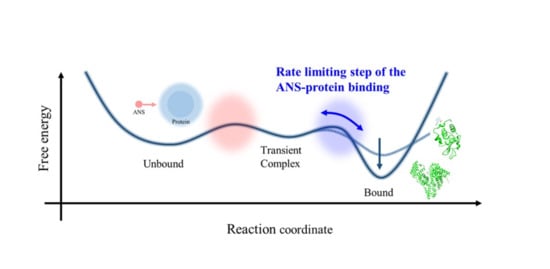
Revisiting the Rate-Limiting Step of the ANS–Protein Binding at the Protein Surface and Inside the Hydrophobic Cavity
Abstract
1. Introduction
2. Results
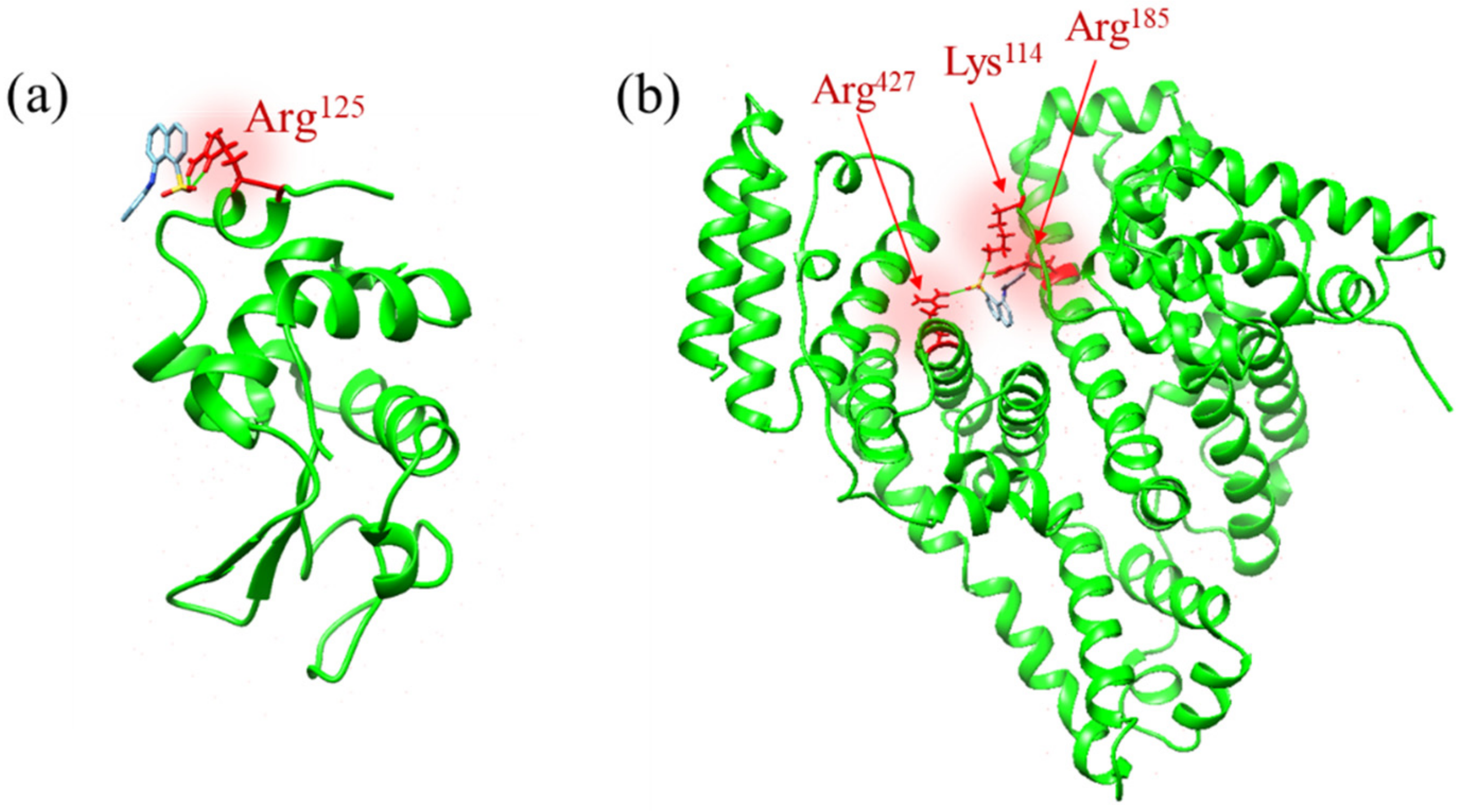
2.1. Molecular Docking Analysis
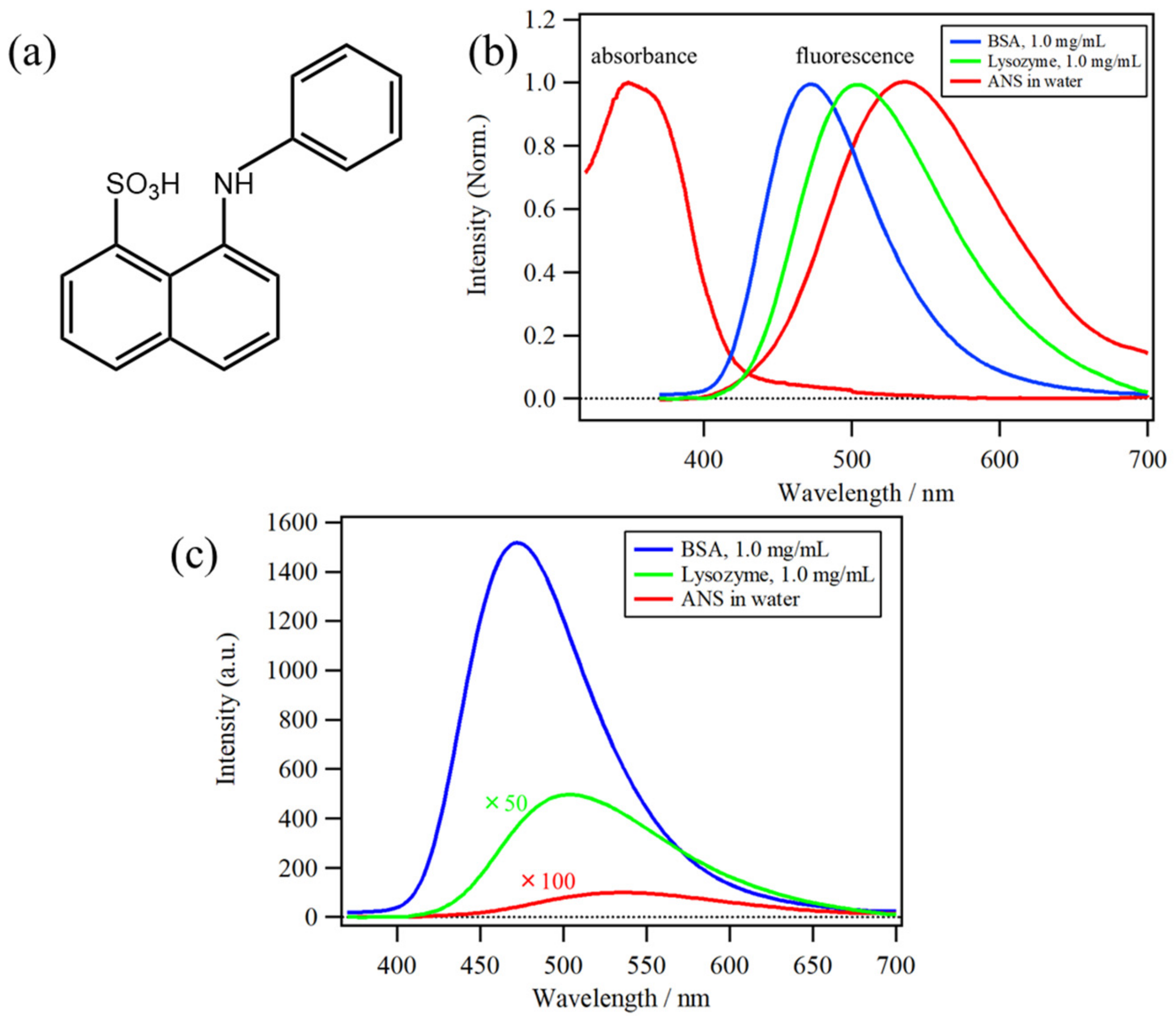
2.2. Protein Concentration Dependence of ANS Fluorescence
2.3. MCR–ALS Analysis of the Protein Concentration Dependence of ANS Fluorescence Spectra
2.4. Estimation of the ANS–Protein Binding Constant
3. Discussion
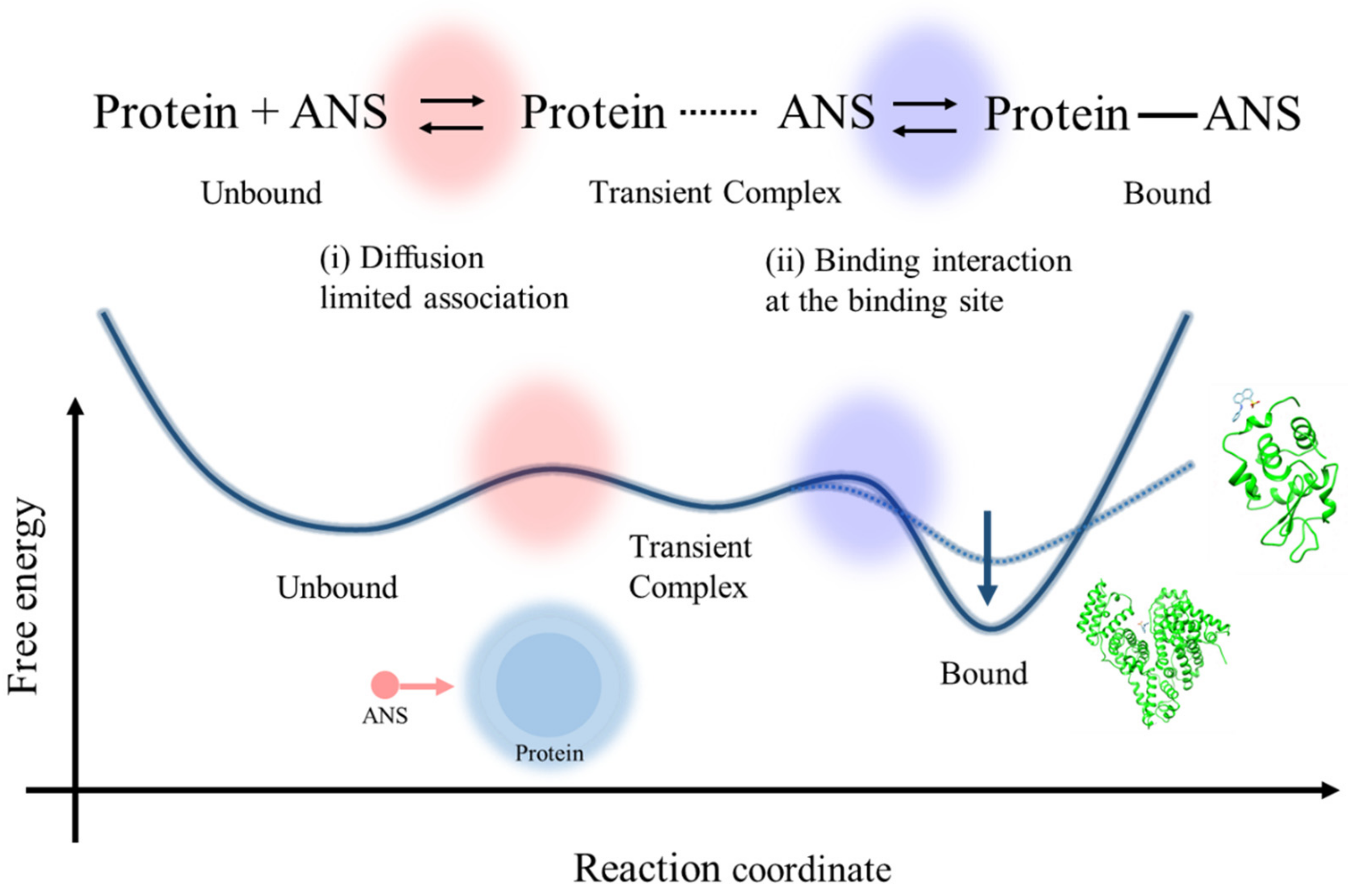
3.1. Effect of the Electrostatic Association on the ANS–Protein Binding Pathway
3.2. Effects of the Binding Interaction on the ANS–Protein Binding Pathway
3.3. Rate-Limiting Step of the Overall ANS–Protein Binding Pathway
4. Experimental Section
4.1. Fluorescence Measurement
4.2. Molecular Docking Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kosower, E.M.; Tanizawa, K. Analysis of Fluorescence Emission and Quenching for Molecules Bearing Latent Donors. Chem. Phys. Lett. 1972, 16, 419–425. [Google Scholar] [CrossRef]
- Kosower, E.M. Intramolecular Donor-Acceptor Systems. 9. Photophysics of (Phenylamino)naphthalenesulfonates: A Paradigm for Excited-State Intramolecular Charge Transfer. Acc. Chem. Res. 1982, 15, 259–266. [Google Scholar] [CrossRef]
- Kosower, E.M.; Huppert, D. Excited State Electron and Proton Transfers. Annu. Rev. Phys. Chem. 1986, 37, 127–156. [Google Scholar] [CrossRef]
- Ota, C.; Takano, K. Spectroscopic Analysis of Protein-Crowded Environments Using the Charge-Transfer Fluorescence Probe 8-Anilino-1-Naphthalenesulfonic Acid. ChemPhysChem 2019, 20, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic Fluorescent Dyes as Tools for Protein Characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef]
- Kayser, V.; Chennamsetty, N.; Voynov, V.; Helk, B.; Trout, B.L. Conformational Stability and Aggregation of Therapeutic Monoclonal Antibodies Studied with ANS and Thioflavin T Binding. mAbs 2011, 3, 408–411. [Google Scholar] [CrossRef]
- Ota, C.; Fukuda, Y.; Tanaka, S.; Takano, K. Spectroscopic Evidence of the Salt-Induced Conformational Change around the Localized Electric Charges on the Protein Surface of Fibronectin Type III. Langmuir 2020, 36, 14243–14254. [Google Scholar] [CrossRef]
- Mukherjee, S.K.; Gautam, S.; Biswas, S.; Kundu, J.; Chowdhury, P.K. Do Macromolecular Crowding Agents Exert Only an Excluded Volume Effect? A Protein Solvation Study. J. Phys. Chem. B 2015, 119, 14145–14156. [Google Scholar] [CrossRef]
- Cardamone, M.; Puri, N.K. Spectrofluorimetric Assessment of the Surface Hydrophobicity of Proteins. Biochem. J. 1992, 282, 589–593. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Sulatskaya, A.I.; Povarova, O.I.; Turoverov, K.K. Reevaluation of ANS Binding to Human and Bovine Serum Albumins: Key Role of Equilibrium Microdialysis in Ligand—Receptor Binding Characterization. PLoS ONE 2012, 7, e40845. [Google Scholar] [CrossRef]
- Haskard, C.A.; Li-Chan, E.C.Y. Hydrophobicity of Bovine Serum Albumin and Ovalbumin Determined Using Uncharged (PRODAN) and Anionic (ANS−) Fluorescent Probes. J. Agric. Food Chem. 1998, 46, 2671–2677. [Google Scholar] [CrossRef]
- Cattoni, D.I.; Kaufman, S.B.; Flecha, F.L.G. Kinetics and Thermodynamics of the Interaction of 1-Anilino-Naphthalene-8-Sulfonate with Proteins. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 1700–1708. [Google Scholar] [CrossRef] [PubMed]
- Cinar, H.; Winter, R. The Effects of Cosolutes and Crowding on the Kinetics of Protein Condensate Formation Based on Liquid-Liquid Phase Separation: A Pressure-Jump Relaxation Study. Sci. Rep. 2020, 10, 7245. [Google Scholar] [CrossRef] [PubMed]
- Matulis, D.; Lovrien, R. 1-Anilino-8-Naphthalene Sulfonate Anion-Protein Binding Depends Primarily on Ion Pair Formation. Biophys. J. 1998, 74, 422–429. [Google Scholar] [CrossRef]
- Ory, J.J.; Banaszak, L.J. Studies of the Ligand Binding Reaction of Adipocyte Lipid Binding Protein Using the Fluorescent Probe 1, 8-Anilinonaphthalene-8-Sulfonate. Biophys. J. 1999, 77, 1107–1116. [Google Scholar] [CrossRef]
- Schonbrunn, E.; Eschenburg, S.; Luger, K.; Kabsch, W.; Amrhein, N. Structural Basis for the Interaction of the Fluorescence Probe 8-Anilino-1-Naphthalene Sulfonate (ANS) with the Antibiotic Target MurA. Proc. Natl. Acad. Sci. USA 2000, 97, 6345–6349. [Google Scholar] [CrossRef]
- Lartigue, A.; Gruez, A.; Spinelli, S.; Rivière, S.; Brossut, R.; Tegoni, M.; Cambillau, C. The Crystal Structure of a Cockroach Pheromone-Binding Protein Suggests a New Ligand Binding and Release Mechanism. J. Biol. Chem. 2003, 278, 30213–30218. [Google Scholar] [CrossRef]
- Gasymov, O.K.; Glasgow, B.J. ANS Fluorescence: Potential to Augment the Identification of the External Binding Sites of Proteins. Biochim. Biophys. Acta Proteins Proteom. 2007, 1774, 403–411. [Google Scholar] [CrossRef]
- Zhou, H.X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef]
- Kim, J.Y.; Meng, F.; Yoo, J.; Chung, H.S. Diffusion-limited association of disordered protein by non-native electrostatic interactions. Nat. Commun. 2018, 9, 4707. [Google Scholar] [CrossRef]
- Meissner, J.; Prause, A.; Bharti, B.; Findenegg, G.H. Characterization of Protein Adsorption onto Silica Nanoparticles: Influence of pH and Ionic Strength. Colloid Polym. Sci. 2015, 293, 3381–3391. [Google Scholar] [CrossRef] [PubMed]
- Kubiak-Ossowska, K.; Cwieka, M.; Kaczynska, A.; Jachimska, B.; Mulheran, P.A. Lysozyme Adsorption at a Silica Surface Using Simulation and Experiment: Effects of pH on Protein Layer Structure. Phys. Chem. Chem. Phys. 2015, 17, 24070–24077. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast Docking Using the CHARMM Force Field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a Protein-Small Molecule Docking Web Service Based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Ota, C.; Furukawa, T.; Shima, K.; Sano, S.; Kuramochi, K.; Tsubaki, K.; Noguchi, S.; Takano, K. Alkyne Tagged Raman Probes for Protein by Chemical Modification Approach. ChemistrySelect 2017, 2, 1267–1270. [Google Scholar] [CrossRef]
- Möller, M.; Denicola, A. Study of Protein-Ligand Binding by Fluorescence. Biochem. Mol. Biol. Educ. 2002, 30, 309–312. [Google Scholar] [CrossRef]
- Togashi, D.M.; Ryder, A.G. A Fluorescence Analysis of ANS Bound to Bovine Serum Albumin: Binding Properties Revisited by Using Energy Transfer. J. Fluoresc. 2008, 18, 519–526. [Google Scholar] [CrossRef]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar]
- Ota, C.; Noguchi, S.; Tsumoto, K. The Molecular Interaction of a Protein in Highly Concentrated Solution Investigated by Raman Spectroscopy. Biopolymers 2015, 103, 237–246. [Google Scholar] [CrossRef]
- Ota, C.; Noguchi, S.; Nagatoishi, S.; Tsumoto, K. Assessment of the Protein-Protein Interactions in a Highly Concentrated Antibody Solution by Using Raman Spectroscopy. Pharm. Res. 2016, 33, 956–969. [Google Scholar] [CrossRef]
- Ota, C.; Takano, K. Behavior of Bovine Serum Albumin Molecules in Molecular Crowding Environments Investigated by Raman Spectroscopy. Langmuir 2016, 32, 7372–7382. [Google Scholar] [CrossRef] [PubMed]
- Ota, C.; Suzuki, H.; Tanaka, S.I.; Takano, K. Spectroscopic Signature of the Steric Strains in an Escherichia coli RNase HI Cavity-Filling Destabilized Mutant Protein. J. Phys. Chem. B 2020, 124, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Kosower, E.M.; Kanety, H. Intramolecular Donor-Acceptor Systems. 10. Multiple Fluorescences from 8-(Phenylamino)-1-Naphthalenesulfonates. J. Am. Chem. Soc. 1983, 105, 6236–6243. [Google Scholar] [CrossRef]
- Chong, S.H.; Ham, S. Anomalous Dynamics of Water Confined in Protein-Protein and Protein-DNA Interfaces. J. Phys. Chem. Lett. 2016, 7, 3967–3972. [Google Scholar] [CrossRef] [PubMed]
- Sterpone, F.; Stirnemann, G.; Laage, D. Magnitude and Molecular Origin of Water Slowdown Next to a Protein. J. Am. Chem. Soc. 2012, 134, 4116–4119. [Google Scholar] [CrossRef]
- Daniel, E.; Yang, J.T. Analysis of the Circular Dichroism of the Complexes of 8-Anilino-1-Naphthalenesulfonate with Bovine Serum Albumin. Biochemistry 1973, 12, 508–512. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Bhowmik, S.; Singh, A.K.; Kodgire, P.; Das, A.K.; Mukherjee, T.K. Direct Evidence of Intrinsic Blue Fluorescence from Oligomeric Interfaces of Human Serum Albumin. Langmuir 2017, 33, 10606–10615. [Google Scholar] [CrossRef]
- Ota, C. Energy Transfer at Heterogeneous Protein-Protein Interfaces to Investigate the Molecular Behaviour in the Crowding Environment. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2017, 175, 145–154. [Google Scholar] [CrossRef]
- Feng, X.-Z.; Lin, Z.; Yang, L.-J.; Wang, C.; Bai, C. Investigation of the Interaction Between Acridine Orange and Bovine Serum Albumin. Talanta 1998, 47, 1223–1229. [Google Scholar] [CrossRef]
- Patra, S.; Santhosh, K.; Pabbathi, A.; Samanta, A. Diffusion of Organic Dyes in Bovine Serum Albumin Solution Studied by Fluorescence Correlation Spectroscopy. RSC Adv. 2012, 2, 6079. [Google Scholar] [CrossRef]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Fersht, A.R. Rapid, Electrostatically Assisted Association of Proteins. Nat. Struct. Biol. 1996, 3, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.-J.; Hage, T.; Sebald, W. Global and Local Determinants for the Kinetics of Interleukin-4/Interleukin-4 Receptor α Chain Interaction. Eur. J. Biochem. 1996, 240, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.E.; Georgiou, C.A.; Koupparis, M.A. Automated Flow Injection Gradient Technique for Binding Studies of Micromolecules to Proteins Using Potentiometric Sensors: Application to Bovine Serum Albumin with Anilinonaphthalenesulfonate Probe and Drugs. Anal. Chem. 1999, 71, 2541–2550. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.T.; Hou, M.C.; Xiao, Z.X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ota, C.; Tanaka, S.-i.; Takano, K. Revisiting the Rate-Limiting Step of the ANS–Protein Binding at the Protein Surface and Inside the Hydrophobic Cavity. Molecules 2021, 26, 420. https://doi.org/10.3390/molecules26020420
Ota C, Tanaka S-i, Takano K. Revisiting the Rate-Limiting Step of the ANS–Protein Binding at the Protein Surface and Inside the Hydrophobic Cavity. Molecules. 2021; 26(2):420. https://doi.org/10.3390/molecules26020420
Chicago/Turabian StyleOta, Chikashi, Shun-ichi Tanaka, and Kazufumi Takano. 2021. "Revisiting the Rate-Limiting Step of the ANS–Protein Binding at the Protein Surface and Inside the Hydrophobic Cavity" Molecules 26, no. 2: 420. https://doi.org/10.3390/molecules26020420
APA StyleOta, C., Tanaka, S.-i., & Takano, K. (2021). Revisiting the Rate-Limiting Step of the ANS–Protein Binding at the Protein Surface and Inside the Hydrophobic Cavity. Molecules, 26(2), 420. https://doi.org/10.3390/molecules26020420