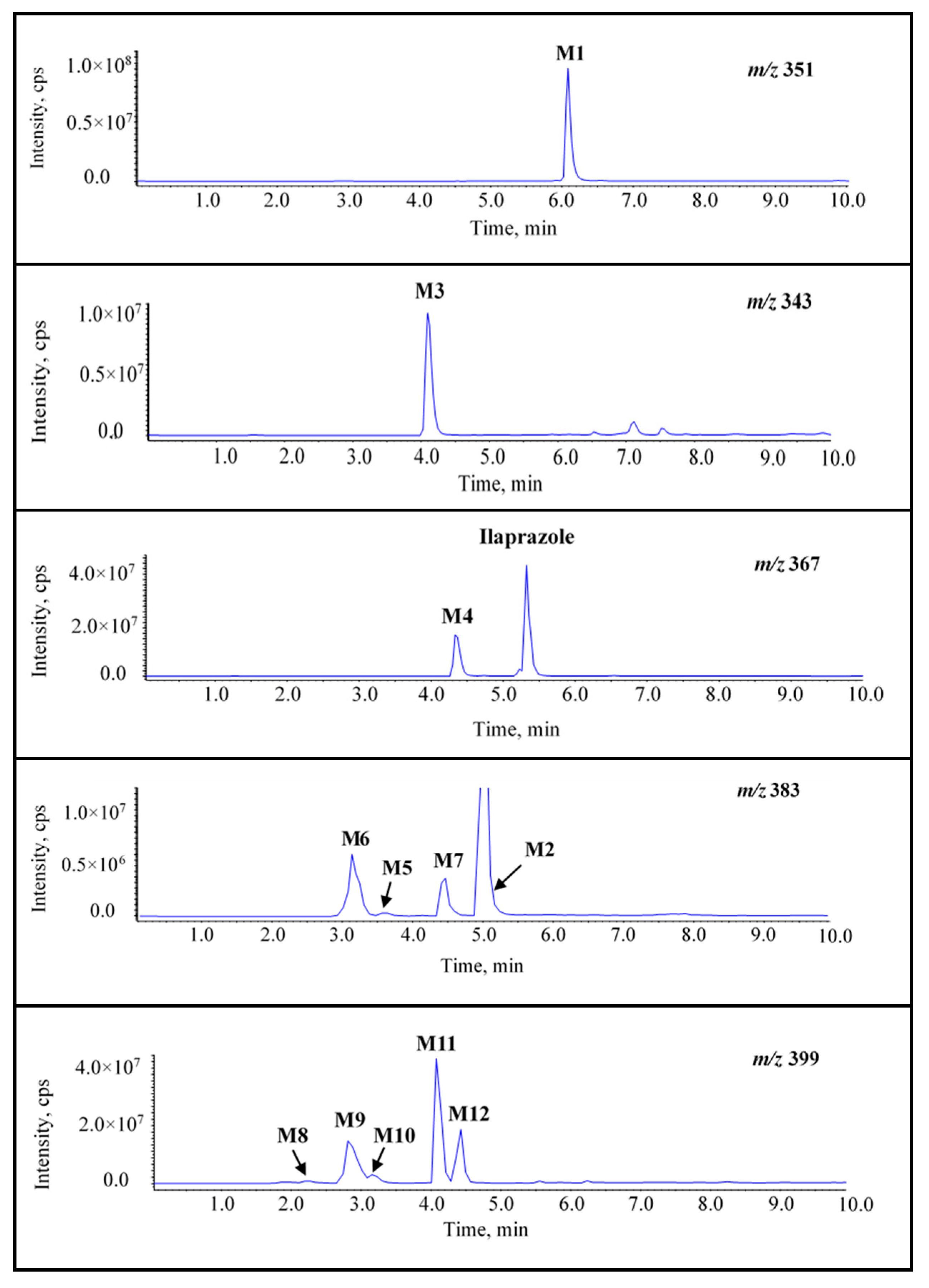
2.1. Detection of Ilaprazole and Its Metabolites
2.2. Characteristics of Ilaprazole, M1, and M2
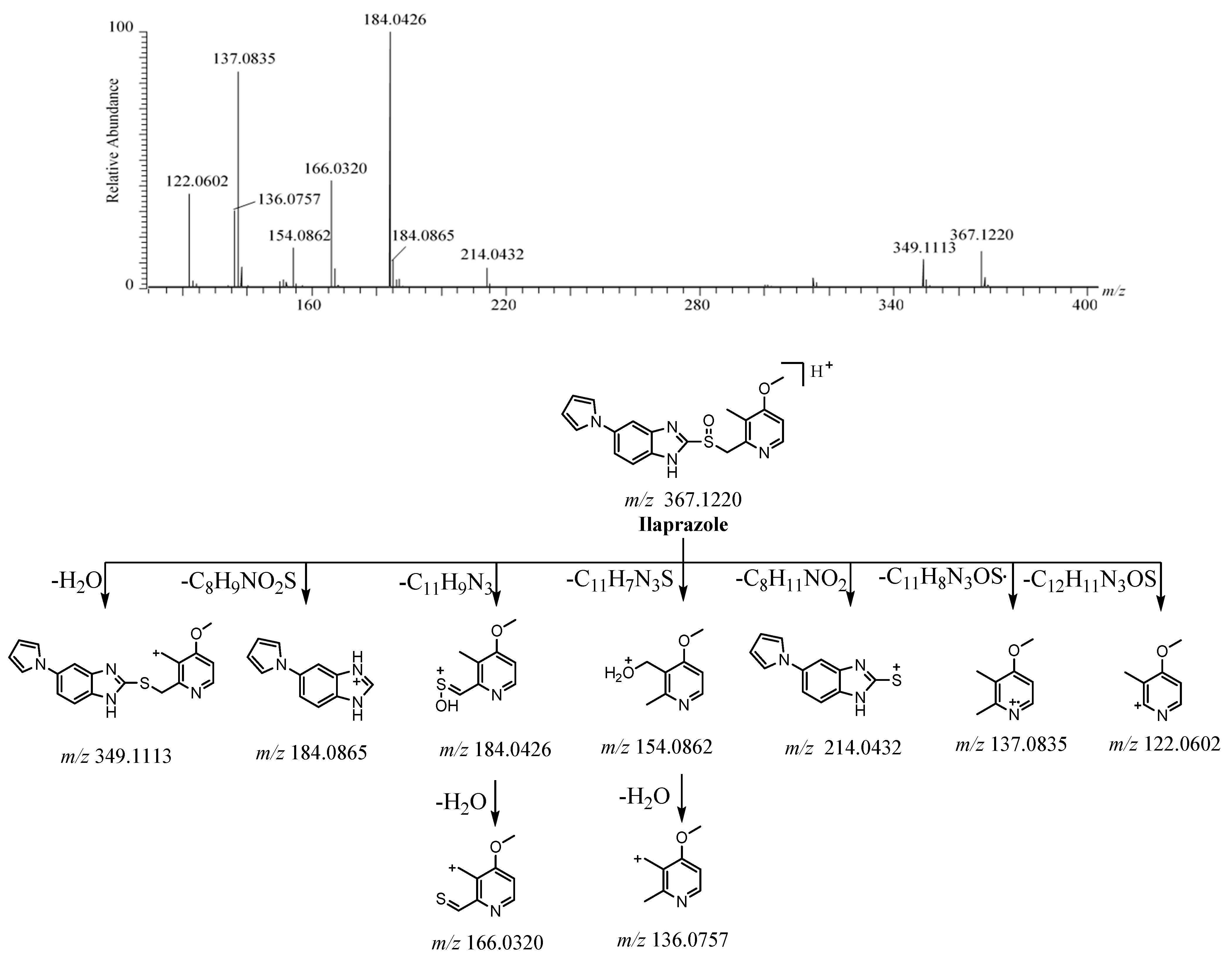
2.2.1. Ilaprazole
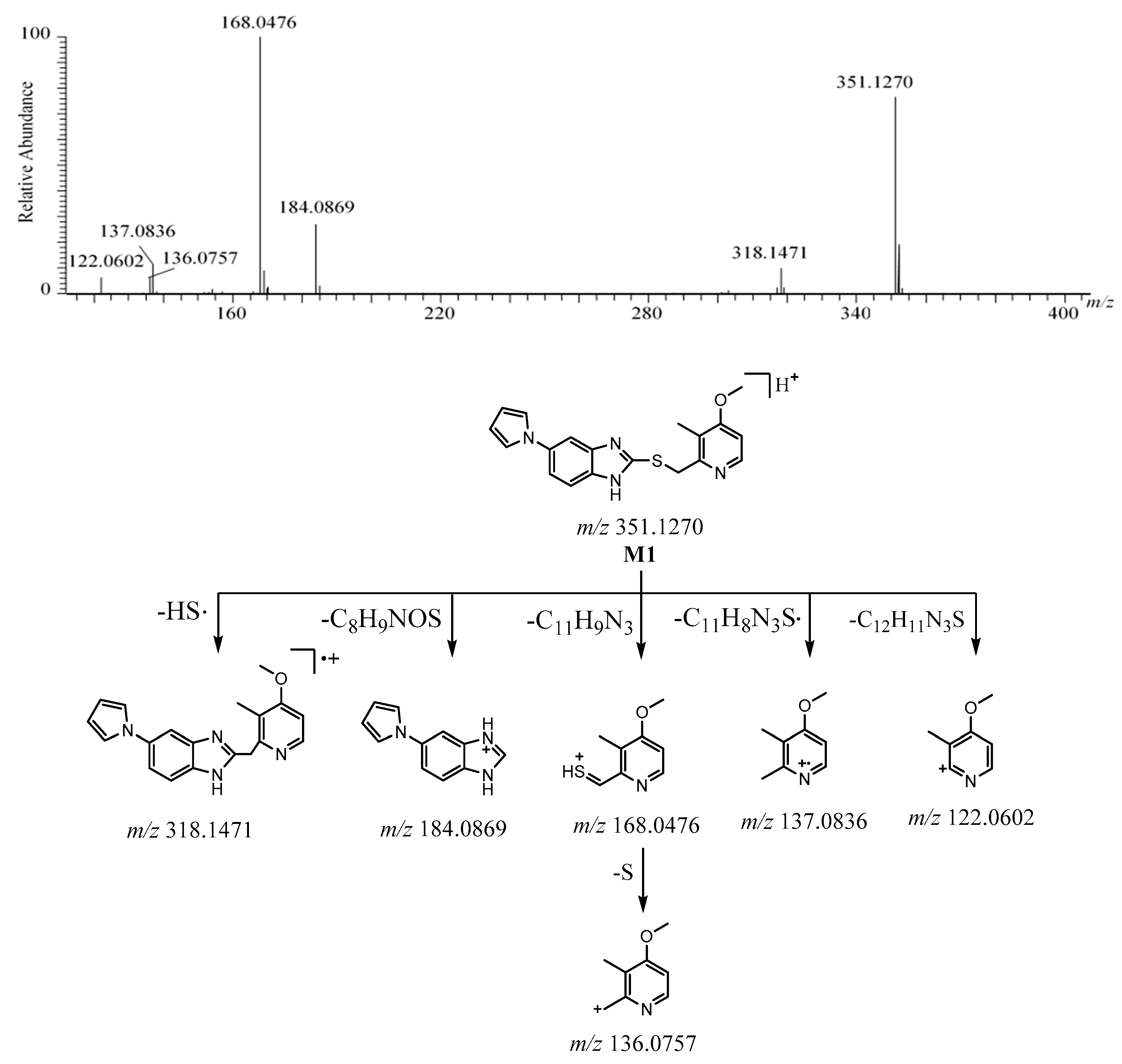
2.2.2. M1
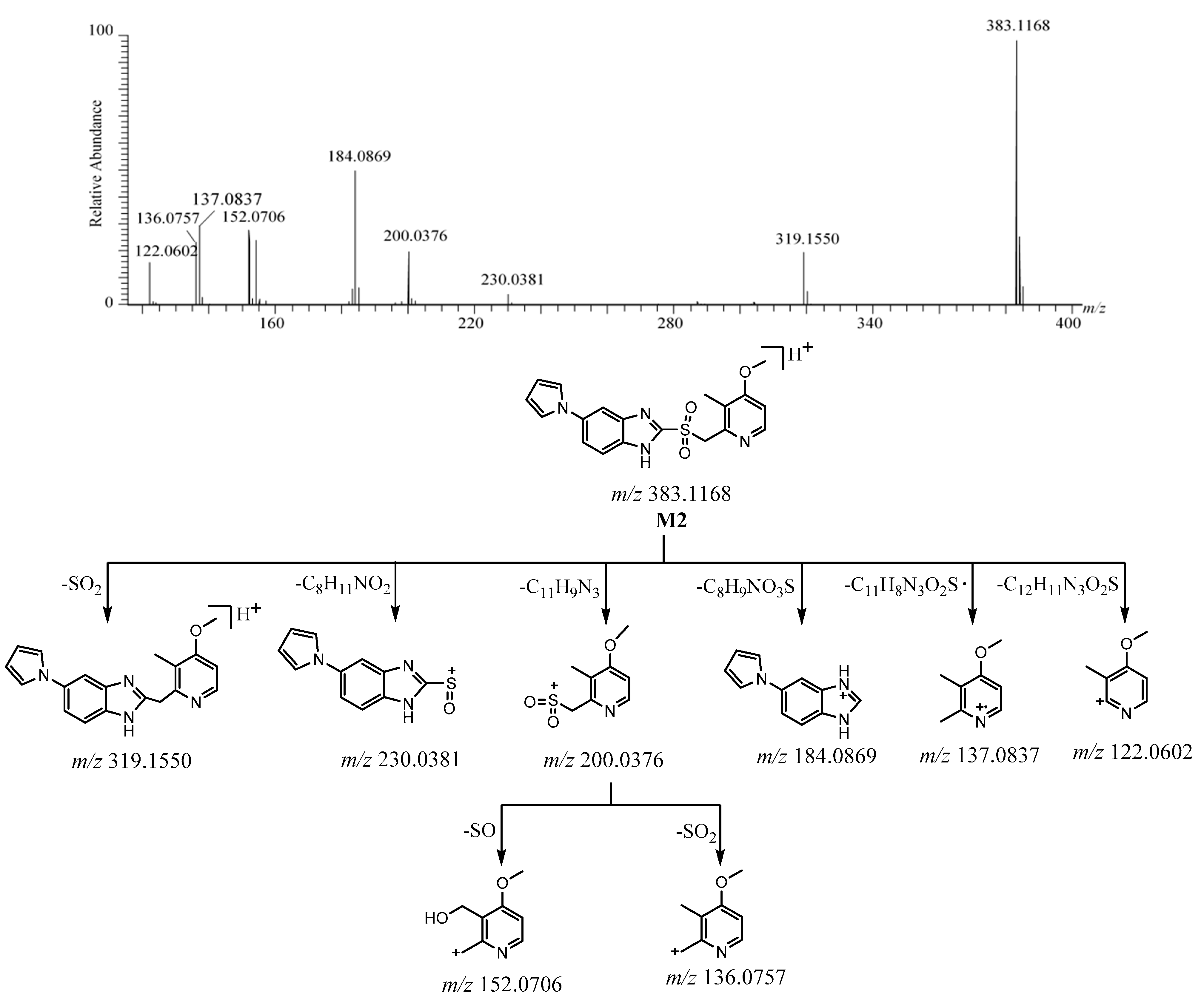
2.2.3. M2
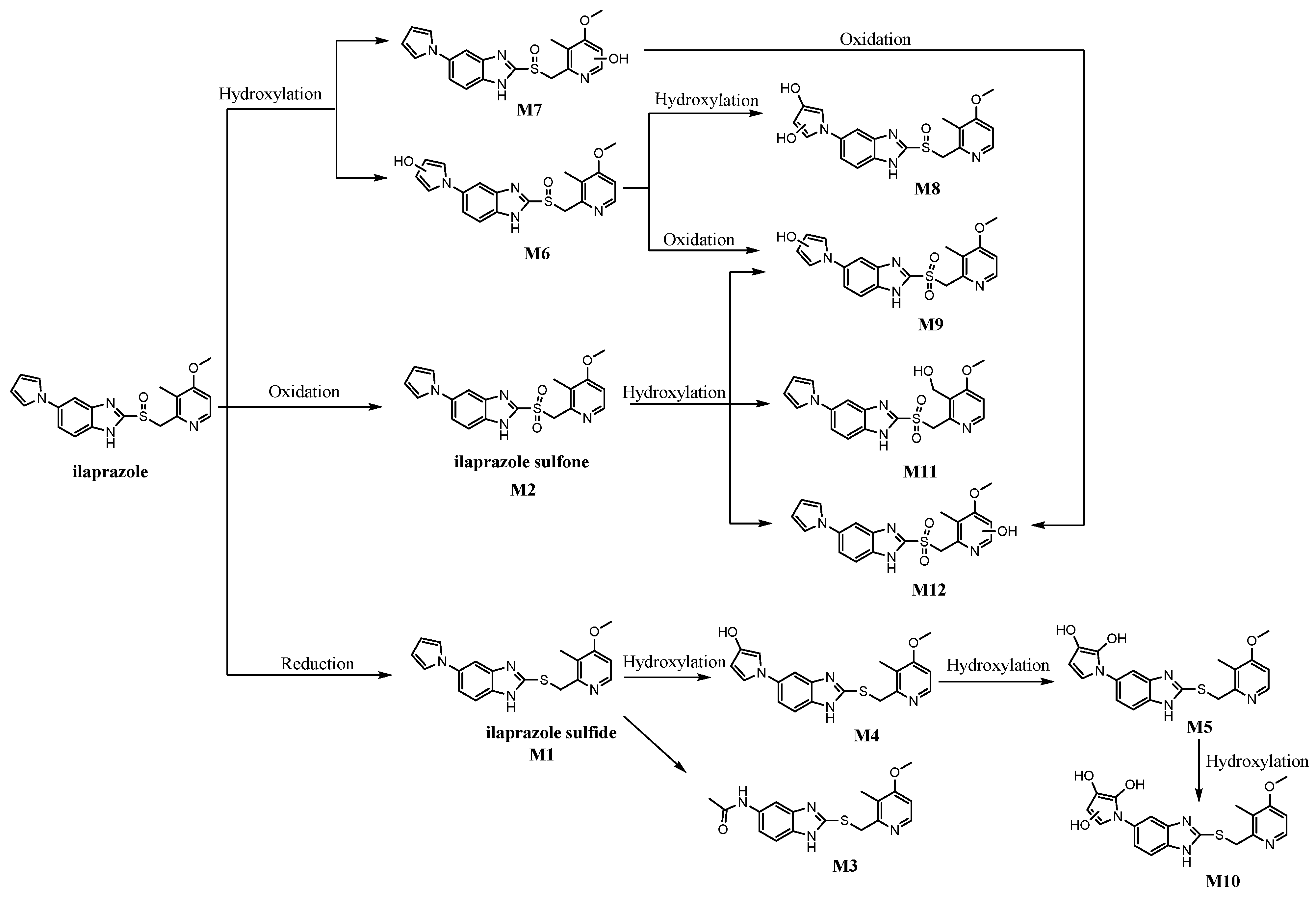
2.3. Identification of the New Metabolites of Ilaprazole
2.3.1. M3
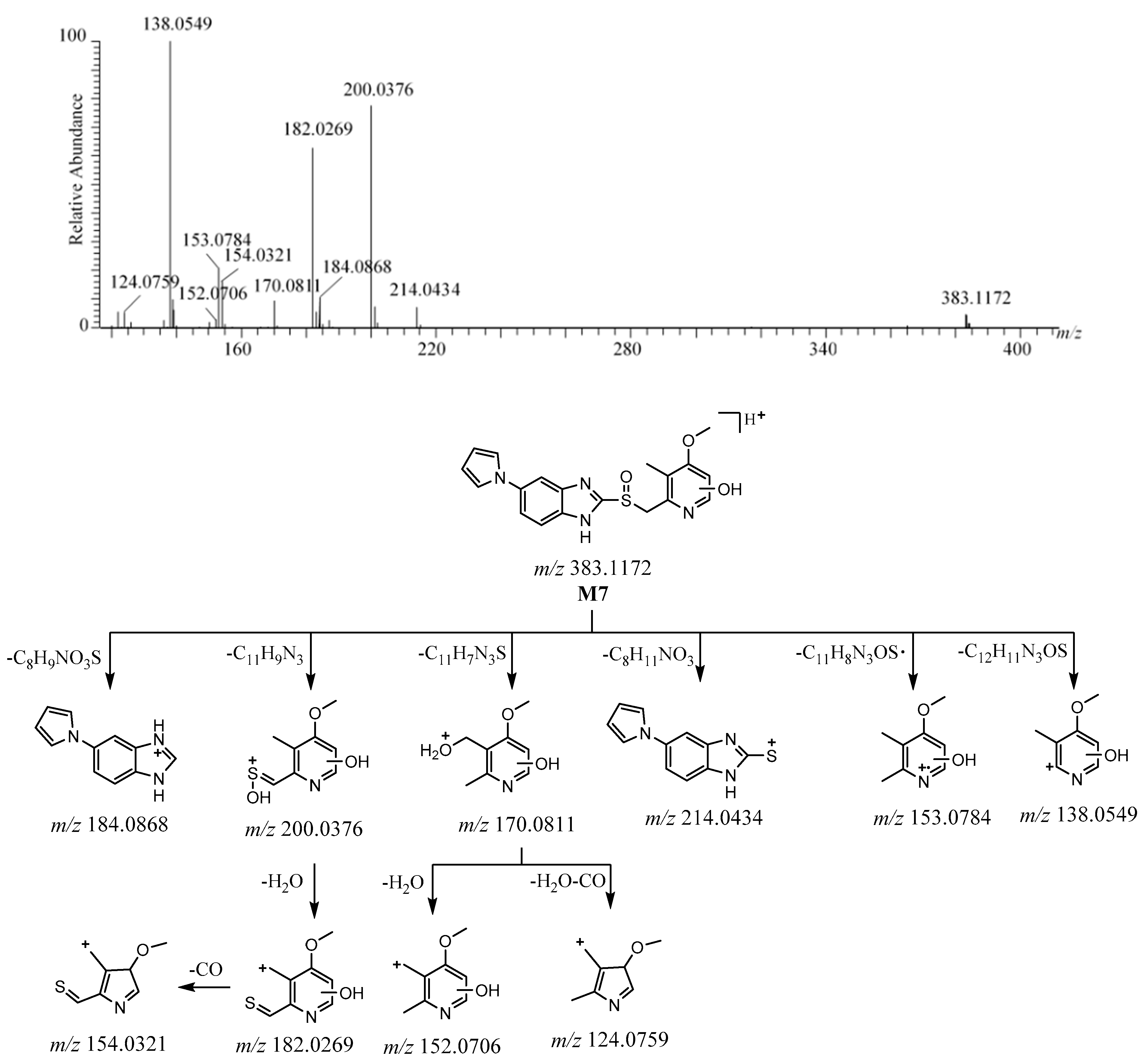
2.3.2. M7
2.3.3. M8
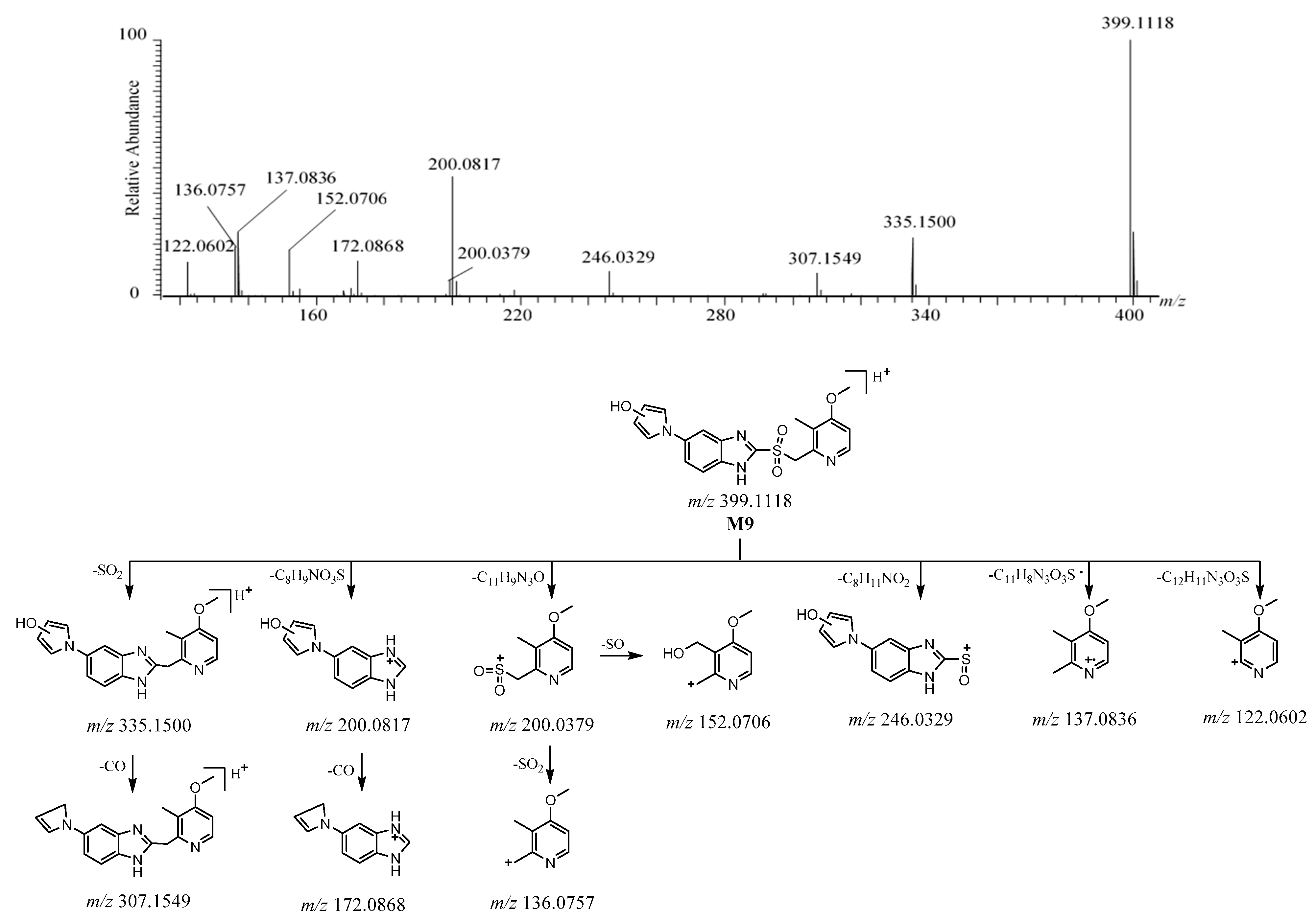
2.3.4. M9
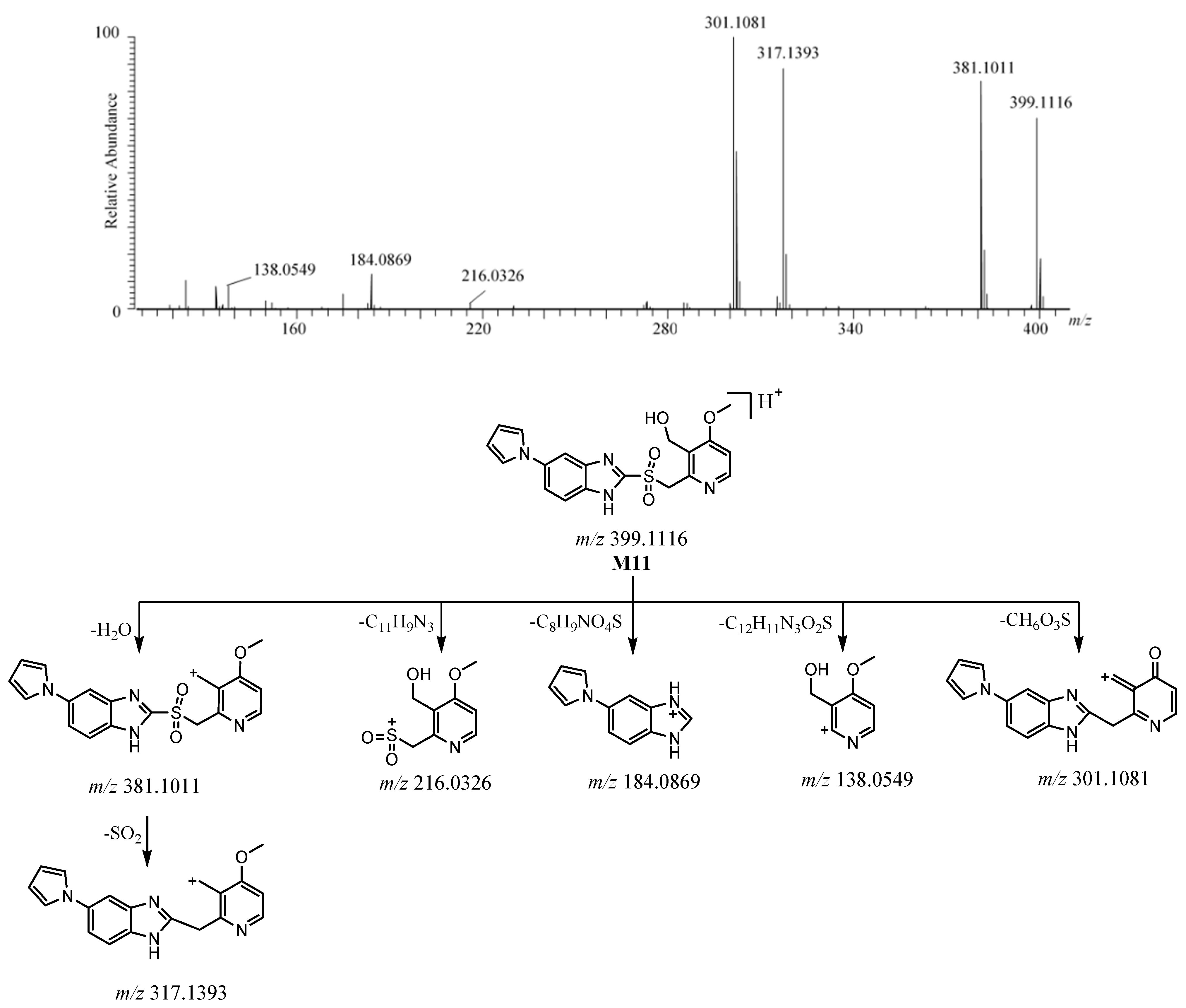
2.3.5. M11
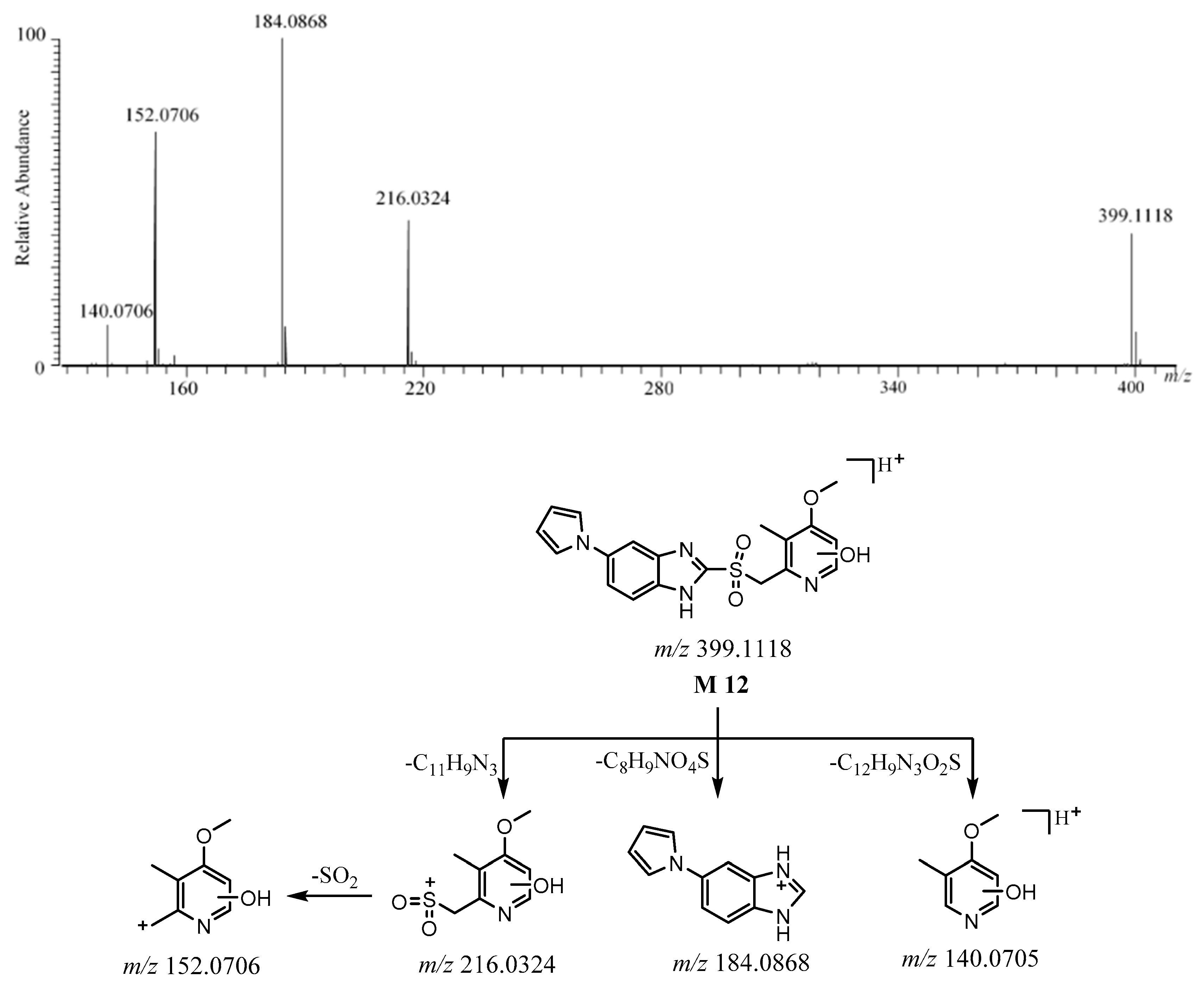
2.3.6. M12
2.4. Identification of the Known Metabolites, M4, M5, M6, and M10
2.4.1. M4
2.4.2. M5
2.4.3. M6
2.4.4. M10
2.5. Metabolic Pathways
2.6. In Silico Bioactivity Prediction of the H+/K+-ATPase Inhibitor
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Instrument and Analysis Conditions
3.2.1. LC-MSn
3.2.2. LC-HRMS
3.3. Animals and Drug Administration
3.4. Pretreatment of the Sample
3.5. In Silico Bioactivity Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Savarino, E.; Ottonello, A.; Martinucci, I.; Dulbecco, P.; Savarino, V. Ilaprazole for the treatment of gastro-esophageal reflux. Expert Opin. Pharmacother. 2016, 17, 2107–2113. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.S.; Shin, W.G.; Seo, S.I.; Choi, M.H.; Jang, H.J.; Park, S.W.; Kae, S.H.; Yang, Y.J.; Shin, S.P.; Baik, G.H.; et al. Effect of ilaprazole on the healing of endoscopic submucosal dissection-induced gastric ulcer: Randomized-controlled, multicenter study. Surg. Endosc. 2019, 33, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.-M.; Nong, G.-H.; Chen, M.-Z.; Huang, X.-P.; Cong, Y.-Y.; Huang, Y.-Y.; Wu, B.-H.; Wei, J.-Q. Modified sequential therapy vs. quadruple therapy as initial therapy in patients with Helicobacter infection. World J. Gastroenterol. 2015, 21, 6310–6316. [Google Scholar] [CrossRef] [PubMed]
- Xuan, J.; Song, R.L.; Xu, G.X.; Lu, W.Q.; Lu, Y.J.; Liu, Z. Modeling the cost-effectiveness of ilaprazole versus omeprazole for the treatment of newly diagnosed duodenal ulcer patients in China. J. Med Econ. 2016, 19, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Chae, J.; Park, C.; Kim, Y.; Lee, S.; Kim, E.; Huh, I.; Kim, N.; Cho, K. Effects of IY-81149, a Newly Developed Proton Pump Inhibitor, on Gastric Acid Secretion in vitro and in vivo. Arzneimittelforschung 2011, 51, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.-Q.; Du, J.-F.; Chen, G.; Chen, G.; Yu, B. Efficacy of ilaprazole in the treatment of duodenal ulcers: A meta-analysis. World J. Gastroenterol. 2014, 20, 5119–5123. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; Lee, J.Y.; Cho, K.-H.; Park, H.L.; Kukulka, M.; Wu, J.-T.; Kim, D.Y.; Park, S.-H. The pharmacokinetics, pharmacodynamics and safety of oral doses of ilaprazole 10, 20 and 40 mg and esomeprazole 40 mg in healthy subjects: A randomised, open-label crossover study. Aliment. Pharmacol. Ther. 2014, 40, 548–561. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wan, H. Drug metabolism and metabolite safety assessment in drug discovery and development. Expert Opin. Drug Metab. Toxicol. 2018, 14, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.-A.; Lee, S.-J.; Kim, K.-B.; Bae, S.K.; Liu, K.-H.; Kim, D.-H.; Shin, J.-G. Ilaprazole, a new proton pump inhibitor, is primarily metabolized to ilaprazole sulfone by CYP3A4 and 3A5. Xenobiotica 2012, 42, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, W.; Guo, N.; Zhou, G.; Zhou, H.; Xiao, Z. Pharmacokinetics of the new proton pump inhibitor ilaprazole in Chinese healthy subjects in relation to CYP3A5 and CYP2C19 genotypes. Clin. Chim. Acta 2008, 391, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Myung, S.-W.; Min, H.-K.; Jin, C.; Kim, M.; Lee, S.M.; Chung, G.J.; Park, S.J.; Kim, D.Y.; Cho, H.-W. Identification of IY81149 and its metabolites in the rat plasma using the on-line HPLC/ESI mass spectrometry. Arch. Pharmacal Res. 1999, 22, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Tan, Z.-R.; Zhang, W.; Ou-Yang, D.-S.; Chen, Y.; Guo, N.; Liu, Y.-Z.; Fan, L.; Deng, H.-W. An improved LC-MS/MS method for quantitative determination of ilaprazole and its metabolites in human plasma and its application to a pharmacokinetic study. Acta Pharmacol. Sin. 2009, 30, 1330–1336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, G.; Shi, S.; Zhang, W.; Tan, Z.; Chen, Y.; Guo, N.; Zhou, H.; Hu, H.; Tan, J. Identification of ilaprazole metabolites in human urine by HPLC-ESI-MS/MS and HPLC-NMR experiments. Biomed. Chromatogr. 2010, 24, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Wang, F.; Tang, W.; Zhu, M. Biotransformation of Ilaprazole in Human Liver Microsomes and Human: Role of CYP3A4 in Ilaprazole Clearance and Drug-Drug Interaction. Drug Metab. Dispos. 2018, 46, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Beccaria, M.; Cabooter, D. Current developments in LC-MS for pharmaceutical analysis. Analyst 2020, 145, 1129–1157. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, Y.; Zhang, Z.; Song, R. Use of LC-QqQ-MS for the detection of emodin metabolites in rat bile and urine. Biomed. Chromatogr. 2017, 31, e3979. [Google Scholar] [CrossRef] [PubMed]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of activity spectra for biologically active substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Xie, Z.; Chen, X. Metabolism of pantoprazole involving conjugation with glutathione in rats. J. Pharm. Pharmacol. 2005, 57, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Pearce, C.M.; Lushnikova, M.V. Microbiological production of omeprazole metabolites by Cunninghamella elegans. J. Mol. Catal. B Enzym. 2006, 41, 87–91. [Google Scholar] [CrossRef]


| Name | Probability of H+/K+-ATPase Inhibitor | Name | Probability of H+/K+-ATPase Inhibitor |
|---|---|---|---|
| Ilaprazole | 94.1% | M8-3 * | 68.4% |
| M3 | 60.9% | M9-1 * | 88.2% |
| M7-1 * | 89.4% | M9-2 * | 73.9% |
| M7-2 * | 76.5% | M11 | 84.3% |
| M8-1 * | 79.1% | M12-1 * | 84.0% |
| M8-2 * | 90.7% | M12-2 * | 66.1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Guo, K.; Wang, P.; Shan, Y.; Ma, C. Identification of the New In Vivo Metabolites of Ilaprazole in Rat Plasma after Oral Administration by LC-MS: In Silico Prediction of the H+/K+-ATPase Inhibitor. Molecules 2021, 26, 459. https://doi.org/10.3390/molecules26020459
Zhang G, Guo K, Wang P, Shan Y, Ma C. Identification of the New In Vivo Metabolites of Ilaprazole in Rat Plasma after Oral Administration by LC-MS: In Silico Prediction of the H+/K+-ATPase Inhibitor. Molecules. 2021; 26(2):459. https://doi.org/10.3390/molecules26020459
Chicago/Turabian StyleZhang, Guiqiu, Kaijing Guo, Pengfei Wang, Yingbo Shan, and Chen Ma. 2021. "Identification of the New In Vivo Metabolites of Ilaprazole in Rat Plasma after Oral Administration by LC-MS: In Silico Prediction of the H+/K+-ATPase Inhibitor" Molecules 26, no. 2: 459. https://doi.org/10.3390/molecules26020459
APA StyleZhang, G., Guo, K., Wang, P., Shan, Y., & Ma, C. (2021). Identification of the New In Vivo Metabolites of Ilaprazole in Rat Plasma after Oral Administration by LC-MS: In Silico Prediction of the H+/K+-ATPase Inhibitor. Molecules, 26(2), 459. https://doi.org/10.3390/molecules26020459






