Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control
Abstract
1. Introduction
2. Signaling Pathways Regulating Skeletal Muscle Mass and Function
2.1. Anabolic Pathways
2.1.1. PI3K/AKT Signaling Pathway
2.1.2. G Protein-Coupled Receptors (GPCRs) and cAMP Signaling
- ß2-Adrenergic Receptors Signaling Pathway
- 2.
- WNT/FZD Signaling Pathway
2.1.3. Calcineurin Signaling Pathway
2.1.4. Hippo Signaling Pathway
2.2. Transforming Growth Factor (TGFs), Pro-Anabolic and Pro-Catabolic Pathways
2.3. Catabolic Pathways
2.3.1. AMPK Signaling Pathway
2.3.2. The NF-κB Signaling Pathway
2.3.3. Glucocorticoid Receptor Signaling Pathway
2.3.4. Angiotensin Signaling Pathway
2.3.5. JAK/STAT Signaling Pathway
2.3.6. Kinin Signaling Pathway
2.3.7. Sphingolipids Signaling Pathway
2.3.8. NOTCH Signaling Pathway
2.3.9. Oxidative Stress Is an Inducer of Skeletal Muscle Atrophy
3. E3 Ligases Involved in the Regulation of Muscle Atrophy
3.1. E3 Ligases Involved in the Regulation of Anabolic Pathways
3.1.1. The CBL-B and FBXO40 E3 Ubiquitin Ligases Target IRS1 to Degradation in Skeletal Muscle
3.1.2. NEDD4-1 E3 Ubiquitin Ligase, Friend or Foe?
3.2. E3 Ubiquitin Ligases Involved in the Regulation of Catabolic Pathways
3.2.1. Regulating the Canonical NF-κB Pathway via the Manipulation of cIAP and TRAF6 E3 Ligases
3.2.2. WWP1 in the Regulation of Muscle Atrophy
3.2.3. TRIM32 in the Regulation of Autophagy
3.2.4. FOXO Transcription Factors Are Regulated by MDM2 and SKP2 E3 Ubiquitin Ligases
3.3. E3 Ubiquitin Ligases Involved in the Regulation of Muscle Mass and Function
3.3.1. MuRF1/TRIM63
3.3.2. MAFbx/Atrogin-1/FBXO32
3.3.3. PARKIN Controls Muscle Mass through the Maintenance of Mitochondrial Homeostasis
3.3.4. MUSA1/FBXO30
3.3.5. FBXL21
3.3.6. Ubiquitin Ring-Type E3 Ligases (UBR)
3.3.7. FBXO21/SMART
3.4. Promising E3 Ubiquitin Ligases Regulating Muscle Mass and Function
4. Current Treatments/Potential Modes of Action
4.1. Indirect Action on E3 Ligases
4.1.1. PI3K-AKT-mTORC1
4.1.2. Glucocorticoids
4.1.3. Il-6
4.1.4. NF-κB
4.1.5. ß2 Adrenergic Receptor (β2-AR)
4.1.6. p38α Mitogen-Activated Protein Kinase (p38α MAPK)
4.1.7. NOTCH
4.1.8. Ion Channels
4.1.9. Acute-Phase Protein Serum Amyloid A1 (SAA1)
4.1.10. TGF-β
4.1.11. Reactive oxygen species (ROS)
4.1.12. Leucine and Its Derivative ß-Hydroxy-ß-Methylbutyrate (HMB)
4.1.13. Plant Derivatives
4.2. E3 Ligases Inhibitors
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | Adenosine 5′-monophosphate-activated (AMP)-activated protein kinase |
| ATG9 | Autophagy related gene 9 |
| BMP | Bone Morphogenic Protein |
| CaMKKß | Ca2+/calmodulin-dependent protein kinase kinase ß |
| cAMP | cyclic Adenosine Monophosphate |
| CHF | Congestive Heart Failure |
| CKD | Chronic Kidney Disease |
| Cn | Calcineurin |
| CSL | CBF1, Suppressor of Hairless, Lag-1 |
| DMD | Duchenne Muscle Dystrophy |
| Dsh | Dishevelled |
| EDL | Extensor digitorum longus |
| ERK | Extracellular signal-regulated kinases |
| Fd | Frizzled |
| FOXO | Forkhead box protein O |
| GC | Glucocorticoids |
| GR | Glucocorticoids Receptor |
| GDF | Growth Differentiation Factor |
| GPCR | G-protein coupled receptors |
| HBM | ß-hydroxy-ß-methylbutyrate |
| HDAC4 | Histone deacetylase 4 |
| HECT | Homologous to E6-Associated Protein C Terminus |
| IGF1 | Insulin-like growth factor 1 |
| IKK | IκB Kinase |
| KD | Knock Down |
| KO | Knock-Out |
| LKB1 | Liver kinase B1 |
| MAFbx/Atrogin-1 | Muscle atrophy F-box |
| MAPK | Mitogen Activated Protein Kinase |
| Mdx | The mdx mouse has a point mutation in its DMD gene (coding for Dystrophin) |
| MSTN | Myostatin |
| mTORC | Mechanistic (or mammalian) target of rapamycin complex |
| MuRF1 | Muscle Ring-Finger 1 Protein |
| MyoD | Myogenic regulatory factor |
| NAC | N-acetyl cysteine |
| NFAT | Nuclear factor of activated T-cells |
| NICD | Notch Intracellular Domain |
| NOX | NADPH oxidase |
| PTEN | Phosphatase and tensin homologue |
| PDK1 | 3-phosphoinositide-dependent protein kinase 1 |
| PGC-1a | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| PI3K | Phosphoinositide 3-kinase |
| PKA | cAMP-dependent protein kinase |
| PQQ | pyrroloquinoline quinone |
| RBR | RING-in-Between-RING |
| RING | Really Interesting New Gene-finger |
| RNS | Reactive Nitrogen Species |
| ROS | Reactive Oxygen Species |
| SDEN | Surgical sympathetic denervation |
| SMAD | Small Mothers Against Decapentaplegic |
| TA | Tibialis Anterior |
| TAK-1 | transforming growth factor ß-activated kinase 1 |
| TAZ/WWTR1 | WW domain containing protein 1 |
| TFs | Transcription factors |
| TGF | Transforming Growth Factor |
| TRADD | TNF receptor associated via death domain |
| TRAF6 | TNF receptor-associated factor 6 |
| TSC | Tuberous Sclerosis Complex |
| ULK1 | uncoordinated 51-like kinase 1 |
| UPS | Ubiquitin-Proteasome System |
| Wnt | Wingless-type mouse mammary tumor virus integration site |
| YAP | Yes-Associated Protein |
References
- Von Haehling, S.; Anker, M.S.; Anker, S.D. Prevalence and clinical impact of cachexia in chronic illness in Europe, USA, and Japan: Facts and numbers update 2016. J. Cachexia Sarcopenia Muscle 2016, 7, 507–509. [Google Scholar] [CrossRef]
- Penna, F.; Ballarò, R.; Beltrà, M.; De Lucia, S.; García Castillo, L.; Costelli, P. The Skeletal Muscle as an Active Player against Cancer Cachexia. Front. Physiol. 2019, 10, 41. [Google Scholar] [CrossRef]
- Blondelle, J.; Biju, A.; Lange, S. The Role of Cullin-RING Ligases in Striated Muscle Development, Function, and Disease. Int. J. Mol. Sci. 2020, 21, 7936. [Google Scholar] [CrossRef]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef]
- Polge, C.; Attaix, D.; Taillandier, D. Role of E2-Ub-conjugating enzymes during skeletal muscle atrophy. Front. Physiol. 2015, 6, 59. [Google Scholar] [CrossRef][Green Version]
- Taillandier, D.; Polge, C. Skeletal muscle atrogenes: From rodent models to human pathologies. Biochimie 2019, 166, 251–269. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends. Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Weissman, A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2001, 2, 169–178. [Google Scholar] [CrossRef]
- Walden, H.; Rittinger, K. RBR ligase–mediated ubiquitin transfer: A tale with many twists and turns. Nat. Struct. Mol. Cell. Biol. 2018, 25, 440–445. [Google Scholar] [CrossRef]
- Dove, K.K.; Klevit, R.E. RING-between-RING E3 Ligases: Emerging Themes amid the Variations. J. Mol. Biol. 2017, 429, 3363–3375. [Google Scholar] [CrossRef]
- Peris-Moreno, D.; Taillandier, D.; Polge, C. MuRF1/TRIM63, Master Regulator of Muscle Mass. Int. J. Mol. Sci. 2020, 21, 6663. [Google Scholar] [CrossRef]
- Frost, R.A.; Nystrom, G.J.; Jefferson, L.S.; Lang, C.H. Hormone, cytokine, and nutritional regulation of sepsis-induced increases in atrogin-1 and MuRF1 in skeletal muscle. Am. J. Physiol.-Endocrinol. Metab. 2007, 292, E501–E512. [Google Scholar] [CrossRef]
- Gopinath, S.D. Inhibition of stat3 signaling ameliorates atrophy of the soleus muscles in mice lacking the vitamin D receptor. Skelet. Muscle 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kline, W.O.; Panaro, F.J.; Yang, H.; Bodine, S.C. Rapamycin inhibits the growth and muscle-sparing effects of clenbuterol. J. Appl. Physiol. 2007, 102, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, J.L.; Lu, J.; Song, Y.; Kwak, K.S.; Jiao, Q.; Rosenfeld, R.; Chen, Q.; Boone, T.; Simonet, W.S.; et al. Reversal of Cancer Cachexia and Muscle Wasting by ActRIIB Antagonism Leads to Prolonged Survival. Cell 2010, 142, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Adams, V.; Gußen, V.; Zozulya, S.; Cruz, A.; Moriscot, A.; Linke, A.; Labeit, S. Small-Molecule Chemical Knockdown of MuRF1 in Melanoma Bearing Mice Attenuates Tumor Cachexia Associated Myopathy. Cells 2020, 9, 2272. [Google Scholar] [CrossRef]
- Adams, V.; Bowen, T.S.; Werner, S.; Barthel, P.; Amberger, C.; Konzer, A.; Graumann, J.; Sehr, P.; Lewis, J.; Provaznik, J.; et al. Small-molecule-mediated chemical knock-down of MuRF1/MuRF2 and attenuation of diaphragm dysfunction in chronic heart failure. J. Cachexia Sarcopenia Muscle 2019, 10, 1102–1115. [Google Scholar] [CrossRef]
- Lala-Tabbert, N.; Lejmi-Mrad, R.; Timusk, K.; Fukano, M.; Holbrook, J.; St-Jean, M.; LaCasse, E.C.; Korneluk, R.G. Targeted ablation of the cellular inhibitor of apoptosis 1 (cIAP1) attenuates denervation-induced skeletal muscle atrophy. Skelet. Muscle 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Saha, A.K.; Xu, X.J.; Lawson, E.; Deoliveira, R.; Brandon, A.E.; Kraegen, E.W.; Ruderman, N.B. Downregulation of AMPK accompanies leucine- and glucose-induced increases in protein synthesis and insulin resistance in rat skeletal muscle. Diabetes 2010, 59, 2426–2434. [Google Scholar] [CrossRef]
- Haar, E.V.; Lee, S.; Bandhakavi, S.; Griffin, T.J.; Kim, D.-H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Luo, L.; Eash, J.; Ibebunjo, C.; Glass, D.J. The SCF-Fbxo40 Complex Induces IRS1 Ubiquitination in Skeletal Muscle, Limiting IGF1 Signaling. Dev. Cell. 2011, 21, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Tremblay, F.; Marette, A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J. Biol. Chem. 2001, 276, 38052–38060. [Google Scholar]
- Huang, J.; Dibble, C.C.; Matsuzaki, M.; Manning, B.D. The TSC1-TSC2 Complex Is Required for Proper Activation of mTOR Complex 2. Mol. Cell. Biol. 2008, 28, 4104–4115. [Google Scholar] [CrossRef]
- Glasgow, C.G.; Steagall, W.K.; Taveira-Dasilva, A.; Pacheco-Rodriguez, G.; Cai, X.; El-Chemaly, S.; Moses, M.; Darling, T.; Moss, J. Lymphangioleiomyomatosis (LAM): Molecular Insights into mTOR Regulation Lead to Targeted Therapies. Respir. Med. 2010, 104, S45–S58. [Google Scholar] [CrossRef]
- Polak, P.; Hall, M.N. mTOR and the control of whole body metabolism. Curr. Opin. Cell Biol. 2009, 21, 209–218. [Google Scholar] [CrossRef]
- Chantranupong, L.; Sabatini, D.M. The TORC1 pathway to protein destruction. Nature 2016, 536, 155–156. [Google Scholar] [CrossRef][Green Version]
- Verhees, K.J.P.; JSchols, A.M.W.; Kelders, M.C.J.M.; Op den Kamp, C.M.H.; van der Velden, J.L.J.; Langen, R.C.J. Glycogen synthase kinase-3β is required for the induction of skeletal muscle atrophy. Am. J. Physiol.-Cell Physiol. 2011, 301, 13. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Kops, G.J.P.L.; Ruiter ND de De Vries-Smits, A.M.M.; Powell, D.R.; Bos, J.L. Burgering BMTh. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 1999, 398, 630–634. [Google Scholar] [CrossRef]
- Rena, G.; Guo, S.; Cichy, S.C.; Unterman, T.G.; Cohen, P. Phosphorylation of the Transcription Factor Forkhead Family Member FKHR by Protein Kinase B. J. Biol. Chem. 1999, 274, 17179–17183. [Google Scholar] [CrossRef] [PubMed]
- Takaishi, H.; Konishi, H.; Matsuzaki, H.; Ono, Y.; Shirai, Y.; Saito, N.; Kitamura, T.; Ogawa, W.; Kasuga, M.; Kikkawa, U. Regulation of nuclear translocation of Forkhead transcription factor AFX by protein kinase B. Proc. Natl. Acad. Sci. USA 1999, 96, 11836–11841. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Inoki, K.; Lee, M.; Wright, E.; Khuong, A.; Khuong, A.; Sugiarto, S.; Garner, M.; Paik, J.; DePinho, R.A.; et al. MTORC1 promotes denervation-induced muscle atrophy through a mechanism involving the activation of FoxO and E3 ubiquitin ligases. Sci. Signal. 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Tang, H.; Inoki, K.; Brooks, S.V.; Okazawa, H.; Lee, M.; Wang, J.; Michael Kim, M.; Catherine L Kennedy, C.L.; Macpherson, P.C.D.; Ji, X.; et al. mTORC1 underlies age-related muscle fiber damage and loss by inducing oxidative stress and catabolism. Aging Cell. 2019, 18, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ham, A.S.; Chojnowska, K.; Tintignac, L.A.; Lin, S.; Schmidt, A.; Ham, D.J.; Sinnreich, M.; Rüegg, M.A. mTORC1 signalling is not essential for the maintenance of muscle mass and function in adult sedentary mice. J. Cachexia Sarcopenia Muscle 2020, 11, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Joassard, O.R.; Durieux, A.C.; Freyssenet, D.G. β2-Adrenergic agonists and the treatment of skeletal muscle wasting disorders. Int. J. Biochem. Cell Biol. 2013, 45, 2309–2321. [Google Scholar] [CrossRef]
- Silveira, W.A.; Gonçalves, D.A.; Machado, J.; Lautherbach, N.; Lustrino, D.; Paula-Gomes, S.; Pereira, M.G.; Miyabara, E.H.; Sandri, M.; Isis C Kettelhut, I.C.; et al. cAMP-dependent protein kinase inhibits FoxO activity and regulates skeletal muscle plasticity in mice. FASEB J. 2020, 34, 12946–12962. [Google Scholar] [CrossRef]
- Arcaro, C.A.; Assis, R.P.; Zanon, N.M.; Paula-Gomes, S.; Navegantes, L.C.C.; Kettelhut, I.C.; Brunetti, I.L.; Baviera, A.M. Involvement of cAMP/EPAC/Akt signaling in the antiproteolytic effects of pentoxifylline on skeletal muscles of diabetic rats. J. App. Physiol. 2018, 124, 704–716. [Google Scholar] [CrossRef]
- Baviera, A.M.; Zanon, N.M.; Navegantes, L.C.C.; Kettelhut, I.C. Involvement of cAMP/Epac/PI3K-dependent pathway in the antiproteolytic effect of epinephrine on rat skeletal muscle. Mol. Cell Endocrinol. 2010, 315, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, Y.; Umeki, D.; Mototani, Y.; Jin, H.; Cai, W.; Shiozawa, K.; Suita, K.; Saeki, Y.; Fujita, T.; Ishikawa, Y.; et al. Role of cyclic AMP sensor Epac1 in masseter muscle hypertrophy and myosin heavy chain transition induced by β2-adrenoceptor stimulation. J. Physiol. 2014, 592, 5461–5475. [Google Scholar] [CrossRef] [PubMed]
- Fedon, Y.; Bonnieu, A.; Gay, S.; Vernus, B.; Bacou, F.; Bernardi, H. Role and Function of Wnts in the Regulation of Myogenesis: When Wnt Meets Myostatin. In Skeletal Muscle-From Myogenesis to Clinical Relations; InTech: London, UK, 2012; p. 13. [Google Scholar] [CrossRef]
- von Maltzahn, J.; Chang, N.C.; Bentzinger, C.F.; Rudnicki, M.A. Wnt signaling in myogenesis. Trends Cell Biol. 2012, 22, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.D.; Wong, V.L.; Esser, K.A. Expression of β-catenin is necessary for physiological growth of adult skeletal muscle. Am. J. Physiol.-Cell Physiol. 2006, 291, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.D.; Esser, K.A. Wnt/β-catenin signaling activates growth-control genes during overload-induced skeletal muscle hypertrophy. Am. J. Physiol.-Cell Physiol. 2005, 289, 853–859. [Google Scholar] [CrossRef]
- Von Maltzahn, J.; Bentzinger, C.F.; Rudnicki, M.A. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat. Cell Biol. 2012, 14, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Poser, C.; von Maltzahn, J. Wnt7a Counteracts Cancer Cachexia. Mol. Ther. Oncolytics 2020, 16, 134–146. [Google Scholar] [CrossRef]
- von Maltzahn, J.; Renaud, J.M.; Parise, G.; Rudnicki, M.A. Wnt7a treatment ameliorates muscular dystrophy. Proc. Natl. Acad. Sci. USA 2012, 109, 20614–20619. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; von Maltzahn, J.; Dumont, N.A.; Stark, D.A.; Wang, Y.X.; Nhan, K.; Frenette, J.; Cornelison, D.D.W.; Rudnicki, A.M. Wnt7a stimulates myogenic stem cell motility and engraftment resulting in improved muscle strength. J. Cell Biol. 2014, 205, 97–111. [Google Scholar] [CrossRef]
- Fischer, M.; Rikeit, P.; Knaus, P.; Coirault, C. YAP-Mediated Mechanotransduction in Skeletal. Muscle Front. Physiol. 2016, 7. [Google Scholar] [CrossRef]
- Kirby, T.J. Mechanosensitive pathways controlling translation regulatory processes in skeletal muscle and implications for adaptation. J. Appl. Physiol. 2019, 127, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Biressi, S.; Miyabara, E.H.; Gopinath, S.D.; MCarlig, P.M.; Rando, T.A. A Wnt-TGF 2 axis induces a fibrogenic program in muscle stem cells from dystrophic mice. Sci. Transl. Med. 2014, 6, 176–267. [Google Scholar] [CrossRef]
- Brack, A.S.; Conboy, M.J.; Roy, S.; Lee, M.; Kuo, C.J.; Keller, C.; Rando, T.A. Increased Wnt Signaling During Aging Alters Muscle Stem Cell Fate and Increases Fibrosis. Science 2007, 317, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Musarò, A.; McCullagh, K.J.A.; Naya, F.J.; Olson, E.N.; Rosenthal, N. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 1999, 400, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Wilson, T.K.; Reddy, R.N.; Bailey, J.L.; Zheng, B.; Ordas, R.; Gooch, J.L.; Price, S.R. Calcineurin signaling and PGC-1α expression are suppressed during muscle atrophy due to diabetes. Biochim. Biophys. Acta-Mol. Cell Res. 2010, 1803, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, K.; Yamaguchi, A. The functional role of calcineurin in hypertrophy, regeneration, and disorders of skeletal muscle. J. Biomed. Biotechnol. 2010, 2010, 721219. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.B.; Woodworth-Hobbs, M.E.; Zheng, B.; Rahnert, J.A.; Blount, M.A.; Gooch, J.L.; Searles, C.D.; Price, S.R. miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am. J. Physiol.-Cell Physiol. 2014, 306, C551–C558. [Google Scholar] [CrossRef]
- Delacroix, C.; Hyzewicz, J.; Lemaitre, M.; Friguet, B.; Li, Z.; Klein, A.; Furling, D.; Agbulut, O.; Ferry, A. Improvement of Dystrophic Muscle Fragility by Short-Term Voluntary Exercise through Activation of Calcineurin Pathway in mdx Mice. Am. J. Pathol. 2018, 188, 2662–2673. [Google Scholar] [CrossRef]
- Lara-Pezzi, E.; Winn, N.; Paul, A.; McCullagh, K.; Slominsky, E.; Santini, M.P.; Mourkioti, F.; Sarathchandra, P.; Fukushima, S.; Suzuki, K.; et al. A naturally occurring calcineurin variant inhibits FoxO activity and enhances skeletal muscle regeneration. J. Cell Biol. 2007, 179, 1205–1218. [Google Scholar] [CrossRef]
- Watt, K.I.; Goodman, C.A.; Hornberger, T.A.; Gregorevic, P. The Hippo Signaling Pathway in the Regulation of Skeletal Muscle Mass and Function. Exerc Sport. Sci. Rev. 2018, 46, 92–96. [Google Scholar] [CrossRef]
- Hulmi, J.J.; Oliveira, B.M.; Silvennoinen, M.; Hoogaars, W.M.H.; Ma, H.; Pierre, P.; Pasternack, A.; Kainulainen, H.; Ritvos, O. Muscle protein synthesis, mTORC1/MAPK/Hippo signaling, and capillary density are altered by blocking of myostatin and activins. Am. J. Physiol.-Endocrinol. Metab. 2013, 304, E41–E50. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.A.; Dietz, J.M.; Jacobs, B.L.; McNally, R.M.; You, J.S.; Hornberger, T.A. Yes-Associated Protein is up-regulated by mechanical overload and is sufficient to induce skeletal muscle hypertrophy. FEBS Lett. 2015, 589, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Watt, K.I.; Turner, B.J.; Hagg, A.; Zhang, X.; Davey, J.R.; Qian, H.; Beyer, C.; Winbanks, C.E.; Harvey, K.F.; Gregorevic, P. The Hippo pathway effector YAP is a critical regulator of skeletal muscle fibre size. Nat Commun. 2015, 6, 6048. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Attisano, L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 47–63. [Google Scholar] [CrossRef]
- Qin, H.; Chan, M.W.; Liyanarachchi, S.; Balch, C.; Potter, D.; Souriraj, I.J.; Cheng, A.S.L.; Agosto-Perez, F.J.; Nikonova, E.V.; Yan, P.S. An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst. Biol. 2009, 3, 73. [Google Scholar] [CrossRef]
- Amirouche, A.; Durieux, A.-C.; Banzet, S.; Koulmann, N.; Bonnefoy, R.; Mouret, C.; Bigard, X.; Peinnequin, A.; Freyssenet, D. Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology 2009, 150, 286–294. [Google Scholar] [CrossRef]
- Bollinger, L.M.; Witczak, C.A.; Houmard, J.A.; Brault, J.J. SMAD3 augments FoxO3-induced MuRF-1 promoter activity in a DNA-binding-dependent manner. Am. J. Physiol.-Cell Physiol. 2014, 307, 278–287. [Google Scholar] [CrossRef]
- Abrigo, J.; Rivera, J.C.; Simon, F.; Cabrera, D.; Cabello-Verrugio, C. Transforming growth factor type beta (TGF-β) requires reactive oxygen species to induce skeletal muscle atrophy. Cell Signal. 2016, 28, 366–376. [Google Scholar] [CrossRef]
- Latres, E.; Mastaitis, J.; Fury, W.; Miloscio, L.; Trejos, J.; Pangilinan, J.; Okamoto, H.; Cavino, K.; Na, E.; Papatheodorou, A.; et al. Activin A more prominently regulates muscle mass in primates than does GDF8. Nat. Commun. 2017, 8, 15153. [Google Scholar] [CrossRef]
- Chen, J.L.; Walton, K.L.; Qian, H.; Colgan, T.D.; Hagg, A.; Watt, M.J.; Harrison, C.A.; Gregorevic, P. Differential Effects of IL6 and Activin A in the Development of Cancer-Associated Cachexia. Cancer Res. 2016, 76, 5372–5382. [Google Scholar] [CrossRef]
- Chen, J.L.; Walton, K.; Winbanks, C.E.; Murphy, K.T.; Thomson, R.E.; Makanji, Y.; Qian, H.; Lynch, G.S.; Harrison, C.A.; Gregorevic, P. Elevated expression of activins promotes muscle wasting and cachexia. FASEB J. 2014, 28, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Zhang, G.; Sin, K.W.T.; Liu, Z.; Lin, R.-K.; Li, M.; Li, Y.-P. Activin A induces skeletal muscle catabolism via p38β mitogen-activated protein kinase. J. Cachexia Sarcopenia Muscle 2017, 8, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Garg, K.; Corona, B.T.; Walters, T.J. Therapeutic strategies for preventing skeletal muscle fibrosis after injury. Front. Pharmacol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.L.; Johnson, K.E.; Harrison, C.A. Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef]
- Ma, Z.-Y.; Zhong, Z.-G.; Qiu, M.-Y.; Zhong, Y.-H.; Zhang, W.-X. TGF-β1 activates the canonical NF-κB signaling to promote cell survival and proliferation in dystrophic muscle fibroblasts in vitro. Biochem. Biophys. Res. Commun. 2016, 471, 576–581. [Google Scholar] [CrossRef]
- Sartori, R.; Schirwis, E.; Blaauw, B.; Bortolanza, S.; Zhao, J.; Enzo, E.; Stantzou, A.; Mouisel, E.; Toniolo, L.; Ferry, A.; et al. BMP signaling controls muscle mass. Nat. Genet. 2013, 45, 1309–1321. [Google Scholar] [CrossRef]
- Sartori, R.; Gregorevic, P.; Sandri, M. TGFβ and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends Endocrinol. Metab. 2014, 25, 464–471. [Google Scholar] [CrossRef]
- Winbanks, C.E.; Chen, J.L.; Qian, H.; Liu, Y.; Bernardo, B.C.; Beyer, C.; Watt, K.I.; Thomson, R.E.; Connor, T.; Turner, B.J.; et al. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J. Cell Biol. 2013, 203, 345–357. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Hitachi, K.; Nakatani, M.; Tsuchida, K. Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle. Non-Coding RNA 2019, 5, 33. [Google Scholar] [CrossRef]
- Neppl, R.L.; Wu, C.-L.; Walsh, K. lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell Biol. 2017, 216, 3497–3507. [Google Scholar] [CrossRef]
- McCarthy, J.J.; Murach, K.A. Anabolic and Catabolic Signaling Pathways That Regulate Skeletal Muscle Mass. In Nutrition and Enhanced Sports Performance: Muscle Building, Endurance, and Strength; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar] [CrossRef]
- Sanchez, A.M.J.; Candau, R.B.; Csibi, A.; Pagano, A.F.; Raibon, A.; Bernardi, H. The role of AMP-activated protein kinase in the coordination of skeletal muscle turnover and energy homeostasis. Am. J. Physiol.-Cell Physiol. 2012, 303, C475–C485. [Google Scholar] [CrossRef]
- Zungu, M.; Schisler, J.C.; Essop, M.F.; McCudden, C.; Patterson, C.; Willis, M.S. Regulation of AMPK by the ubiquitin proteasome system. Am. J. Pathol. 2011, 178, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Candau, R.; Bernardi, H. Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise. Cells 2019, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Egawa, T.; Goto, A.; Ohno, Y.; Yokoyama, S.; Ikuta, A.; Suzuki, M.; Sugiura, T.; Ohira, Y.; Yoshioka, T.; Hayashi, T.; et al. Involvement of AMPK in regulating slow-twitch muscle atrophy during hindlimb unloading in mice. Am. J. Physiol.-Endocrinol. Metab. 2015, 309, E651–E662. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.M. The Role of AMPK in the Regulation of Skeletal Muscle Size, Hypertrophy, and Regeneration. Int. J. Mol. Sci. 2018, 19, 3125. [Google Scholar] [CrossRef]
- Cai, D.; Frantz, J.D.; Tawa, N.E.; Melendez, P.A.; Oh, B.C.; Lidov, H.G.W.; Hasselgren, P.-O.; Frontera, W.R.; Lee, J.; Glass, D.J.; et al. IKKβ/NF-κB activation causes severe muscle wasting in mice. Cell 2004, 119, 285–298. [Google Scholar] [CrossRef]
- Enwere, E.K.; Boudreault, L.; Holbrook, J.; Timusk, K.; Earl, N.; LaCasse, E.; Renaud, J.-M.; Korneluk, R.G. Loss of cIAP1 attenuates soleus muscle pathology and improves diaphragm function in mdx mice. Hum. Mol. Genet. 2013, 22, 867–878. [Google Scholar] [CrossRef]
- Enwere, E.K.; Holbrook, J.; Lejmi-Mrad, R.; Vineham, J.; Timusk, K.; Sivaraj, B.; Isaac, M.; Uehling, D.; Al-awar, R.; LaCasse, E.; et al. TWEAK and cIAP1 Regulate Myoblast Fusion Through the Noncanonical NF- B Signaling Pathway. Sci. Signal. 2012, 5, ra75. [Google Scholar] [CrossRef]
- Li, H.; Malhotra, S.; Kumar, A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J. Mol. Med. 2008, 86, 1113–1126. [Google Scholar] [CrossRef]
- Li, Y.P.; Reid, M.B. NF-κB mediates the protein loss induced by TNF-α in differentiated skeletal muscle myotubes. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2000, 279, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Ogura, Y.; Kumar, A. TWEAK/Fn14 Signaling Axis Mediates Skeletal Muscle Atrophy and Metabolic Dysfunction. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Shih, V.F.S.; Tsui, R.; Caldwell, A.; Hoffmann, A. A single NFκB system for both canonical and non-canonical signaling. Cell Res. 2011, 21, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Mourkioti, F.; Kratsios, P.; Luedde, T.; Song, Y.H.; Delafontaine, P.; Adami, R.; Parente, V.; Bottinelli, R.; Pasparakis, M.; Rosenthal, N. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J. Clin. Investig. 2006, 116, 2945–2954. [Google Scholar] [CrossRef]
- Hunter, R.B.; Kandarian, S.C. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J. Clin. Investig. 2004, 114, 1504–1511. [Google Scholar] [CrossRef]
- Agustí, A.; Morlá, M.; Sauleda, J.; Saus, C.; Busquets, X. NF-κB activation and iNOS upregulation in skeletal muscle of patients with COPD and low body weight. Thorax 2004, 59, 483–487. [Google Scholar] [CrossRef]
- Adams, V.; Späte, U.; Kränkel, N.; Schulze, P.C.; Linke, A.; Schuler, G.; Hambrecht, R. Nuclear factor-kappa B activation in skeletal muscle of patients with chronic heart failure: Correlation with the expression of inducible nitric oxide synthase. Eur. J. Cardiovasc. Prev. Rehabil. 2003, 10, 273–277. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 1–9. [Google Scholar] [CrossRef]
- Lamothe, B.; Besse, A.; Campos, A.D.; Webster, W.K.; Wu, H.; Darnay, B.G. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J. Biol. Chem. 2007, 282, 4102–4112. [Google Scholar] [CrossRef] [PubMed]
- Hindi, S.M.; Sato, S.; Choi, Y.; Kumar, A. Distinct roles of TRAF6 at early and late stages of muscle pathology in the mdx model of duchenne muscular dystrophy. Hum. Mol. Genet. 2014, 23, 1492–1505. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Enwere, E.K.; Lacasse, E.C.; Adam, N.J.; Korneluk, R.G. Role of the TWEAK-Fn14-cIAP1-NF-κB Signaling Axis in the Regulation of Myogenesis and Muscle Homeostasis. Front. Immunol. 2014, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Strickson, S. The role of hybrid ubiquitin chains in the MyD88 and other innate immune signalling pathways. Cell Death Differ. 2017, 24, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Shared Principles in NF-κB Signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Gensler, L.S. Glucocorticoids: Complications to Anticipate and Prevent. Neurohospitalist 2013, 3, 92–97. [Google Scholar] [CrossRef]
- Hardy, R.S.; Raza, K.; Cooper, M.S. Therapeutic glucocorticoids: Mechanisms of actions in rheumatic diseases. Nat. Rev. Rheumatol. 2020, 16, 133–144. [Google Scholar] [CrossRef]
- Revollo, J.R.; Cidlowski, J.A. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann. N. Y. Acad. Sci. 2009, 1179, 167–178. [Google Scholar] [CrossRef]
- Bodine, S.C.; Furlow, J.D. Glucocorticoids and Skeletal Muscle. Adv. Exp. Med. Biol. 2015, 872, 145–176. [Google Scholar]
- Braun, T.P.; Marks, D.L. The regulation of muscle mass by endogenous glucocorticoids. Front. Physiol. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- 112. Fappi, A.; Neves, J.D.; Sanches, L.N.; Massaroto e Silva, P.V.; Sikusawa, G.Y.; Brandão, T.P.; Chadi, G.; Zanoteli, E. Skeletal Muscle Response to Deflazacort, Dexamethasone and Methylprednisolone. Cells 2019, 8, 406. [Google Scholar] [CrossRef]
- Fry, C.S.; Nayeem, S.Z.; Dillon, E.L.; Sarkar, P.S.; Tumurbaatar, B.; Urban, R.J.; Wright, T.J.; Sheffield-Moore, S.; Tilton, R.G.; Choudhary, S. Glucocorticoids increase skeletal muscle NF-κB inducing kinase (NIK): Links to muscle atrophy. Physiol. Rep. 2016, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.Y.; Richardson, D.; Cregor, M.; Davis, H.M.; Au, E.D.; McAndrews, K.; Zimmers, T.A.; Organ, J.M.; Peacock, M.; Plotkin, L.I.; et al. Glucocorticoids induce bone and muscle atrophy by tissue-specific mechanisms upstream of E3 ubiquitin ligases. Endocrinology 2017, 158, 664–677. [Google Scholar] [PubMed]
- Cea, L.A.; Balboa, E.; Puebla, C.; Vargas, A.A.; Cisterna, B.A.; Escamilla, R.; Regueira, T.; Sáez, J.C. Dexamethasone-induced muscular atrophy is mediated by functional expression of connexin-based hemichannels. Biochim. Biophys. Acta-Mol. Basis Dis. 2016, 1862, 1891–1899. [Google Scholar] [CrossRef]
- Adhikary, S.; Kothari, P.; Choudhary, D.; Tripathi, A.K.; Trivedi, R. Glucocorticoid aggravates bone micro-architecture deterioration and skeletal muscle atrophy in mice fed on high-fat diet. Steroids 2019, 149, 108416. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Agon, K.W.; Capel, A.J.; Fleming, J.W.; Player, D.J.; Martin, N.R.W.; Lewis, M.P. Mechanical loading of tissue engineered skeletal muscle prevents dexamethasone induced myotube atrophy. J. Muscle Res. Cell Motil. 2020. [Google Scholar] [CrossRef]
- Powers, S.K.; Morton, A.B.; Hyatt, H.; Hinkley, M.J. The Renin-Angiotensin System and Skeletal Muscle Exerc. Sport Sci. Rev. 2018, 46, 205–214. [Google Scholar]
- Du Bois, P.; Tortola, C.P.; Lodka, D.; Kny, M.; Schmidt, F.; Song, K.; Schmidt, S.; Bassel-Duby, R.; Olson, E.N.; Fielitz, J. Angiotensin II Induces Skeletal Muscle Atrophy by Activating TFEB-Mediated MuRF1 Expression. Circ. Res. 2015, 117, 424–436. [Google Scholar] [CrossRef]
- Rezk, B.M.; Yoshida, T.; Semprun-Prieto, L.; Higashi, Y.; Sukhanov, S.; Delafontaine, P. Angiotensin II infusion induces marked diaphragmatic skeletal muscle atrophy. PLoS ONE 2012, 7, e30276. [Google Scholar] [CrossRef]
- Sugiyama, M.; Yamaki, A.; Furuya, M.; Inomata, N.; Minamitake, Y.; Ohsuye, K.; Kangawa, K. Ghrelin improves body weight loss and skeletal muscle catabolism associated with angiotensin II-induced cachexia in mice. Regul. Pept. 2012, 178, 21–28. [Google Scholar] [CrossRef]
- Tabony, A.M.; Yoshida, T.; Galvez, S.; Higashi, Y.; Sukhanov, S.; Chandrasekar, B.; Mitch, W.E.; Delafontaine, P. Angiotensin II Upregulates Protein Phosphatase 2Cα and Inhibits AMP-Activated Protein Kinase Signaling and Energy Balance Leading to Skeletal Muscle Wasting. Hypertension 2011, 58, 643–649. [Google Scholar] [CrossRef]
- Yoshida, T.; Semprun-Prieto, L.; Sukhanov, S.; Delafontaine, P. IGF-1 prevents ANG II-induced skeletal muscle atrophy via Akt- and Foxo-dependent inhibition of the ubiquitin ligase atrogin-1 expression. Am. J. Physiol.-Heart Circ. Physiol. 2010, 298, H1565–H1570. [Google Scholar] [CrossRef] [PubMed]
- Aravena, J.; Abrigo, J.; Gonzalez, F.; Aguirre, F.; Gonzalez, A.; Simon, F.; Cabello-Verrugio, C. Angiotensin (1-7) decreases myostatin-induced NF-kb signaling and skeletal muscle atrophy. Int. J. Mol. Sci. 2020, 21, 1167. [Google Scholar] [CrossRef] [PubMed]
- Meneses, C.; Morales, M.G.; Abrigo, J.; Simon, F.; Brandan, E.; Cabello-Verrugio, C. The angiotensin-(1–7)/Mas axis reduces myonuclear apoptosis during recovery from angiotensin II-induced skeletal muscle atrophy in mice. Pflug. Arch.-Eur. J. Physiol. 2015, 467, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.G.; Abrigo, J.; Acuña, M.J.; Santos, R.A.; Bader, M.; Brandan, E.; Simon, F.; Olguin, H.; Cabrera, D.; Cabello-Verrugio, C. Angiotensin-(1-7) attenuates disuse skeletal muscle atrophy in mice via its receptor. Mas. Dis. Model. Mech. 2016, 9, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Echeverría-Rodríguez, O.; Gallardo-Ortíz, I.A.; Valle-Mondragón, L.D.; Villalobos-Molina, R. Angiotensin-(1-7) participates in enhanced skeletal muscle insulin sensitivity after a bout of exercise. J. Endocr. Soc. 2020, 4, 1–11. [Google Scholar] [CrossRef]
- Ábrigo, J.; Simon, F.; Cabrera, D.; Cabello-Verrugio, C. Angiotensin-(1-7) Prevents Skeletal Muscle Atrophy Induced by Transforming Growth Factor Type Beta (TGF-β) via Mas Receptor Activation. Cell. Physiol. Biochem. 2016, 40, 27–38. [Google Scholar] [CrossRef]
- Yan, F.; Yuan, Z.; Wang, N.; Carey, R.M.; Aylor, K.W.; Chen, L.; Zhou, X.; Liu, Z. Direct activation of angiotensin II type 2 receptors enhances muscle microvascular perfusion, oxygenation, and insulin delivery in male rats. Endocrinology 2018, 159, 685–695. [Google Scholar] [CrossRef]
- Bahat, G. Covid-19 and the Renin Angiotensin System: Implications for the Older Adults. J. Nutr. Health Aging 2020, 24, 699–704. [Google Scholar] [CrossRef]
- Sanders, P.M.; Russell, S.T.; Tisdale, M.J. Angiotensin II directly induces muscle protein catabolism through the ubiquitin-proteasome proteolytic pathway and may play a role in cancer cachexia. Br. J. Cancer 2005, 93, 425–434. [Google Scholar] [CrossRef]
- Song, Y.-H.; Li, Y.; Du, J.; Mitch, W.E.; Rosenthal, N.; Delafontaine, P. Muscle-specific expression of IGF-1 blocks angiotensin II-induced skeletal muscle wasting. J. Clin. Investig. 2005, 115, 451–458. [Google Scholar] [CrossRef]
- Belizário, J.E.; Fontes-Oliveira, C.C.; Borges, J.P.; Kashiabara, J.A.; Vannier, E. Skeletal muscle wasting and renewal: A pivotal role of myokine IL-6. SpringerPlus 2016, 5, 619. [Google Scholar] [CrossRef] [PubMed]
- Moresi, V.; Adamo, S.; Berghella, L. The JAK/STAT pathway in skeletal muscle pathophysiology. Front. Physiol. 2019, 30, 500. [Google Scholar] [CrossRef]
- Guadagnin, E.; Mázala, D.; Chen, Y.W. STAT3 in skeletal muscle function and disorders. Int. J. Mol. Sci. 2018, 19, 2265. [Google Scholar] [CrossRef] [PubMed]
- Mashili, F.; Chibalin, A.V.; Krook, A.; Zierath, J.R. Constitutive STAT3 phosphorylation contributes to skeletal muscle insulin resistance in type 2 diabetes. Diabetes 2013, 62, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Choi, S.E.; Ha, E.S.; Jung, J.G.; Han, S.J.; Kim, H.J.; Kim, D.J.; Kang, Y.; Lee, K.W. IL-6 induction of TLR-4 gene expression via STAT3 has an effect on insulin resistance in human skeletal muscle. Acta Diabetol. 2013, 50, 189–200. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, J.; Dong, Y.; Tweardy, D.J.; Dong, Y.; Garibotto, G.; Mitch, W.E. Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab. 2013, 18, 368–379. [Google Scholar] [CrossRef]
- Silva, K.A.S.; Dong, J.; Dong, Y.; Dong, Y.; Schor, N.; Tweardy, D.J.; Zhang, L.; Mitch, W.E. Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J. Biol. Chem. 2015, 290, 11177–11187. [Google Scholar] [CrossRef]
- Abid, H.; Ryan, Z.C.; Delmotte, P.; Sieck, G.C.; Lanza, I.R. Extramyocellular interleukin-6 influences skeletal muscle mitochondrial physiology through canonical JAK/STAT signaling pathways. FASEB J. 2020, 34, 14458–14472. [Google Scholar] [CrossRef]
- Calixto, J.B.; Medeiros, R.; Fernandes, E.S.; Ferreira, J.; Cabrini, D.A.; Campos, M.M. Kinin B 1 receptors: Key G-protein-coupled receptors and their role in inflammatory and painful processes. Br. J. Pharmacol. 2004, 143, 803–818. [Google Scholar] [CrossRef]
- Parreiras-e-Silva, L.T.; Reis, R.I.; Santos, G.A.; Pires-Oliveira, M.; Pesquero, J.B.; Gomes, M.D.; Godinho, R.O.; Costa-Neto, C.M. The kinin B1 receptor regulates muscle-specific E3 ligases expression and is involved in skeletal muscle mass control. Clin. Sci. 2014, 127, 185–194. [Google Scholar] [CrossRef]
- de Picoli Souza, K.; Batista, E.C.; Silva, E.D.; Reis, F.C.; Silva, S.M.A.; Araujo, R.C.; Luz, J.; Santos, E.L.; Pesquero, J.B. Effect of kinin B2 receptor ablation on skeletal muscle development and myostatin gene expression. Neuropeptides 2010, 44, 209–214. [Google Scholar] [CrossRef]
- Popadic Gacesa, J.Z.; Momcilovic, M.; Veselinovic, I.; Brodie, D.A.; Grujic, N.G. Bradykinin type 2 receptor -9/-9 genotype is associated with triceps brachii muscle hypertrophy following strength training in young healthy men. BMC Musculoskelet. Disord. 2012, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.A.; Knotts, T.A.; Wang, L.P.; Li, G.; Dobrowsky, R.T.; Florant, G.L.; Summers, S.A. A role for ceramide, but not diacylglycerol, in the antagonism of insulin signal transduction by saturated fatty acids. J. Biol. Chem. 2003, 278, 10297–10303. [Google Scholar] [CrossRef]
- Hyde, R.; Hajduch, E.; Powell, D.J.; Taylor, P.M.; Hundal, H.S. Ceramide down-regulates System A amino acid transport and protein synthesis in rat skeletal muscle cells. FASEB J. 2005, 19, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Orsini, M.; Chateauvieux, S.; Rhim, J.; Gaigneaux, A.; Cheillan, D.; Christov, C.; Dicato, M.; Morceau, F.; Diederich, M. Sphingolipid-mediated inflammatory signaling leading to autophagy inhibition converts erythropoiesis to myelopoiesis in human hematopoietic stem/progenitor cells. Cell Death Diff. 2019, 26, 1796–1812. [Google Scholar] [CrossRef] [PubMed]
- Tardif, N.; Salles, J.; Guillet, C.; Tordjman, J.; Reggio, S.; Landrier, J.; Giraudet, C.; Patrac, V.; Bertrand-Michel, J.; Migne, C. Muscle ectopic fat deposition contributes to anabolic resistance in obese sarcopenic old rats through e IF 2α activation. Aging Cell 2014, 13, 1001–1011. [Google Scholar] [CrossRef]
- De Larichaudy, J.; Zufferli, A.; Serra, F.; Isidori, A.M.; Naro, F.; Dessalle, K.; Desgeorges, M.; Piraud, M.; Cheillan, D.; Vidal, H.; et al. TNF-α- and tumor-induced skeletal muscle atrophy involves sphingolipid metabolism. Skelet. Muscle 2012, 2, 1–19. [Google Scholar] [CrossRef]
- Rivas, D.A.; McDonald, D.J.; Rice, N.P.; Haran, P.H.; Dolnikowski, G.G.; Fielding, R.A. Diminished anabolic signaling response to insulin induced by intramuscular lipid accumulation is associated with inflammation in aging but not obesity. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2016, 310, R561–R569. [Google Scholar] [CrossRef]
- Rivas, D.A.; Morris, E.P.; Haran, P.H.; Pasha, E.P.; Da Silva Morais, M.; Dolnikowski, G.G.; Phillips, E.M.; Fielding, R.A. Increased ceramide content and NFκB signaling may contribute to the attenuation of anabolic signaling after resistance exercise in aged males. J. Appl. Physiol. 2012, 113, 1727–1736. [Google Scholar] [CrossRef]
- Zanin, M.; Germinario, E.; Dalla Libera, L.; Sandonà, D.; Sabbadini, R.A.; Betto, R.; Danieli-Betto, D. Trophic action of sphingosine 1-phosphate in denervated rat soleus muscle. Am. J. Physiol.-Cell Physiol. 2008, 294, 36–46. [Google Scholar] [CrossRef]
- Pierucci, F.; Frati, A.; Battistini, C.; Matteini, F.; Iachini, M.C.; Vestri, A.; Penna, F.; Costelli, P.; Meacci, E. Involvement of released sphingosine 1-phosphate/sphingosine 1-phosphate receptor axis in skeletal muscle atrophy. Biochim. Biophys. Acta-Mol. Basis Dis. 2018, 1864, 3598–3614. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Agarwal, R.; March, D.; Rothenberg, A.; Voigt, C.; Tebbets, J.; Huard, J.; Weiss, K. Notch Signaling Mediates Skeletal Muscle Atrophy in Cancer Cachexia Caused by Osteosarcoma. Sarcoma 2016, 3758162. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Shan, L.; Deng, J.X.; Luo, L.L.; Huang, Q.S. Role of the Notch Signaling Pathway in Fibrosis of Denervated Skeletal Muscle. Curr. Med. Sci. 2019, 39, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Yao, S.; Qiao, R.F.; Levine, A.C.; Kirschenbaum, A.; Pan, J.; Wu, Y.; Qin, W.; Bauman, W.A.; Cardozo, C.P. Nandrolone reduces activation of Notch signaling in denervated muscle associated with increased Numb expression. Biochem. Biophys. Res. Commun. 2011, 414, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, Y.; Zhao, W.; Wu, Y.; Pan, J.; Bauman, W.A.; Cardozo, C. Effects of nandrolone on denervation atrophy depend upon time after nerve transection. Muscle Nerve 2008, 37, 42–49. [Google Scholar] [CrossRef]
- Khayrullin, A.; Smith, L.; Mistry, D.; Dukes, A.; Pan, Y.A.; Hamrick, M.W. Chronic alcohol exposure induces muscle atrophy (myopathy) in zebrafish and alters the expression of microRNAs targeting the Notch pathway in skeletal muscle. Biochem. Biophys. Res. Commun. 2016, 479, 590–595. [Google Scholar] [CrossRef]
- Domingues-Faria, C.; Chanet, A.; Salles, J.; Berry, A.; Giraudet, C.; Patrac, V.; Denis, P.; Bouton, K.; Goncalves-Mendes, N.; Vasson, M.-P.; et al. Vitamin D deficiency down-regulates Notch pathway contributing to skeletal muscle atrophy in old wistar rats. Nutr. Metab. 2014, 11, 1–13. [Google Scholar] [CrossRef]
- Hori, K.; Sen, A.; Artavanis-Tsakonas, S. Notch signaling at a glance. J. Cell Sci. 2013, 126, 2135–2140. [Google Scholar] [CrossRef]
- von Grabowiecki, Y.; Licona, C.; Palamiuc, L.; Abreu, P.; Vidimar, V.; Coowar, D.; Mellitzer, G.; Gaiddon, C. Regulation of a Notch3-Hes1 pathway and protective effect by a tocopherol-omega alkanol chain derivative in muscle atrophy. J. Pharmacol. Exp. Therap. 2015, 352, 23–32. [Google Scholar] [CrossRef]
- Powers, S.K.; Morton, A.B.; Ahn, B.; Smuder, A.J. Redox control of skeletal muscle atrophy. Free Radic. Biol. Med. 2016, 98, 208–217. [Google Scholar] [CrossRef]
- Abrigo, J.; Elorza, A.A.; Riedel, C.A.; Vilos, C.; Simon, F.; Cabrera, D.; Estrada, L.; Cabello-Verrugio, C. Role of oxidative stress as key regulator of muscle wasting during cachexia. Oxid Med. Cell. Longev. 2018, 28, 2063179. [Google Scholar] [CrossRef]
- Passey, S.L.; Hansen, M.J.; Bozinovski, S.; McDonald, C.F.; Holland, A.E.; Vlahos, R. Emerging therapies for the treatment of skeletal muscle wasting in chronic obstructive pulmonary disease. Pharmacol. Therapeut. 2016, 166, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Leitner, L.M.; Wilson, R.J.; Yan, Z.; Gödecke, A. Reactive Oxygen Species/Nitric Oxide Mediated Inter-Organ Communication in Skeletal Muscle Wasting Diseases. Antioxid. Redox Signal. 2017, 26, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Pomiès, P.; Blaquière, M.; Maury, J.; Mercier, J.; Gouzi, F.; Hayot, M. Involvement of the FoxO1/MuRF1/Atrogin-1 Signaling Pathway in the Oxidative Stress-Induced Atrophy of Cultured Chronic Obstructive Pulmonary Disease Myotubes. PLoS ONE 2016, 11, e0160092. [Google Scholar] [CrossRef]
- Beyfuss, K.; Hood, D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018, 23, 100–117. [Google Scholar] [CrossRef]
- Mastrocola, R.; Reffo, P.; Penna, F.; Tomasinelli, C.E.; Boccuzzi, G.; Baccino, F.M.; Aragno, M.; Costelli, P. Muscle wasting in diabetic and in tumor-bearing rats: Role of oxidative stress. Free Radic. Biol. Med. 2008, 44, 584–593. [Google Scholar] [CrossRef]
- Rosa-Caldwell, M.E.; Greene, N.P. Muscle metabolism and atrophy: Let’s talk about sex. Biol. Sex Differ. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Xu, X.; Sarikas, A.; Dias-Santagata, D.C.; Dolios, G.; Lafontant, P.J.; Tsai, S.-C.; Zhu, W.; Nakajima, H.; Nakajima, H.-O.; Field, L.J.; et al. The CUL7 E3 Ubiquitin Ligase Targets Insulin Receptor Substrate 1 for Ubiquitin-Dependent Degradation. Mol. Cell. 2008, 30, 403–414. [Google Scholar] [CrossRef]
- Nakao, R.; Hirasaka, K.; Goto, J.; Ishidoh, K.; Yamada, C.; Ohno, A.; Okumura, Y.; Nonaka, I.; Yasutomo, K.; Baldwin, K.M.; et al. Ubiquitin Ligase Cbl-b Is a Negative Regulator for Insulin-Like Growth Factor 1 Signaling during Muscle Atrophy Caused by Unloading. Mol. Cell. Biol. 2009, 29, 4798–4811. [Google Scholar] [CrossRef]
- Nikawa, T.; Ishidoh, K.; Hirasaka, K.; Ishihara, I.; Ikemoto, M.; Kano, M.; Kominami, E.; Nonaka, I.; Ogawa, T.; Adams, G.R.; et al. Skeletal muscle gene expression in space-flown rats. FASEB J. 2004, 18, 522–524. [Google Scholar] [CrossRef]
- Uchida, T.; Sakashita, Y.; Kitahata, K.; Yamashita, Y.; Tomida, C.; Kimori, Y.; Komatsu, A.; Hirasaka, K.; Ohno, A.; Nakao, R.; et al. Reactive oxygen species upregulate expression of muscle atrophy-associated ubiquitin ligase Cbl-b in rat L6 skeletal muscle cells. Am. J. Physiol.-Cell Physiol. 2018, 314, C721–C731. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, Y.; Xu, J.; Zhang, Q.; Zhu, D. FBXO40, a gene encoding a novel muscle-specific F-box protein, is upregulated in denervation-related muscle atrophy. Gene 2007, 404, 53–60. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Z.; Wang, Y.; Tweardy, D.J.; Mitch, W.E. Stat3 activation induces insulin resistance via a muscle-specific E3 ubiquitin ligase Fbxo40. Am. J. Physiol.-Endocrinol. Metab. 2020, 318, E625–E635. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, P.; Gu, J.; Yu, Q.; Zhang, D. NEDD4-1 protects against ischaemia/reperfusion-induced cardiomyocyte apoptosis via the PI3K/Akt pathway. Apoptosis 2017, 22, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Trotman, L.C.; Koppie, T.; Alimonti, A.; Chen, Z.; Gao, Z.; Wang, J.; Erdjument-Bromage, H.; Tempst, P.; Cordon-Cardo, C.; et al. NEDD4-1 Is a Proto-Oncogenic Ubiquitin Ligase for PTEN. Cell 2007, 128, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Plant, P.J.; Correa, J.; Bain, A.; Takeda, M.; Kawabe, H.; Rotin, D.; Bain, J.R.; Batt, J.A.E. The Ubiquitin Ligase Nedd4-1 Participates in Denervation-Induced Skeletal Muscle Atrophy in Mice. PLoS ONE 2012, 7, e46427. [Google Scholar] [CrossRef]
- Paul, P.K.; Bhatnagar, S.; Mishra, V.; Srivastava, S.; Darnay, B.G.; Choi, Y.; Kumar, A. The E3 Ubiquitin Ligase TRAF6 Intercedes in Starvation-Induced Skeletal Muscle Atrophy through Multiple Mechanisms. Mol. Cell. Biol. 2012, 32, 1248–1259. [Google Scholar] [CrossRef]
- Paul, P.K.; Gupta, S.K.; Bhatnagar, S.; Panguluri, S.K.; Darnay, B.G.; Choi, Y.; Kumar, A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J. Cell Biol. 2010, 191, 1395–1411. [Google Scholar] [CrossRef]
- Hirata, Y.; Nomura, K.; Senga, Y.; Okada, Y.; Kobayashi, K.; Okamoto, S.; Minokoshi, Y.; Imamura, M.; Takeda, S.; Hosooka, T.; et al. Hyperglycemia induces skeletal muscle atrophy via a WWP1/KLF15 axis. JCI Insight 2019, 4, e124952. [Google Scholar] [CrossRef]
- Kudryashova, E.; Wu, J.; Havton, L.A.; Spencer, M.J. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 2009, 18, 1353–1367. [Google Scholar] [CrossRef]
- Kudryashova, E.; Struyk, A.; Mokhonova, E.; Cannon, S.C.; Spencer, M.J. The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum. Mol. Genet. 2011, 20, 3925–3932. [Google Scholar] [CrossRef]
- Peker, N.; Donipadi, V.; Sharma, M.; McFarlane, C.; Kambadur, R. Loss of Parkin impairs mitochondrial function and leads to muscle atrophy. Am. J. Physiol.-Cell Physiol. 2018, 315, C164–C185. [Google Scholar] [CrossRef] [PubMed]
- Leduc-Gaudet, J.P.; Reynaud, O.; Hussain, S.N.; Gouspillou, G. Parkin overexpression protects from ageing-related loss of muscle mass and strength. J. Physiol. 2019, 597, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Leduc-Gaudet, J.-P.; Mayaki, D.; Reynaud, O.; Broering, F.E.; Chaffer, T.J.; Hussain, S.N.A.; Gouspillou, G. Parkin Overexpression Attenuates Sepsis-Induced Muscle Wasting. Cells 2020, 9, 1454. [Google Scholar] [CrossRef]
- Milan, G.; Romanello, V.; Pescatore, F.; Armani, A.; Paik, J.-H.; Frasson, L.; Seydel, A.; Zhao, J.; Abraham, R.; Goldberg, A.L.; et al. Regulation of autophagy and the ubiquitin–proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 2015, 6, 6670. [Google Scholar] [CrossRef] [PubMed]
- Wirianto, M.; Yang, J.; Kim, E.; Gao, S.; Paudel, K.R.; Choi, J.M.; Choe, J.; Gloston, G.F.; Ademoji, P.; Parakramaweera, R.; et al. The GSK-3β-FBXL21 Axis Contributes to Circadian TCAP Degradation and Skeletal Muscle Function. Cell Rep. 2020, 32, 108140. [Google Scholar] [CrossRef]
- Hunt, L.C.; Stover, J.; Haugen, B.; Shaw, T.I.; Li, Y.; Pagala, V.R.; Finkelstein, D.; Berton, E.R.; Fan, Y.; Labelle, M.; et al. A Key Role for the Ubiquitin Ligase UBR4 in Myofiber Hypertrophy in Drosophila and Mice. Cell Rep. 2019, 28, 1268–1281.e6. [Google Scholar] [CrossRef]
- Hughes, D.C.; Turner, D.C.; Baehr, L.M.; Seaborne, R.A.; Viggars, M.; Jarvis, J.C.; Gorski, P.P.; Stewart, C.E.; Owens, D.J.; Bodine, S.C.; et al. Knockdown of the E3 Ubiquitin ligase UBR5 and its role in skeletal muscle anabolism. Am. J. Physiol.-Cell Physiol. 2020, 320, C45–C56. [Google Scholar] [CrossRef]
- Stana, F.; Vujovic, M.; Mayaki, D.; Leduc-Gaudet, J.P.; Leblanc, P.; Huck, L.; Hussain, S.N.A. Differential Regulation of the Autophagy and Proteasome Pathways in Skeletal Muscles in Sepsis. Crit. Care Med. 2017, 45, e971–e979. [Google Scholar] [CrossRef]
- Batt, J.; Bain, J.; Goncalves, J.; Michalski, B.; Plant, P.; Fahnestock, M.; Woodgett, J. Differential gene expression profiling of short and long term denervated muscle. FASEB J. 2006, 20, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Koncarevic, A.; Jackman, R.W.; Kandarian, S.C. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J. 2007, 21, 427–437. [Google Scholar] [CrossRef][Green Version]
- Yue, F.; Song, C.; Huang, D.; Narayanan, N.; Qiu, J.; Jia, Z.; Yuan, Z.; Oprescus, S.N.; Roseguini, B.T.; Deng, M.; et al. PTEN Inhibition Ameliorates Muscle Degeneration and Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Therap. 2020. [Google Scholar] [CrossRef]
- Kim, B.G.; Lee, J.H.; Yasuda, J.; Ryoo, H.M.; Cho, J.Y. Phospho-Smad1 modulation by nedd4 e3 ligase in BMP/TGF-β signaling. J. Bone Min. Res. 2011, 26, 1411–1424. [Google Scholar] [CrossRef]
- Li, J.; Yi, X.; Yao, Z.; Chakkalakal, J.V.; Xing, L.; Boyce, B.F. TNF Receptor-Associated Factor 6 Mediates TNFα-Induced Skeletal Muscle Atrophy in Mice During Aging. J. Bone Min. Res. 2020, 35, 1535–1548. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Gong, Y.; Qiu, J.; Chen, Y.; Ding, F.; Zhao, Q. TRAF6 inhibition rescues dexamethasone-induced muscle atrophy. Int. J. Mol. Sci. 2014, 15, 11126–11141. [Google Scholar] [CrossRef]
- Sun, H.; Qiu, J.; Chen, Y.; Yu, M.; Ding, F.; Gu, X. Proteomic and bioinformatic analysis of differentially expressed proteins in denervated skeletal muscle. Int. J. Mol. Med. 2014, 33, 1586–1596. [Google Scholar] [CrossRef]
- He, Q.; Qiu, J.; Dai, M.; Fang, Q.; Sun, X.; Gong, Y.; Ding, F.; Sun, H. MicroRNA-351 inhibits denervation-induced muscle atrophy by targeting TRAF6. Exp. Therap. Med. 2016, 12, 4029–4034. [Google Scholar] [CrossRef][Green Version]
- Qiu, J.; Wang, L.; Wang, Y.; Zhang, Q.; Ma, W.; Fang, Q.; Sun, H.; Ding, F. MicroRNA351 targeting TRAF6 alleviates dexamethasone-induced myotube atrophy. J. Thorac. Dis. 2018, 10, 6238–6246. [Google Scholar] [CrossRef]
- Qiu, J.; Zhu, J.; Zhang, R.; Liang, W.; Ma, W.; Zhang, Q.; Huang, Z.; Ding, F.; Sun, H. miR-125b-5p targeting TRAF6 relieves skeletal muscle atrophy induced by fasting or denervation. Ann. Transl. Med. 2019, 7, 456. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.K.; Kumar, A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy 2011, 7, 555–556. [Google Scholar] [CrossRef]
- Sun, Y.S.; Ye, Z.Y.; Qian, Z.Y.; Xu, X.D.; Hu, J.F. Expression of TRAF6 and ubiquitin mRNA in skeletal muscle of gastric cancer patients. J. Exp. Clin. Cancer Res. 2012, 31, 1–5. [Google Scholar] [CrossRef]
- Imamura, M.; Nakamura, A.; Mannen, H.; Takeda, S. Characterization of WWP1 protein expression in skeletal muscle of muscular dystrophy chickens. J. Biochem. 2016, 159, 171–179. [Google Scholar] [CrossRef]
- Shimizu, N.; Yoshikawa, N.; Ito, N.; Maruyama, T.; Suzuki, Y.; Takeda, S.I.; Nakae, J.; Tagata, Y.; Nishitani, S.; Takehana, K.; et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011, 13, 170–182. [Google Scholar] [CrossRef]
- Lee, J.O.; Lee, S.K.; Kim, N.; Kim, J.H.; You, G.Y.; Moon, J.W.; Jie, S.; Kim, S.J.; Lee, Y.W.; Kang, H.J.; et al. E3 ubiquitin ligase, WWP1, interacts with AMPKα2 and down-regulates its expression in skeletal muscle C2C12 cells. J. Biol. Chem. 2013, 288, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Frosk, P.; Weiler, T.; Nylen, E.; Sudha, T.; Greenberg, C.R.; Morgan, K.; Fujiwara, T.M.; Wrogemann, K. Limb-Girdle Muscular Dystrophy Type 2H Associated with Mutation in TRIM32, a Putative E3-Ubiquitin–Ligase Gene. Am. J. Hum. Genet. 2002, 70, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Mokhonova, E.I.; Avliyakulov, N.K.; Kramerova, I.; Kudryashova, E.; Haykinson, M.J.; Spencer, M.J. The E3 ubiquitin ligase TRIM32 regulates myoblast proliferation by controlling turnover of NDRG2. Hum. Mol. Genet. 2015, 24, 2873–2883. [Google Scholar] [CrossRef]
- Kudryashova, E.; Kramerova, I.; Spencer, M.J. Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J. Clin. Investig. 2012, 122, 1764–1776. [Google Scholar] [CrossRef] [PubMed]
- Servián-Morilla, E.; Cabrera-Serrano, M.; Rivas-Infante, E.; Carvajal, A.; Lamont, P.J.; Pelayo-Negro, A.L.; Ravenscroft, G.; Junckerstorff, R.; Dyke, J.M.; Fletcher, S.; et al. Altered myogenesis and premature senescence underlie human TRIM32-related myopathy. Acta Neuropathol. Commun. 2019, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, M.; Antonioli, M.; Fusco, C.; Liu, Y.; Mari, M.; Orhon, I.; Refolo, G.; Germani, F.; Corazzari, M.; Romagnoli, A.; et al. Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci. Adv. 2019. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Yang, J.; Li, G.; You, Q.; Han, W.; Li, T.; Gao, D.; Xie, X.; Lee, B.-H.; Du, J.; et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017, 27, 657–674. [Google Scholar] [CrossRef]
- Overå, K.S.; Garcia-Garcia, J.; Bhujabal, Z.; Jain, A.; Øvervatn, A.; Larsen, K.B.; Johansen, T.; Lamark, T.; Sjøttem, E. TRIM32, but not its muscular dystrophy-associated mutant, positively regulates and is targeted to autophagic degradation by p62/SQSTM1. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Alamdari, N.; Aversa, Z.; Castillero, E.; Hasselgren, P.-O. Acetylation and deacetylation—Novel factors in muscle wasting. Metabolism 2013, 62, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bertaggia, E.; Coletto, L.; Sandri, M. Posttranslational modifications control FoxO3 activity during denervation. Am. J. Physiol.-Cell Physiol. 2012, 302, C587–C596. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, J.-S.; Jeong, H.-J. Arginine methylation as a key post-translational modification in skeletal muscle homeostasis: A review. Precis. Future Med. 2019, 3, 139–145. [Google Scholar] [CrossRef]
- Brown, A.K.; Webb, A.E. Regulation of FOXO Factors in Mammalian Cells. Curr. Top. Dev. Biol. 2018, 127, 165–192. [Google Scholar]
- Eijkelenboom, A.; Burgering, B.M.T. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell. Biol. 2013, 14, 83–97. [Google Scholar] [CrossRef]
- Jamart, C.; Naslain, D.; Gilson, H.; Francaux, M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am. J. Physiol.-Endocrinol. Metab. 2013, 305, 964–974. [Google Scholar] [CrossRef]
- Louis, E.; Raue, U.; Yang, Y.; Jemiolo, B.; Trappe, S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J. Appl. Physiol. 2007, 103, 1744–1751. [Google Scholar] [CrossRef]
- Pasiakos, S.M.; McClung, H.L.; McClung, J.P.; Urso, M.L.; Pikosky, M.A.; Cloutier, G.J.; Fielding, R.A.; Young, A.J. Molecular responses to moderate endurance exercise in skeletal muscle. Int. J. Sport Nutr. Exerc. Metab. 2010, 20, 282–290. [Google Scholar] [CrossRef]
- Sanchez, A.M.J.; Candau, R.B.; Bernardi, H. FoxO transcription factors: Their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 2014, 71, 1657–1671. [Google Scholar] [CrossRef]
- Labeit, S.; Kohl, C.H.; Witt, C.C.; Labeit, D.; Jung, J.; Granzier, H. Modulation of muscle atrophy, fatigue and MLC phosphorylation by MuRF1 as indicated by hindlimb suspension studies on MuRF1-KO mice. J. Biomed. Biotechnol. 2010, 693741. [Google Scholar] [CrossRef]
- Baehr, L.M.; Furlow, J.D.; Bodine, S.C. Muscle sparing in muscle RING finger 1 null mice: Response to synthetic glucocorticoids. J. Physiol. 2011, 589, 4759–4776. [Google Scholar] [CrossRef] [PubMed]
- Koyama, S.; Hata, S.; Witt, C.C.; Ono, Y.; Lerche, S.; Ojima, K.; Chiba, T.; Doi, N.; Kitamura, F.; Tanaka, K.; et al. Muscle RING-Finger Protein-1 (MuRF1) as a Connector of Muscle Energy Metabolism and Protein Synthesis. J. Mol. Biol. 2008, 376, 1224–1236. [Google Scholar] [CrossRef]
- Files, D.C.; D’Alessio, F.R.; Johnston, L.F.; Kesari, P.; Aggarwal, N.R.; Garibaldi, B.T.; Mock, J.R.; Simmers, J.L.; DeGorordo, A.; Murdoch, J.; et al. A Critical Role for Muscle Ring Finger-1 in Acute Lung Injury–associated Skeletal Muscle Wasting. Am. J. Respir. Crit. Care Med. 2012, 185, 825–834. [Google Scholar] [CrossRef]
- Fielitz, J.; Kim, M.-S.; Shelton, J.M.; Latif, S.; Spencer, J.A.; Glass, D.J.; Richardson, J.A.; Bassel-Duby, R.; Olson, R.N. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J. Clin. Investig. 2007, 117, 2486–2495. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Heng, A.; Jarzaguet, M.; Ventadour, S.; Claustre, A.; Combaret, L.; Béchet, D.; Matondo, M.; Uttenweiler-Joseph, S.; Monsarrat, B.; et al. Muscle actin is polyubiquitinylated in vitro and in vivo and targeted for breakdown by the E3 ligase MuRF1. FASEB J. 2011, 25, 3790–3802. [Google Scholar] [CrossRef] [PubMed]
- Kedar, V.; McDonough, H.; Arya, R.; Li, H.-H.; Rockman, H.A.; Patterson, C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. USA 2004, 101, 18135–18140. [Google Scholar] [CrossRef]
- Polge, C.; Cabantous, S.; Deval, C.; Claustre, A.; Hauvette, A.; Bouchenot, C.; Aniort, J.; Béchet, D.; Combaret, L.; Attaix, D.; et al. A muscle-specific MuRF1-E2 network requires stabilization of MuRF1-E2 complexes by telethonin, a newly identified substrate. J. Cachexia Sarcopenia Muscle 2018, 9, 129–145. [Google Scholar] [CrossRef]
- Rudolf, R.; Bogomolovas, J.; Strack, S.; Choi, K.R.; Khan, M.M.; Wagner, A.; Brohm, K.; Hanashima, A.; Gasch, A.; Labeit, D.; et al. Regulation of nicotinic acetylcholine receptor turnover by MuRF1 connects muscle activity to endo/lysosomal and atrophy pathways. Age 2013, 35, 1663–1674. [Google Scholar] [CrossRef]
- Khan, M.M.; Strack, S.; Wild, F.; Hanashima, A.; Gasch, A.; Brohm, K.; Reischl, M.; Carnio, S.; Labeit, D.; Sandri, M.; et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy 2014, 10, 123–136. [Google Scholar] [CrossRef]
- Li, H.-H.; Du, J.; Fan, Y.-N.; Zhang, M.-L.; Liu, D.-P.; Li, L.; Lockyer, P.; Kang, E.Y.; Patterson, C.; Willis, M.S. The Ubiquitin Ligase MuRF1 Protects Against Cardiac Ischemia/Reperfusion Injury by Its Proteasome-Dependent Degradation of Phospho-c-Jun. Am. J. Pathol. 2011, 178, 1043–1058. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol.-Endocrinol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Leibovitch, M.P.; Cornille, K.; Tintignac, L.A.; Leibovitch, S.A. MAFbx/Atrogin-1 Controls the Activity of the Initiation Factor eIF3-f in Skeletal Muscle Atrophy by Targeting Multiple C-terminal Lysines. J. Biol. Chem. 2009, 284, 4413–4421. [Google Scholar] [CrossRef] [PubMed]
- Jogo, M.; Shiraishi, S.; Tamura, T.A. Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Lett. 2009, 583, 2715–2719. [Google Scholar] [CrossRef]
- Lagirand-Cantaloube, J.; Cornille, K.; Csibi, A.; Batonnet-Pinchon, S.; Leibovitch, M.P.; Leibovitch, S.A. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLoS ONE 2009, 4, e4973. [Google Scholar] [CrossRef] [PubMed]
- Wardle, F.C. Master control: Transcriptional regulation of mammalian Myod. J. Muscle Res. Cell Motil. 2019, 40, 211–226. [Google Scholar] [CrossRef]
- Lokireddy, S.; Wijesoma, I.W.; Sze, S.K.; McFarlane, C.; Kambadur, R.; Sharma, M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am. J. Physiol.-Cell Physiol. 2012, 303, C512–C529. [Google Scholar] [CrossRef]
- Romanello, V.; Sandri, M. Mitochondrial quality control and muscle mass maintenance. Front. Physiol. 2016, 2, 422. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, J.J.; Zhou, S.J.; Chen, J.; Hu, Q.; Pu, J.X.; Lu, J.-L. Diosgenin inhibits the expression of nedd4 in prostate cancer cells. Am. J. Transl. Res. 2019, 11, 3461–3471. [Google Scholar]
- Leermakers, P.A.; Schols, A.M.W.J.; Kneppers, A.E.M.; Kelders, M.C.J.M.; de Theije, C.C.; Lainscak, M.; Gosker, H.R. Molecular signalling towards mitochondrial breakdown is enhanced in skeletal muscle of patients with chronic obstructive pulmonary disease (COPD). Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Deval, C.; Calonne, J.; Coudy-Gandilhon, C.; Vazeille, E.; Bechet, D.; Polge, C.; Taillandier, D.; Attaix, D.; Combaret, L. Mitophagy and Mitochondria Biogenesis Are Differentially Induced in Rat Skeletal Muscles during Immobilization and/or Remobilization. Int. J. Mol. Sci. 2020, 21, 3691. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Yeo, D.; Ji, L.L. Muscle immobilization activates mitophagy and disrupts mitochondrial dynamics in mice. Acta Physiol. 2016, 218, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Balan, E.; Schwalm, C.; Naslain, D.; Nielens, H.; Francaux, M.; Deldicque, L. Regular Endurance Exercise Promotes Fission, Mitophagy, and Oxidative Phosphorylation in Human Skeletal Muscle Independently of Age. Front. Physiol. 2019, 10, 1088. [Google Scholar] [CrossRef] [PubMed]
- Ehrlicher, S.E.; Stierwalt, H.D.; Miller, B.F.; Newsom, S.A.; Robinson, M.M. Mitochondrial adaptations to exercise do not require Bcl2-mediated autophagy but occur with BNIP3/Parkin activation. FASEB J. 2020, 34, 4602–4618. [Google Scholar] [CrossRef]
- Drummond, M.J.; Addison, O.; Brunker, L.; Hopkins, P.N.; McClain, D.A.; LaStayo, P.C.; Marcus, R.L. Downregulation of E3 Ubiquitin Ligases and Mitophagy-Related Genes in Skeletal Muscle of Physically Inactive, Frail Older Women: A Cross-Sectional Comparison. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 1040–1048. [Google Scholar] [CrossRef]
- Russ, D.W.; Wills, A.M.; Boyd, I.M.; Krause JWeakness, S.R. function and stress in gastrocnemius muscles of aged male rats. Exp. Gerontol. 2014, 50, 40–44. [Google Scholar] [CrossRef]
- Marzetti, E.; Lorenzi, M.; Landi, F.; Picca, A.; Rosa, F.; Tanganelli, F.; Galli, M.; Doglietto, G.B.; Pacelli, F.; Cesari, M.; et al. Altered mitochondrial quality control signaling in muscle of old gastric cancer patients with cachexia. Exp. Gerontol. 2017, 87, 92–99. [Google Scholar] [CrossRef]
- Chen, C.C.W.; Erlich, A.T.; Crilly, M.J.; Hood, D.A. Parkin is required for exercise-induced mitophagy in muscle: Impact of aging. Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E404–E415. [Google Scholar] [CrossRef]
- Gouspillou, G.; Godin, R.; Piquereau, J.; Picard, M.; Mofarrahi, M.; Mathew, J.; Purves-Smith, F.M.; Sgarioto, N.; Hepple, R.T.; Burelle, Y.; et al. Protective role of Parkin in skeletal muscle contractile and mitochondrial function: Parkin is essential for optimal muscle and mitochondrial functions. J. Physiol. 2018, 596, 2565–2579. [Google Scholar] [CrossRef]
- Ramesh, M.; Campos, J.C.; Lee, P.; Song, Y.; Hernandez, G.; Sin, J.; Tucker, K.C.; Saadaeijahromi, H.; Gurney, M.; Ferreira, J.C.B.; et al. Mitophagy protects against statin-mediated skeletal muscle toxicity. FASEB J. 2019, 33, 11857–11869. [Google Scholar] [CrossRef]
- Furuya, N.; Ikeda, S.-I.; Sato, S.; Soma, S.; Ezaki, J.; Trejo, J.A.O.; Takeda-Ezaki, M.; Fujimura, T.; Arikawa-Hirasawa, E.; Tada, N.; et al. PARK2/Parkin-mediated mitochondrial clearance contributes to proteasome activation during slow-twitch muscle atrophy via NFE2L1 nuclear translocation. Autophagy 2014, 10, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.C.; Whitworth, A.J.; Kuo, I.; Andrews, L.A.; Feany, M.B.; Pallanck, L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 2003, 100, 4078–4083. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Chen, J.A.; Xu, J.; Cao, J.; Wang, Y.; Thomas, S.S.; Hu, Z. Suppression of muscle wasting by the plant-derived compound ursolic acid in a model of chronic kidney disease. J. Cachexia Sarcopenia Muscle 2017, 8, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Kny, M.; Pablo-Tortola, C.; Todiras, M.; Willenbrock, M.; Schmidt, S.; Schmoeckel, K.; Jorde, I.; Nowak, M.; Jarosch, E.; et al. Serum amyloid A1 mediates myotube atrophy via Toll-like receptors. J. Cachexia Sarcopenia Muscle 2020, 11, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-H.; Mohawk, J.A.; Siepka, S.M.; Shan, Y.; Huh, S.K.; Hong, H.-K.; Kornblum, I.; Kumar, V.; Koike, N.; Xu, M.; et al. Competing E3 Ubiquitin Ligases Govern Circadian Periodicity by Degradation of CRY in Nucleus and Cytoplasm. Cell 2013, 152, 1091–1105. [Google Scholar] [CrossRef]
- Lucas, X.; Ciulli, A. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr. Opin. Struct. Biol. 2017, 44, 101–110. [Google Scholar] [CrossRef]
- Kwak, K.S.; Zhou, X.; Solomon, V.; Baracos, V.E.; Davis, J.; Bannon, A.W.; Boyle, W.J.; Lacey, D.L.; Han, H.Q. Regulation of protein catabolism by muscle-specific and cytokine-inducible ubiquitin ligase E3alpha-II during cancer cachexia. Cancer Res. 2004, 64, 8193–8198. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Hughes, D.C.; Turner, D.C.; Owens, D.J.; Baehr, L.M.; Gorski, P.; Semenova, E.A.; Borisov, O.V.; Larin, A.K.; Popov, D.V.; et al. UBR5 is a novel E3 ubiquitin ligase involved in skeletal muscle hypertrophy and recovery from atrophy. J. Physiol. 2019, 597, 3727–3749. [Google Scholar] [CrossRef]
- Besche, H.C.; Haas, W.; Gygi, S.P.; Goldberg, A.L. Isolation of Mammalian 26S Proteasomes and p97/VCP Complexes Using the Ubiquitin-like Domain from HHR23B Reveals Novel Proteasome-Associated Proteins. Biochemistry 2009, 48, 2538–2549. [Google Scholar] [CrossRef]
- Morén, A.; Imamura, T.; Miyazono, K.; Heldin, C.H.; Moustakas, A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 2005, 280, 22115–22123. [Google Scholar] [CrossRef]
- Ebisawa, T.; Fukuchi, M.; Murakami, G.; Chiba, T.; Tanaka, K.; Imamura, T.; Miyazono, K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001, 276, 12477–12480. [Google Scholar] [CrossRef] [PubMed]
- Tényi, Á.; Cano, I.; Marabita, F.; Kiani, N.; Kalko, S.G.; Barreiro, E.; de Atauri, P.; Cascante, M.; Gomez-Cabrero, D.; Roca, J. Network modules uncover mechanisms of skeletal muscle dysfunction in COPD patients. J. Transl. Med. 2018, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Xu, X.; Li, L.; Ning, H.; Rong, Y.; Shang, Y.; Wang, Y.; Fu, X.-Y.; Chang, Z. CHIP controls the sensitivity of transforming growth factor-β signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J. Biol. Chem. 2005, 280, 20842–20850. [Google Scholar] [CrossRef] [PubMed]
- Li, R.F.; Shang, Y.; Liu, D.; Ren, Z.S.; Chang, Z.; Sui, S.F. Differential Ubiquitination of Smad1 Mediated by CHIP: Implications in the Regulation of the Bone Morphogenetic Protein Signaling Pathway. J. Mol. Biol. 2007, 374, 777–790. [Google Scholar] [CrossRef]
- Schisler, J.C.; Patterson, C.; Willis, M.S. Skeletal Muscle Mitochondrial Alterations in Carboxyl Terminus of HSC70 Interacting Protein (CHIP)−/− Mice. Afr. J. Cell. Pathol. 2016, 6, 28–36. [Google Scholar]
- Chen, N.; Balasenthil, S.; Reuther, J.; Frayna, A.; Wang, Y.; Chandler, D.S.; Abruzzo, L.V.; Rashid, A.; Rodriguez, J.; Lozano, G.; et al. DEAR1 is a chromosome 1p35 tumor suppressor and master regulator of TGF-β-driven epithelial-mesenchymal transition. Cancer Discov. 2013, 3, 1172–1189. [Google Scholar] [CrossRef]
- Schmidt, F.; Kny, M.; Zhu, X.; Wollersheim, T.; Persicke, K.; Langhans, C.; Lodka, D.; Kleber, C.; Weber-Carstens, S.; Fielitz, J. The E3 ubiquitin ligase TRIM62 and inflammationinduced skeletal muscle atrophy. Crit. Care 2014, 18, 1–12. [Google Scholar] [CrossRef]
- He, B.; Tang, R.H.; Weisleder, N.; Xiao, B.; Yuan, Z.; Cai, C.; Zhu, H.; Lin, P.; Qiao, C.; Li, J.; et al. Enhancing muscle membrane repair by gene delivery of MG53 ameliorates muscular dystrophy and heart failure inδ-sarcoglycan-deficient hamsters. Mol. Ther. 2012, 20, 727–735. [Google Scholar] [CrossRef]
- Gushchina, L.V.; Bhattacharya, S.; McElhanon, K.E.; Choi, J.H.; Manring, H.; Beck, E.X.; Alloush, J.; Weisleder, N. Treatment with Recombinant Human MG53 Protein Increases Membrane Integrity in a Mouse Model of Limb Girdle Muscular Dystrophy 2B. Mol. Ther. 2017, 25, 2360–2371. [Google Scholar] [CrossRef]
- Cao, C.M.; Zhang, Y.; Weisleder, N.; Ferrante, C.; Wang, X.; Lv, F.; Zhang, Y.; Song, R.; Hwang, M.; Jin, L.; et al. MG53 constitutes a primary determinant of cardiac ischemic preconditioning. Circulation 2010, 121, 2565–2574. [Google Scholar] [CrossRef]
- Gonçalves, D.A.P.; Silveira, W.A.; Lira, E.C.; Graça, F.A.; Paula-Gomes, S.; Zanon, N.M.; Kettelhut, I.C.; Navegantes, L.C.C. Clenbuterol suppresses proteasomal and lysosomal proteolysis and atrophy-related genes in denervated rat soleus muscles independently of Akt. Am. J. Physiol.-Endocrinol. Metab. 2012, 302, E123–E133. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, K.; Fallon, K.; Ruut, T.; Lane, D.; McKay, R.; Shadbolt, B.; Ang, S.; Cook, M.; Platten, J.; Pavli, P.; et al. Infliximab reverses inflammatory muscle wasting (sarcopenia) in Crohn’s disease. Aliment. Pharmacol. Ther. 2015, 41, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Yakabe, M.; Ogawa, S.; Ota, H.; Iijima, K.; Eto, M.; Ouchi, Y.; Akishita, M. Inhibition of interleukin-6 decreases atrogene expression and ameliorates tail suspension-induced skeletal muscle atrophy. PLoS ONE 2018, 13, e0191318. [Google Scholar] [CrossRef]
- Salazar-Degracia, A.; Busquets, S.; Argilés, J.M.; Bargalló-Gispert, N.; López-Soriano, F.J.; Barreiro, E. Effects of the beta2 agonist formoterol on atrophy signaling, autophagy, and muscle phenotype in respiratory and limb muscles of rats with cancer-induced cachexia. Biochimie 2018, 149, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.I.; Gómez-SanMiguel, A.B.; Priego, T.; López-Calderón, A. Formoterol treatment prevents the effects of endotoxin on muscle TNF/NF-kB, Akt/mTOR, and proteolytic pathways in a rat model. Role of IGF-I and miRNA 29b. Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E705–E714. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Talukder, M.A.H.; Gurjar, A.; Lee, J.I.; Noble, M.; Dirksen, R.T.; Chakkalakal, J.; Elfar, J.C. 4-Aminopyridine attenuates muscle atrophy after sciatic nerve crush injury in mice. Muscle Nerve 2019, 60, 192–201. [Google Scholar] [CrossRef]
- Wang, H.; Lai, Y.-J.; Chan, Y.-L.; Li, T.-L.; Wu, C.-J. Epigallocatechin-3-gallate effectively attenuates skeletal muscle atrophy caused by cancer cachexia. Cancer Lett. 2011, 305, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Pötsch, M.S.; Tschirner, A.; Palus, S.; Haehling, S.; von Doehner, W.; Beadle, J.; Coats, A.J.S.; Anker, S.D.; Springer, J. The anabolic catabolic transforming agent (ACTA) espindolol increases muscle mass and decreases fat mass in old rats. J. Cachexia Sarcopenia Muscle 2014, 5, 149–158. [Google Scholar] [CrossRef]
- Gómez-SanMiguel, A.B.; Gomez-Moreira, C.; Nieto-Bona, M.P.; Fernández-Galaz, C.; Villanúa, M.Á.; Martín, A.I.; López-Calderón, A. Formoterol decreases muscle wasting as well as inflammation in the rat model of rheumatoid arthritis. Am. J. Physiol.-Endocrinol. Metab. 2016, 310, E925–E937. [Google Scholar] [CrossRef]
- Noh, K.K.; Chung, K.W.; Choi, Y.J.; Park, M.H.; Jang, E.J.; Park, C.H.; Yoon, C.; Kim, N.D.; Kim, M.K.; Chung, H.Y. β–Hydroxy β–Methylbutyrate Improves Dexamethasone-Induced Muscle Atrophy by Modulating the Muscle Degradation Pathway in SD Rat. PLoS ONE 2014, 9, e102947. [Google Scholar] [CrossRef] [PubMed]
- Baptista, I.L.; Leal, M.L.; Artioli, G.G.; Aoki, M.S.; Fiamoncini, J.; Turri, A.O.; Curi, R.; Miyabara, E.H.; Moriscot, A.S. Leucine attenuates skeletal muscle wasting via inhibition of ubiquitin ligases. Muscle Nerve 2010, 41, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, L.; Wan, L.; Huo, Y.; Huang, J.; Li, J.; Lu, J.; Xin, B.; Yang, Q.; Guo, C. Matrine improves skeletal muscle atrophy by inhibiting E3 ubiquitin ligases and activating the Akt/mTOR/FoxO3α signaling pathway in C2C12 myotubes and mice. Oncol. Rep. 2019, 42, 479–494. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Fang, Q.; Xu, T.; Wu, C.; Xu, L.; Wang, L.; Yang, X.; Yu, S.; Zhang, Q.; Ding, F.; et al. Mechanistic role of reactive oxygen species and therapeutic potential of antioxidants in denervation-or fasting-induced skeletal muscle atrophy. Front. Physiol. 2018, 9, 215. [Google Scholar] [CrossRef]
- Ryu, Y.; Lee, D.; Jung, S.H.; Lee, K.-J.; Jin, H.; Kim, S.J.; Lee, H.M.; Kim, B.; Won, K.-J. Sabinene Prevents Skeletal Muscle Atrophy by Inhibiting the MAPK-MuRF-1 Pathway in Rats. Int. J. Mol. Sci. 2019, 20, 4955. [Google Scholar] [CrossRef]
- Powers, S.K.; Hudson, M.B.; Nelson, W.B.; Talbert, E.E.; Min, K.; Szeto, H.H.; Kavazis, A.N.; Smuder, A.J. Mitochondria-targeted antioxidants protect against mechanical ventilation-induced diaphragm weakness. Crit. Care Med. 2011, 39, 1749–1759. [Google Scholar] [CrossRef]
- Guillory, B.; Chen, J.; Patel, S.; Luo, J.; Splenser, A.; Mody, A.; Ding, M.; Baghaie, S.; Anderson, B.; Iankova, B.; et al. Deletion of ghrelin prevents aging-associated obesity and muscle dysfunction without affecting longevity. Aging Cell. 2017, 16, 859–869. [Google Scholar] [CrossRef]
- Hsieh, S.K.; Lin, H.Y.; Chen, C.J.; Jhuo, C.F.; Liao, K.Y.; Chen, W.Y.; Tzen, J.T.C. Promotion of myotube differentiation and attenuation of muscle atrophy in murine C2C12 myoblast cells treated with teaghrelin. Chem. Biol. Interact. 2020, 315, 108893. [Google Scholar] [CrossRef]
- Wu, C.-S.; Wei, Q.; Wang, H.; Kim, D.M.; Balderas, M.; Wu, G.; Lawler, J.; Safe, S.; Guo, S.; Devaraj, S.; et al. Protective Effects of Ghrelin on Fasting-Induced Muscle Atrophy in Aging Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 621–630. [Google Scholar] [CrossRef]
- Servais, S.; Letexier, D.; Favier, R.; Duchamp, C.; Desplanches, D. Prevention of unloading-induced atrophy by vitamin E supplementation: Links between oxidative stress and soleus muscle proteolysis? Free Radic. Biol. Med. 2007, 42, 627–635. [Google Scholar] [CrossRef]
- Belova, S.P.; Mochalova, E.P.; Kostrominova, T.Y.; Shenkman, B.S.; Nemirovskaya, T.L. P38α-MAPK Signaling Inhibition Attenuates Soleus Atrophy during Early Stages of Muscle Unloading. Int. J. Mol. Sci. 2020, 21, 2756. [Google Scholar] [CrossRef]
- Bowen, T.S.; Adams, V.; Werner, S.; Fischer, T.; Vinke, P.; Brogger, M.; Mangner, N.; Linke, A.; Sehr, P.; Lewis, J.; et al. Small-molecule inhibition of MuRF1 attenuates skeletal muscle atrophy and dysfunction in cardiac cachexia: Inhibition of MuRF1 prevents skeletal muscle wasting. J. Cachexia Sarcopenia Muscle 2017, 8, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Eddins, M.J.; Marblestone, J.G.; Suresh Kumar, K.G.; Leach, C.A.; Sterner, D.E.; Mattern, M.R.; Nicholson, B. Targeting the ubiquitin E3 ligase MuRF1 to inhibit muscle atrophy. Cell Biochem. Biophys. 2011, 60, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, K.; Okubo, K.; Yoda, M.; Otsu, K.; Ishii, Y.; Nakamura, M.; Itoh, Y.; Horiuchi, K. Targeted ablation of p38α MAPK suppresses denervation-induced muscle atrophy. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Tricarico, D.; Selvaggi, M.; Passantino, G.; De Palo, P.; Dario, C.; Centoducati, P.; Tateo, A.; Curci, A.; Maqoud, F.; Mele, A.; et al. ATP Sensitive Potassium Channels in the Skeletal Muscle Function: Involvement of the KCNJ11(Kir6.2) Gene in the Determination of Mechanical Warner Bratzer Shear Force. Front. Physiol. 2016, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.D.; Tang, X.; Campbell, M.D.; Reyes, G.; Kramer, P.A.; Stuppard, R.; Keller, A.; Zhang, H.; Rabinovitch, P.S.; Marcinek, D.J.; et al. Mitochondrial protein interaction landscape of SS-31. Proc. Natl. Acad. Sci. USA 2020, 117, 15363–15373. [Google Scholar] [CrossRef]
- Rittig, N.; Bach, E.; Thomsen, H.H.; Møller, A.B.; Hansen, J.; Johannsen, M.; Jensen, E.; Serena, A.; Jørgensen, J.O.; Richelsen, B.; et al. Anabolic effects of leucine-rich whey protein, carbohydrate, and soy protein with and without β-hydroxy-β-methylbutyrate (HMB) during fasting-induced catabolism: A human randomized crossover trial. Clin. Nutr. 2017, 36, 697–705. [Google Scholar] [CrossRef]
- Girón, M.D.; Vílchez, J.D.; Salto, R.; Manzano, M.; Sevillano, N.; Campos, N.; Argilés, J.M.; Rueda, R.; López-Pedrosa, J.M. Conversion of leucine to β-hydroxy-β-methylbutyrate by α-keto isocaproate dioxygenase is required for a potent stimulation of protein synthesis in L6 rat myotubes. J. Cachexia Sarcopenia Muscle 2016, 7, 68–78. [Google Scholar] [CrossRef]
- Baptista, I.L.; Silvestre, J.G.; Silva, W.J.; Labeit, S.; Moriscot, A.S. FoxO3a suppression and VPS34 activity are essential to anti-atrophic effects of leucine in skeletal muscle. Cell Tissue Res. 2017, 369, 381–394. [Google Scholar] [CrossRef]
- van den Hoek, A.M.; Zondag, G.C.M.; Verschuren, L.; de Ruiter, C.; Attema, J.; de Wit, E.C.; Schwerk, A.M.K.; Guigas, B.; Lek, S.; Rietman, A.; et al. A novel nutritional supplement prevents muscle loss and accelerates muscle mass recovery in caloric-restricted mice. Metabolism 2019, 97, 57–67. [Google Scholar] [CrossRef]
- Balasubramaniam, A.; Joshi, R.; Su, C.; Friend, L.A.; Sheriff, S.; Kagan, R.J.; James, J.H. Ghrelin inhibits skeletal muscle protein breakdown in rats with thermal injury through normalizing elevated expression of E3 ubiquitin ligases MuRF1 and MAFbx. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2009, 296, R893–R901. [Google Scholar] [CrossRef]
- Clarke, B.A.; Drujan, D.; Willis, M.S.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 Ligase MuRF1 Degrades Myosin Heavy Chain Protein in Dexamethasone-Treated Skeletal Muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef]
- Ochi, A.; Abe, T.; Nakao, R.; Yamamoto, Y.; Kitahata, K.; Takagi, M.; Hirasaka, K.; Ohno, A.; Teshima-Kondo, S.; Taesik, G.; et al. N-myristoylated ubiquitin ligase Cbl-b inhibitor prevents on glucocorticoid-induced atrophy in mouse skeletal muscle. Arch. Biochem. Biophys. 2015, 570, 23–31. [Google Scholar] [CrossRef]
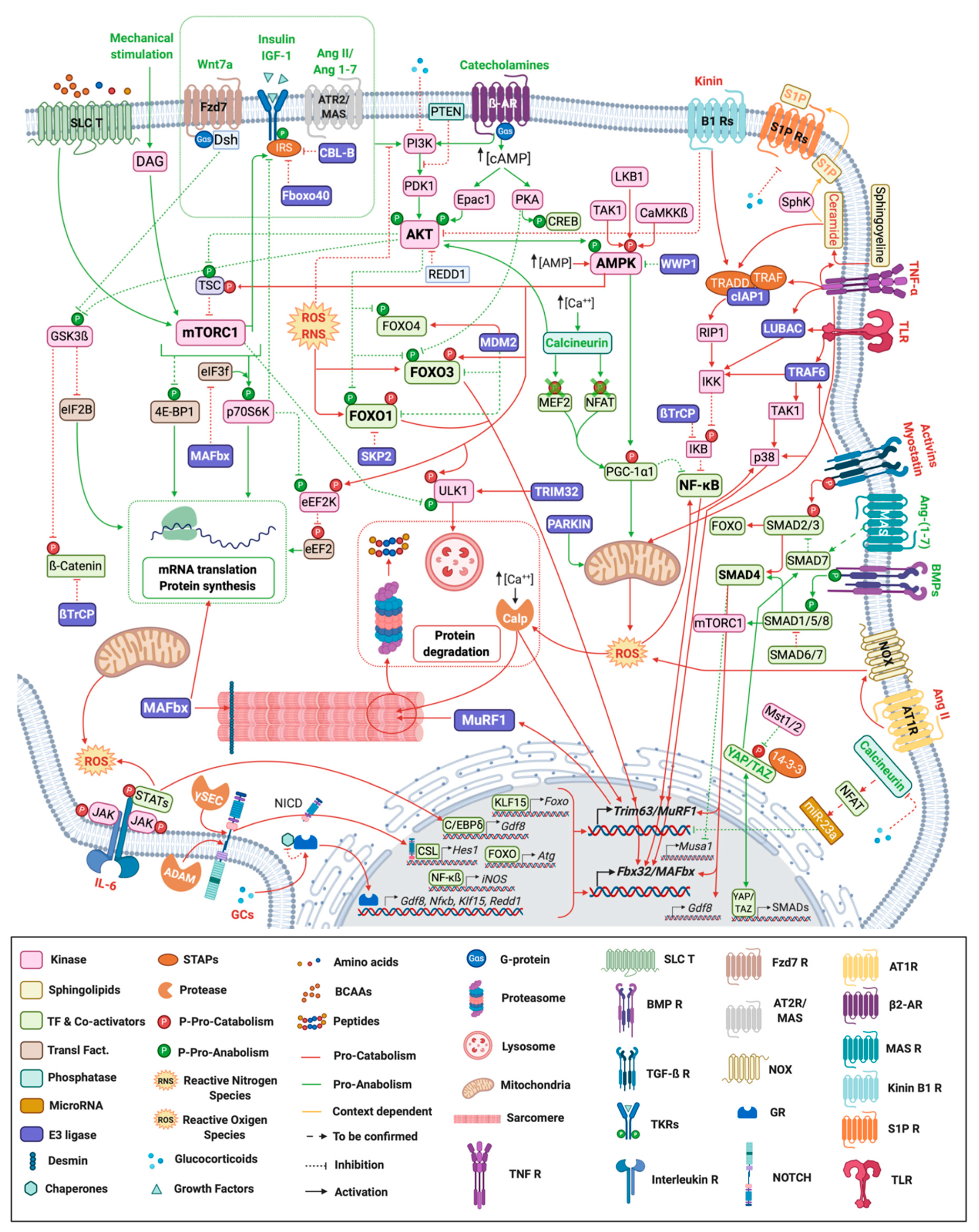
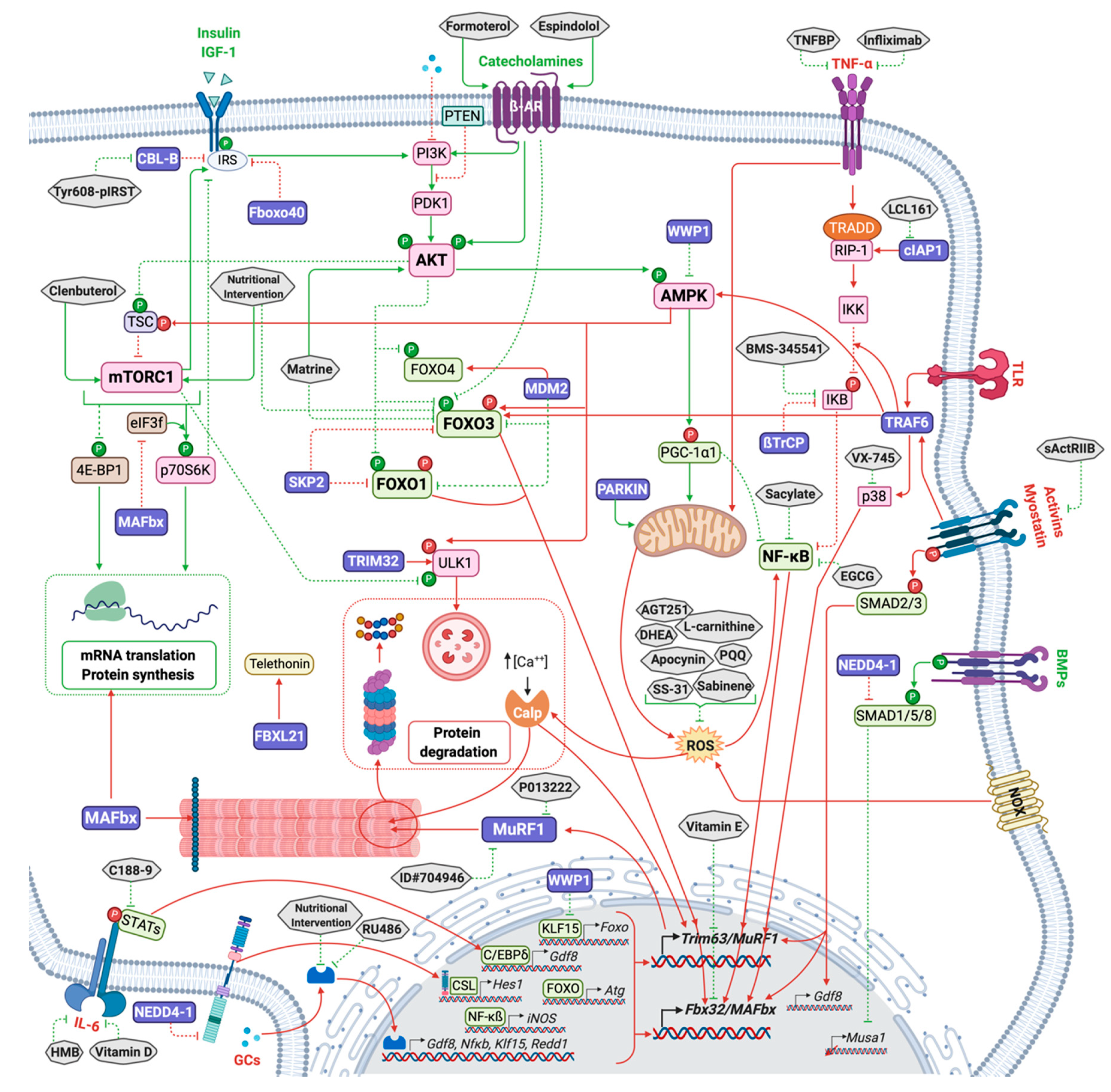
| Gene Product | E3 Family | Mouse Model | Phenotype | References |
|---|---|---|---|---|
| E3 ligases regulating the anabolic pathways | ||||
| CBL-B | RING | KO | Protection from unloading-induced muscle atrophy and dysfunction | [171] |
| FBXO40 | RING | KD | Myofibers hypertrophy | [22] |
| KO | Muscle hypertrophy | |||
| NEDD4-1 | HECT | OX | Myocardial activation of AKT during I/R | [176,177] |
| KO | Partially resistant to denervation-induced skeletal muscle atrophy | [178] | ||
| E3 ligases regulating the catabolic pathways | ||||
| TRAF6 | RING | m.KO | Resistance to starvation induced muscle atrophy | [179] |
| m.KO | Resistance to denervation-induced loss of muscle mass and function | [180] | ||
| cIAP1 | RING | KO | Limitation of denervation-induced muscle atrophy | [19] |
| OX | Myotube atrophy | |||
| WWP1 | HECT | KD | Muscle fiber atrophy | [181] |
| TRIM32 | RING | KO | Muscular dystrophy | [182] |
| DN | Muscular dystrophy | [183] | ||
| Other E3 ligases involved in the control of muscle mass and function | ||||
| MuRF1 | RING | KO | Resistance to catabolic-induced muscle atrophy | [4] |
| MAFbx | RING | KO | Resistance to catabolic-induced muscle atrophy | [4] |
| PARKIN | RBR | KO | Impaired mitochondrial function and muscle atrophy | [184] |
| OX | Increased muscle mass and function in young and old mice | [185] | ||
| OX | Prevention of sepsis-induced muscle atrophy | [186] | ||
| SMART/FBXO21 | RING | KD | Resistance to denervation-induced muscle atrophy | [187] |
| MUSA1/FBXO30 | RING | KD | Resistance to denervation-induced muscle atrophy | [77] |
| FBXL21 | RING | HM | Impaired muscle functions | [188] |
| UBR4 | HECT | KD | Muscle hypertrophy | [189] |
| UBR5 | HECT | KD | Muscle atrophy | [190] |
| E3 Ligases Inhibited | Molecule | Mode of Inhibition | Signal inhibited/Activated | Efficiency on E3 Ligases | Efficiency on Muscle Mass | References |
|---|---|---|---|---|---|---|
| Indirect inhibition of E3 ligases | ||||||
| MuRF1/MAFbx Expression | 4-aminopyridine (4-AP) | K+-channels blockade | K+-channels blocking | Yes | Yes | [278] |
| MuRF1 Expression | AGT251 | Notch1, Notch3 expression inhibition | NOTCH | Yes | Yes | [161] |
| MuRF1/MAFbx/MuSA1 Expression | Anti-TLR2 | IKK2 (NF-κB) | TLRs Serum Amyloib A1 | Yes | Yes | [256] |
| MuRF1 Expression | Anti-TLR4 | IKK2 (NF-κB) | TLRs Serum Amyloib A1 | Yes | Yes | [256] |
| MuRF1/MAFbx/MuSA1 Expression | BMS-345541 | IKK2 (NF-κB) | TLRs Serum Amyloib A1 | Yes | Yes | [256] |
| MuRF1 expression | C188-9 | STAT3 inhibition | STAT3 signaling | ND | Partially | [14] |
| MuRF1/MAFbx Expression | Clenbuterol | AKT-FOXO axis | Activation of PI3K-AKT | Yes | Yes | [15] |
| MuRF1 not MAFbx | Dehydroepiandrosterone (DHEA) | ND | ND | Yes | Yes | [168] |
| MuRF1 Expression | Epigallocatechin-3-gallate/EGCG | ND | NF-κB | Yes | Yes | [279] |
| MuRF1 Expression | Espindolol | ND | Myostatin and NF-κB | Yes | Yes | [280] |
| MuRF1/MAFbx Expression | Formoterol | ND | ND | Yes | Yes | [276] |
| MuRF1/MAFbx Expression | Formoterol | AKT/mTORC1/FOXO1 | ß2 Adrenergic receptor? | Yes | Yes | [277] |
| MuRF1/MAFbx Expression | Formoterol | ND | AKT and NF-κB | Yes | Yes | [281] |
| MuRF1/MAFbx Expression | HMB | IL-6 receptor inhibition | NF-κB | Yes | Partially | [275] |
| MuRF1 expression | HMB or Leucine | FOXO1 nuclear translocation | Glucocorticoid | Yes | No | [282] |
| Cbl-b activity | IRS1 peptide mimetic | Cbl-b targeting | Activation of PI3K-AKT | Yes | Yes | [171] |
| MuRF1/MAFbx Expression | Leucine | ND | FOXO3a and VPS34 nuclear translocation | Yes | Yes, myotube diameter | [283] |
| MuRF1/MAFbx Expression | Matrine | AKT/mTORC1/FOXO3α | FOXO3a and VPS34 nuclear translocation | Yes | Yes | [284] |
| MuRF1 expression | MR16-1 | Anti-IL-6 receptor | NF-κB | Mitigated | No | [275] |
| MuRF1 expression | N-acetyl cysteine | ROS | TGF-ß | Yes | Yes | [69] |
| MuRF1/MAFbx Expression | Pyrroloquinoline quinone (PQQ) | ROS | ND | Yes | Yes | [285] |
| MuRF1/MAFbx Expression | RU486 | GR | Glucocorticoid | Yes | ND | [13] |
| MuRF1 Expression | Sabinene | ROS | ERK, p38 MAPK | Yes | Yes | [286] |
| MuRF1/MAFbx Expression | sActRIIB | ActRIIB antagonist | SMADs | Yes | Yes | [16] |
| MuRF1/MAFbx/MuSA1 Expression | Salicylate | IKK2 (NF-κB) | NF-κB | Yes | Yes but toxic | [89] |
| MuRF1/MAFbx Expression | SS-31 | ROS | No | Yes | Yes | [287] |
| MuRF1/MAFbx Expression | Teaghrelin | ND | Myogenin | Yes | Moderate | [288,289,290] |
| MuRF1/MAFbx Expression | TNF-BP | TNF binding | TNF | Yes | ND | [13] |
| MuRF1/MAFbx/MuSA1 Expression | Ursolic acid | ND | Myostatin and inflammatory cytokines | Yes | Moderate | [255] |
| MuRF1/MAFbx Expression | Vitamin E | ND but seems ROS independent | Unknown | Yes | Moderate | [291] |
| MuRF1/MAFbx Expression | Vitamin-D | IL-6 receptor inhibition | NF-κB | Yes | Partially | [275] |
| MuRF1 expression | VX-745/Neflamapimod | p38α MAPK | p38α MAPK | Partially | Moderate | [292] |
| Direct inhibition of E3 ligases | ||||||
| MuRF1 expression | ID#704946/MyoMed-946 | ND | MuRF1 Expression | Yes | Partially | [293] |
| MuRF1 Expression | ID#704946/MyoMed-946 | ND | MuRF1 and MuRF2 Expression | Yes | Partially | [17,18] |
| MuRF1 and MuRF2 Expression | MyoMed-205 | ND | MuRF1 expression | [17] | ||
| MuRF1 activity | P013222 | MuRF1 targeting | -- | Yes | ND | [294] |
| cIAP1 (activity??) | LCL161 | cIAP1 | NF-κB | Yes | Very moderate | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peris-Moreno, D.; Cussonneau, L.; Combaret, L.; Polge, C.; Taillandier, D. Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control. Molecules 2021, 26, 407. https://doi.org/10.3390/molecules26020407
Peris-Moreno D, Cussonneau L, Combaret L, Polge C, Taillandier D. Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control. Molecules. 2021; 26(2):407. https://doi.org/10.3390/molecules26020407
Chicago/Turabian StylePeris-Moreno, Dulce, Laura Cussonneau, Lydie Combaret, Cécile Polge, and Daniel Taillandier. 2021. "Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control" Molecules 26, no. 2: 407. https://doi.org/10.3390/molecules26020407
APA StylePeris-Moreno, D., Cussonneau, L., Combaret, L., Polge, C., & Taillandier, D. (2021). Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control. Molecules, 26(2), 407. https://doi.org/10.3390/molecules26020407








