Sulforaphane Inhibits Osteoclastogenesis via Suppression of the Autophagic Pathway
Abstract
1. Introduction
2. Results
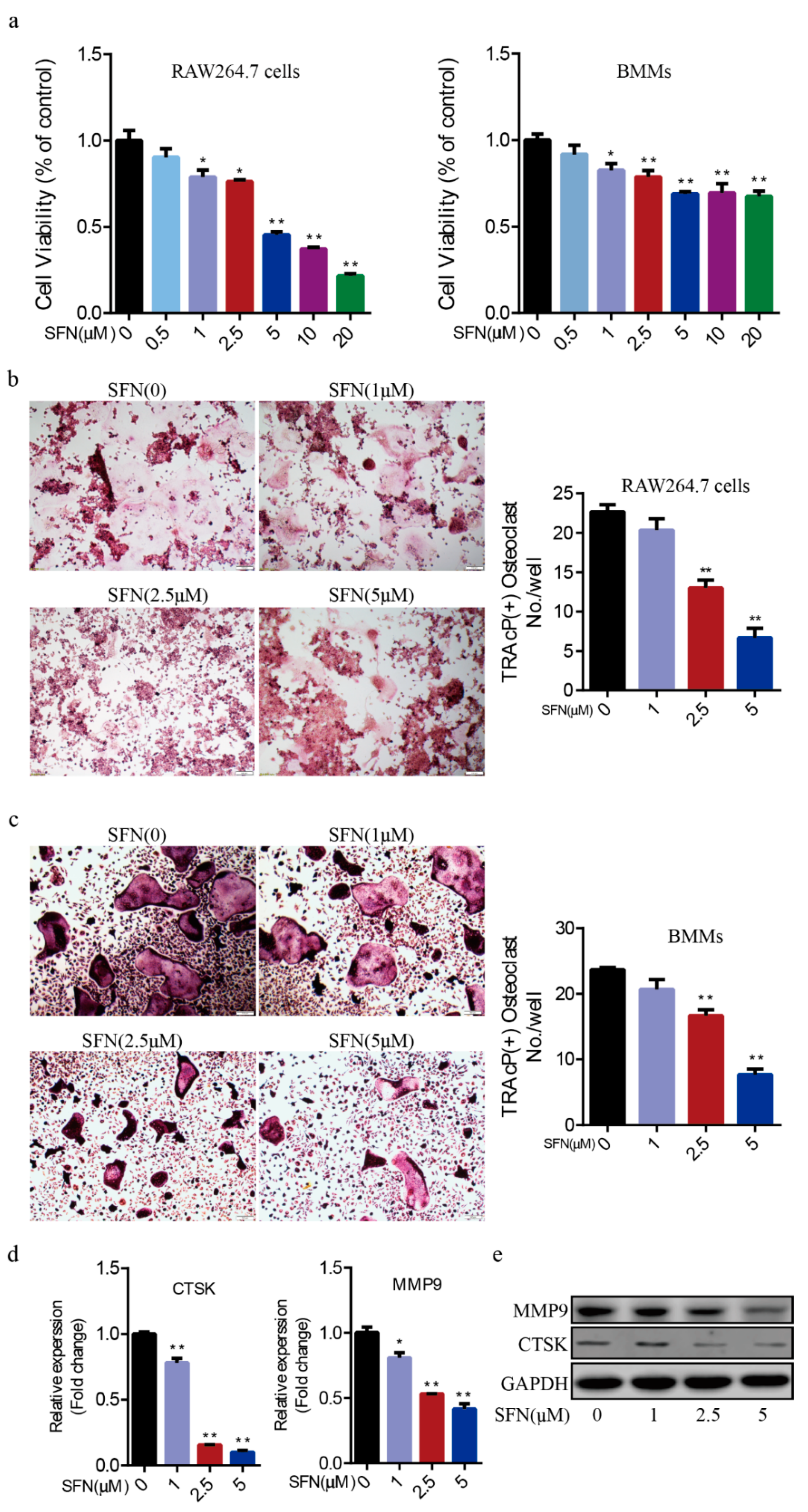
2.1. SFN Inhibited Osteoclastogenesis in a Dose-Dependent Manner
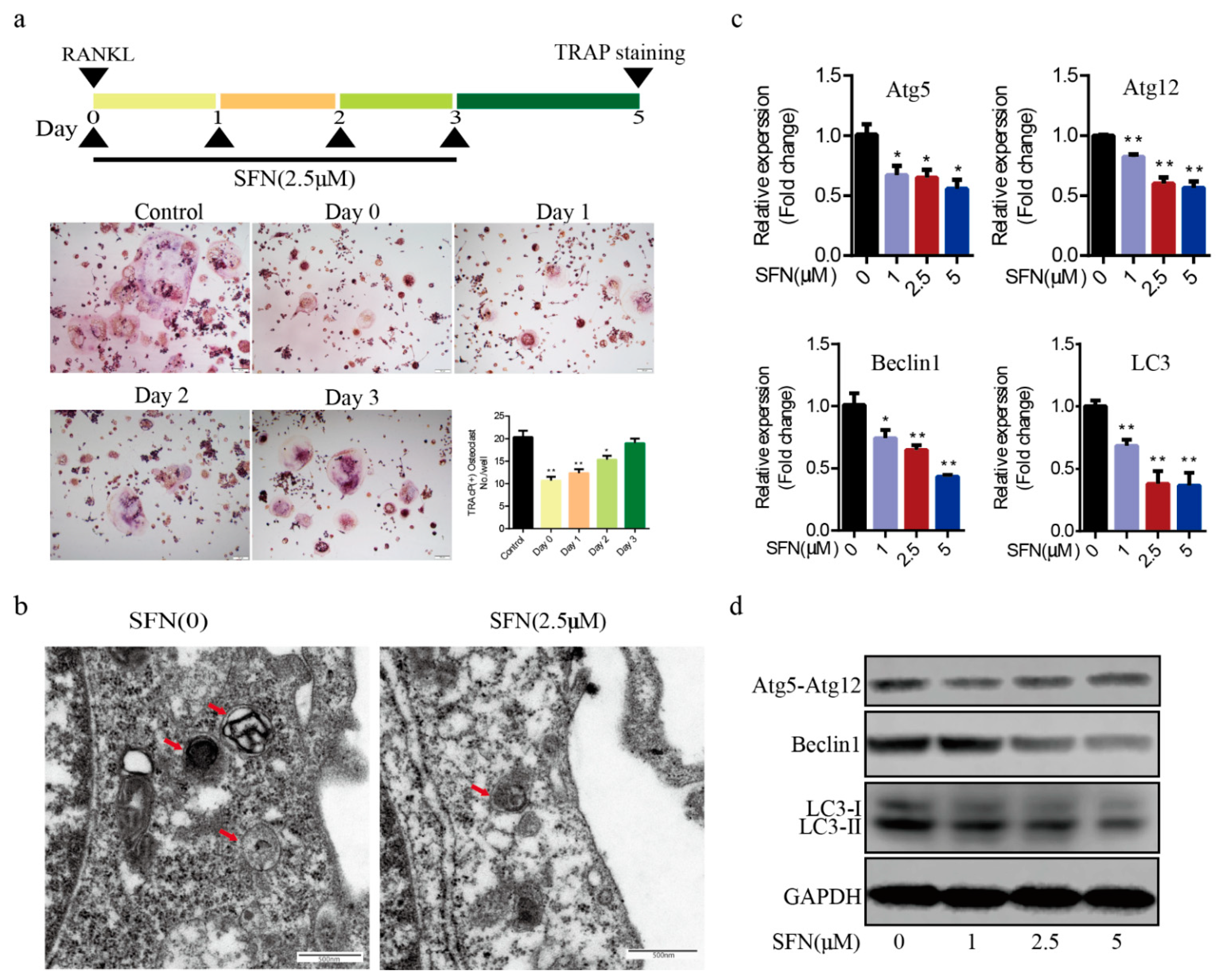
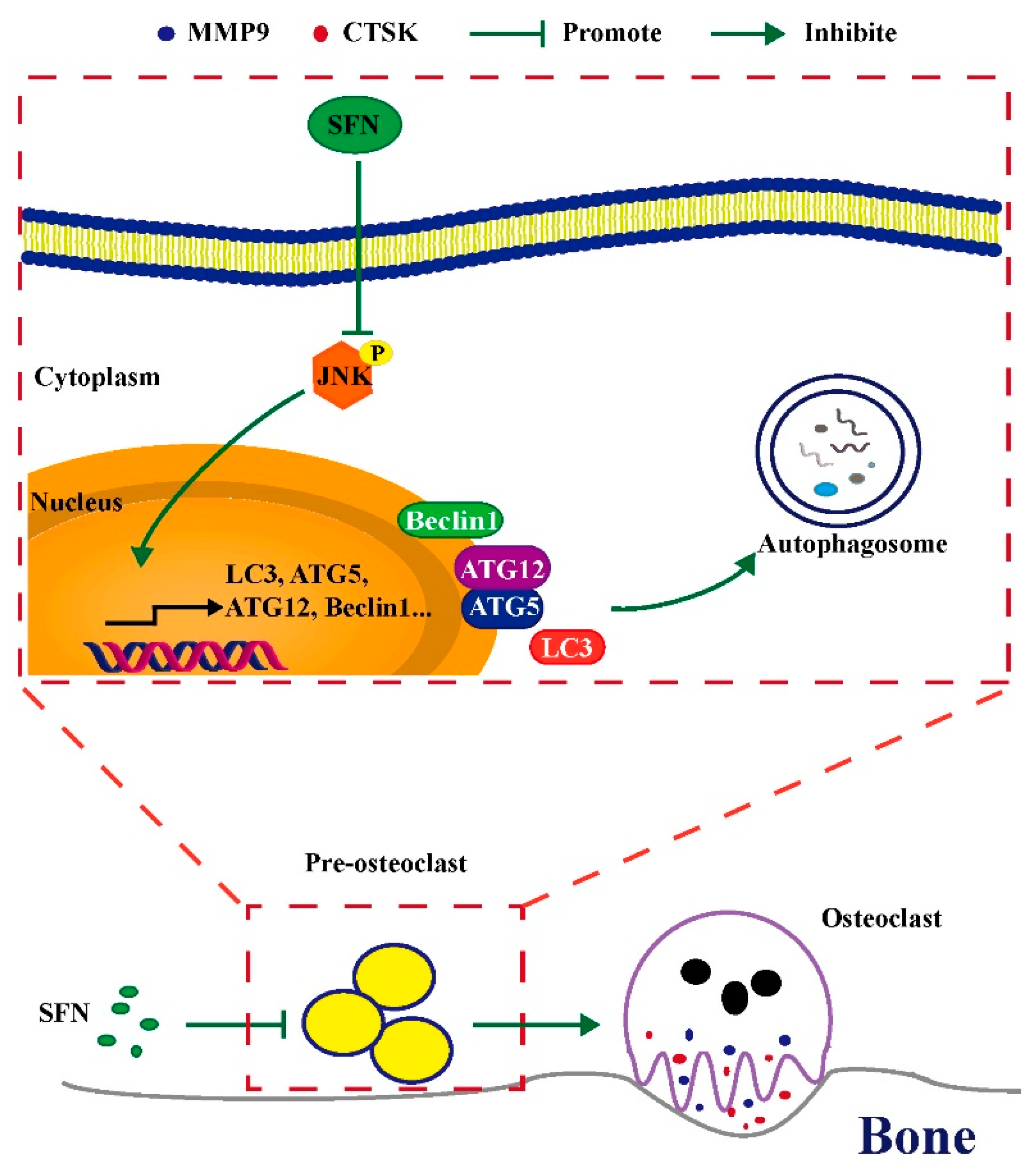
2.2. SFN Treatment Blocked the Autophagic Pathway in Pre-Osteoclasts
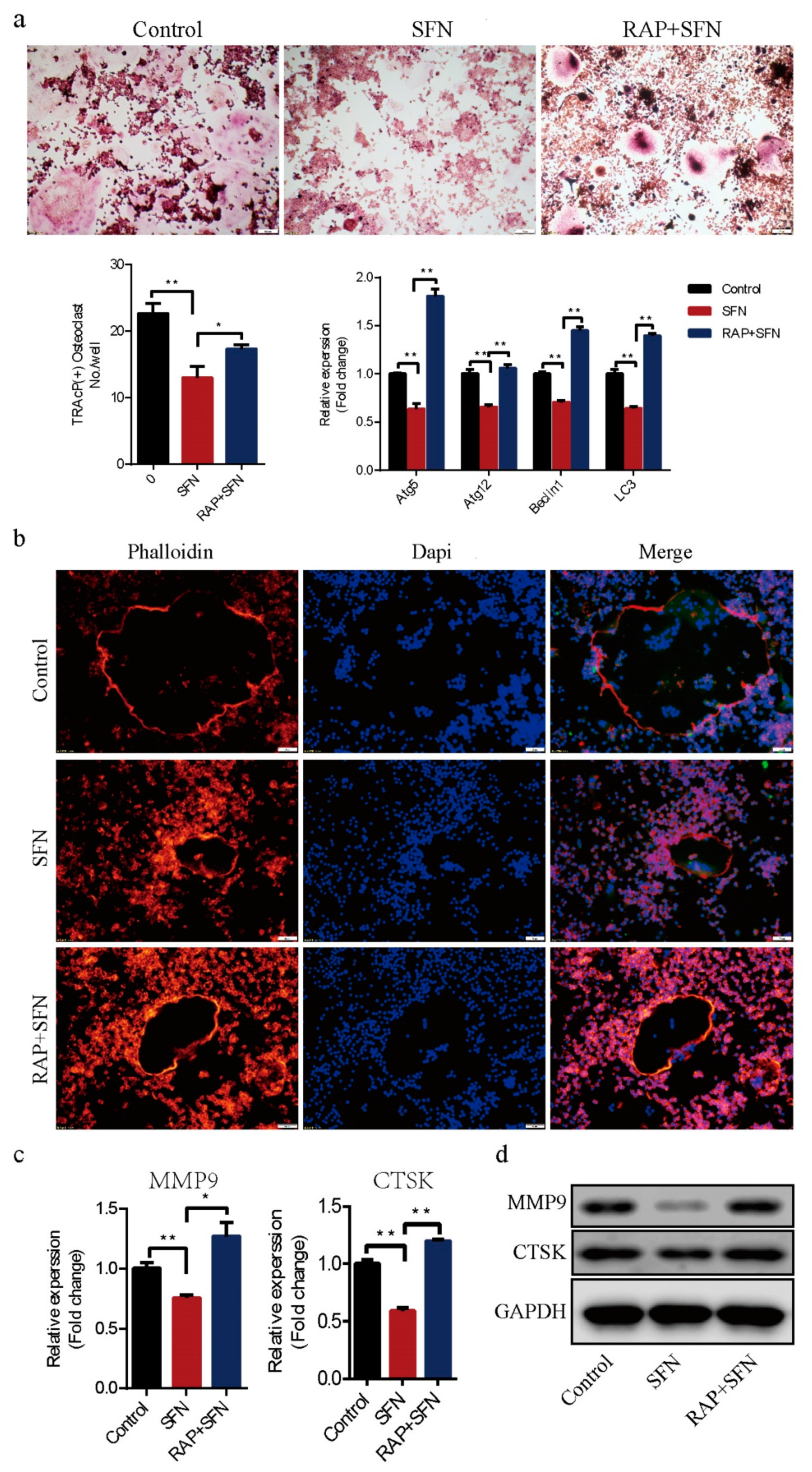
2.3. Rapamycin Reversed SFN-Mediated Anti-Osteoclatogenic Effects
2.4. c-Jun N-Terminal Kinase (JNK) Signaling Pathway Appeared to Participate in SFN-Mediated Autophagy Inhibition in the Early Period of Osteoclastogenesis
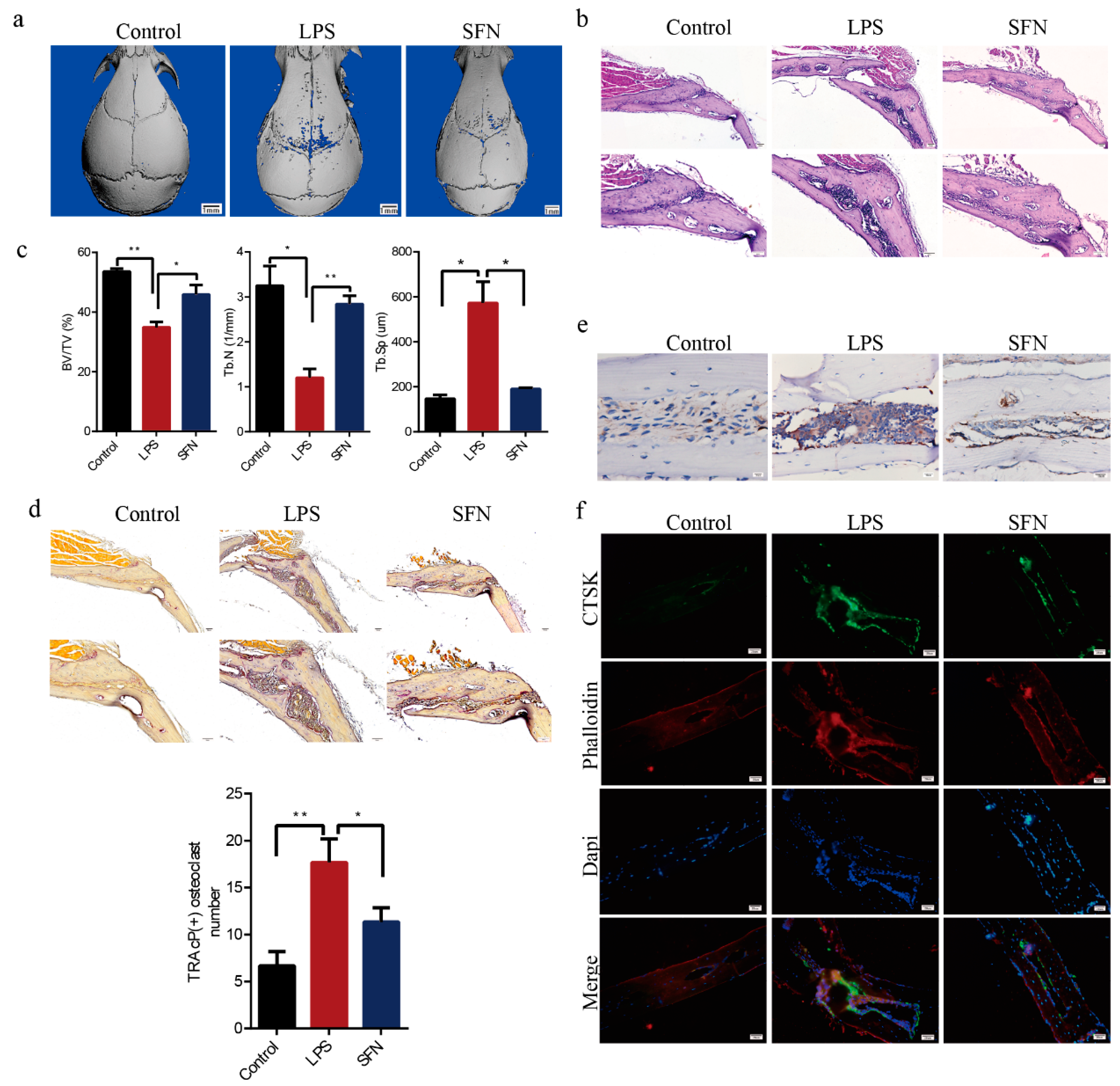
2.5. SFN Protected Against LPS-Induced Calvarial Bone Destruction in Mice
3. Discussion
4. Materials and Methods
4.1. Reagents and Mice
4.2. Preparation of Bone Marrow Macrophages (BMMs)
4.3. Proliferation Viability Assay
4.4. Osteoclast Differentiation Assay and Fibrous Actin (F-actin) Ring Observation
4.5. Immunofluorescence Analysis
4.6. Transmission Electron Microscopy (TEM)
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.8. Western Blot Assay
4.9. LPS-Mediated Calvarial Bone Erosion Experiment
4.10. MicroCT Scanning and Analysis
4.11. Bone Histological Analysis
4.12. Immunohistochemical and Immunofluorescence Analysis
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Inoue, K.; Hu, X.; Zhao, B. Regulatory network mediated by RBP-J/NFATc1-miR182 controls inflammatory bone resorption. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 2392–2407. [Google Scholar] [CrossRef] [PubMed]
- Kittaka, M.; Yoshimoto, T.; Schlosser, C.; Rottapel, R.; Kajiya, M.; Kurihara, H.; Reichenberger, E.J.; Ueki, Y. Alveolar Bone Protection by Targeting the SH3BP2-SYK Axis in Osteoclasts. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2020, 35, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Interactions between cancer cells and bone microenvironment promote bone metastasis in prostate cancer. Cancer Commun. 2019, 39, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Talalay, P.; Cho, C.G.; Posner, G.H. A major inducer of anticarcinogenic protective enzymes from broccoli: Isolation and elucidation of structure. Proc. Natl. Acad. Sci. USA 1992, 89, 2399–2403. [Google Scholar] [CrossRef] [PubMed]
- Houghton, C.A.; Fassett, R.G.; Coombes, J.S. Sulforaphane: Translational research from laboratory bench to clinic. Nutr. Rev. 2013, 71, 709–726. [Google Scholar] [CrossRef]
- Ko, J.Y.; Choi, Y.J.; Jeong, G.J.; Im, G.I. Sulforaphane-PLGA microspheres for the intra-articular treatment of osteoarthritis. Biomaterials 2013, 34, 5359–5368. [Google Scholar] [CrossRef]
- Patel, B.; Mann, G.E.; Chapple, S.J. Concerted redox modulation by sulforaphane alleviates diabetes and cardiometabolic syndrome. Free Radic. Biol. Med. 2018, 122, 150–160. [Google Scholar] [CrossRef]
- Matsui, T.A.; Sowa, Y.; Yoshida, T.; Murata, H.; Horinaka, M.; Wakada, M.; Nakanishi, R.; Sakabe, T.; Kubo, T.; Sakai, T. Sulforaphane enhances TRAIL-induced apoptosis through the induction of DR5 expression in human osteosarcoma cells. Carcinogenesis 2006, 27, 1768–1777. [Google Scholar] [CrossRef]
- Thaler, R.; Maurizi, A.; Roschger, P.; Sturmlechner, I.; Khani, F.; Spitzer, S.; Rumpler, M.; Zwerina, J.; Karlic, H.; Dudakovic, A.; et al. Anabolic and Antiresorptive Modulation of Bone Homeostasis by the Epigenetic Modulator Sulforaphane, a Naturally Occurring Isothiocyanate. J. Biol. Chem. 2016, 291, 6754–6771. [Google Scholar] [CrossRef]
- Takagi, T.; Inoue, H.; Takahashi, N.; Katsumata-Tsuboi, R.; Uehara, M. Sulforaphane inhibits osteoclast differentiation by suppressing the cell-cell fusion molecules DC-STAMP and OC-STAMP. Biochem. Biophys. Res. Commun. 2017, 483, 718–724. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Hu, S. Recent insights into the role of autophagy in the pathogenesis of rheumatoid arthritis. Rheumatology 2016, 55, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Ren, H.; Shang, Q.; Qiu, T.; Yu, X.; Zhang, Z.; Huang, J.; Zhao, W.; Zhang, Y.; Liang, D.; et al. Autophagy as a target for glucocorticoid-induced osteoporosis therapy. Cell. Mol. Life Sci. CMLS 2018, 75, 2683–2693. [Google Scholar] [CrossRef]
- Abdelaziz, D.M.; Stone, L.S.; Komarova, S.V. Osteolysis and pain due to experimental bone metastases are improved by treatment with rapamycin. Breast Cancer Res. Treat. 2014, 143, 227–237. [Google Scholar] [CrossRef] [PubMed]
- DeSelm, C.J.; Miller, B.C.; Zou, W.; Beatty, W.L.; van Meel, E.; Takahata, Y.; Klumperman, J.; Tooze, S.A.; Teitelbaum, S.L.; Virgin, H.W. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell 2011, 21, 966–974. [Google Scholar] [CrossRef]
- Arai, A.; Kim, S.; Goldshteyn, V.; Kim, T.; Park, N.H.; Wang, C.Y.; Kim, R.H. Beclin1 Modulates Bone Homeostasis by Regulating Osteoclast and Chondrocyte Differentiation. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2019, 34, 1753–1766. [Google Scholar] [CrossRef]
- Lee, J.H.; Jeong, J.K.; Park, S.Y. Sulforaphane-induced autophagy flux prevents prion protein-mediated neurotoxicity through AMPK pathway. Neuroscience 2014, 278, 31–39. [Google Scholar] [CrossRef]
- Yang, F.; Wang, F.; Liu, Y.; Wang, S.; Li, X.; Huang, Y.; Xia, Y.; Cao, C. Sulforaphane induces autophagy by inhibition of HDAC6-mediated PTEN activation in triple negative breast cancer cells. Life Sci. 2018, 213, 149–157. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.M.; Jeong, J.U.; Shin, J.H.; Shin, J.M.; Bang, K.T. Nrf2-heme oxygenase-1 modulates autophagy and inhibits apoptosis triggered by elevated glucose levels in renal tubule cells. Kidney Res. Clin. Pract. 2019, 38, 318–325. [Google Scholar] [CrossRef]
- Zheng, K.; Ma, J.; Wang, Y.; He, Z.; Deng, K. Sulforaphane inhibits autophagy and induces exosome-mediated paracrine senescence via regulating mtor/tfe3. Mol. Nutr. Food Res. 2020, 64, e1901231. [Google Scholar] [CrossRef]
- Tope, A.M.; Rogers, P.F. Evaluation of protective effects of sulforaphane on DNA damage caused by exposure to low levels of pesticide mixture using comet assay. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2009, 44, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Wang, X.; Wang, W.; He, C.; Bao, Y. p38 MAPK plays a distinct role in sulforaphane-induced up-regulation of ARE-dependent enzymes and down-regulation of COX-2 in human bladder cancer cells. Oncol. Rep. 2010, 23, 1133–1138. [Google Scholar] [CrossRef]
- Lan, F.; Pan, Q.; Yu, H.; Yue, X. Sulforaphane enhances temozolomide-induced apoptosis because of down-regulation of miR-21 via Wnt/beta-catenin signaling in glioblastoma. J. Neurochem. 2015, 134, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Bertl, E.; Bartsch, H.; Gerhauser, C. Inhibition of angiogenesis and endothelial cell functions are novel sulforaphane-mediated mechanisms in chemoprevention. Mol. Cancer Ther. 2006, 5, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Pore, S.K.; Hahm, E.R.; Kim, S.H.; Singh, K.B.; Nyiranshuti, L.; Latoche, J.D.; Anderson, C.J.; Adamik, J.; Galson, D.L.; Weiss, K.R.; et al. A Novel Sulforaphane-Regulated Gene Network in Suppression of Breast Cancer-Induced Osteolytic Bone Resorption. Mol. Cancer Ther. 2020, 19, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wei, B.; Li, G.; Zheng, J.; Sun, J.; Chu, J.; Zeng, R.; Niu, Y. Sulforaphane reverses glucocorticoid-induced apoptosis in osteoblastic cells through regulation of the Nrf2 pathway. Drug Design Dev. Ther. 2014, 8, 973–982. [Google Scholar] [CrossRef]
- Javaheri, B.; Poulet, B.; Aljazzar, A.; de Souza, R.; Piles, M.; Hopkinson, M.; Shervill, E.; Pollard, A.; Chan, B.; Chang, Y.M.; et al. Stable sulforaphane protects against gait anomalies and modifies bone microarchitecture in the spontaneous STR/Ort model of osteoarthritis. Bone 2017, 103, 308–317. [Google Scholar] [CrossRef]
- Xue, P.; Hu, X.; Powers, J.; Nay, N.; Chang, E.; Kwon, J.; Wong, S.W.; Han, L.; Wu, T.H.; Lee, D.J.; et al. CDDO-Me, Sulforaphane and tBHQ attenuate the RANKL-induced osteoclast differentiation via activating the NRF2-mediated antioxidant response. Biochem. Biophys. Res. Commun. 2019, 511, 637–643. [Google Scholar] [CrossRef]
- Naumann, P.; Fortunato, F.; Zentgraf, H.; Buchler, M.W.; Herr, I.; Werner, J. Autophagy and cell death signaling following dietary sulforaphane act independently of each other and require oxidative stress in pancreatic cancer. Int. J. Oncol. 2011, 39, 101–109. [Google Scholar] [CrossRef]
- Jo, C.; Kim, S.; Cho, S.J.; Choi, K.J.; Yun, S.M.; Koh, Y.H.; Johnson, G.V.; Park, S.I. Sulforaphane induces autophagy through ERK activation in neuronal cells. FEBS Lett. 2014, 588, 3081–3088. [Google Scholar] [CrossRef]
- Van Limbergen, J.; Stevens, C.; Nimmo, E.R.; Wilson, D.C.; Satsangi, J. Autophagy: From basic science to clinical application. Mucosal Immunol. 2009, 2, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, E.; Lippens, S.; Vanden Berghe, T.; Romagnoli, A.; Fimia, G.M.; Piacentini, M.; Vandenabeele, P. Beclin1: A role in membrane dynamics and beyond. Autophagy 2012, 8, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Fujita, N.; Kanno, E.; Yamamoto, A.; Yoshimori, T.; Fukuda, M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell 2008, 19, 2916–2925. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Ren, T.; Huang, Y.; Sun, K.; Bao, X.; Wang, S.; Zheng, B.; Guo, W. Apatinib promotes autophagy and apoptosis through VEGFR2/STAT3/BCL-2 signaling in osteosarcoma. Cell Death Dis. 2017, 8, e3015. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.J.; Kong, F.Q.; Cai, W.; Xu, T.; Zhou, Z.M.; Wang, Z.B.; Xu, A.D.; Yang, Y.Q.; Chen, J.; Tang, P.Y.; et al. GIT1 contributes to autophagy in osteoclast through disruption of the binding of Beclin1 and Bcl2 under starvation condition. Cell Death Dis. 2018, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.Y.; Yang, B.; Shi, Y.X.; Zhang, W.L.; Liu, F.; Zhao, W.; Yang, M.W. High glucose downregulates the effects of autophagy on osteoclastogenesis via the AMPK/mTOR/ULK1 pathway. Biochem. Biophys. Res. Commun. 2018, 503, 428–435. [Google Scholar] [CrossRef]
- Ke, D.; Fu, X.; Xue, Y.; Wu, H.; Zhang, Y.; Chen, X.; Hou, J. IL-17A regulates the autophagic activity of osteoclast precursors through RANKL-JNK1 signaling during osteoclastogenesis in vitro. Biochem. Biophys. Res. Commun. 2018, 497, 890–896. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Gestwicki, J.E.; Murphy, L.O.; Klionsky, D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007, 6, 304–312. [Google Scholar] [CrossRef]
- Lee, K.; Chung, Y.H.; Ahn, H.; Kim, H.; Rho, J.; Jeong, D. Selective Regulation of MAPK Signaling Mediates RANKL-dependent Osteoclast Differentiation. Int. J. Biol. Sci. 2016, 12, 235–245. [Google Scholar] [CrossRef]
- Zhou, J.; Fan, Y.; Zhong, J.; Huang, Z.; Huang, T.; Lin, S.; Chen, H. TAK1 mediates excessive autophagy via p38 and ERK in cisplatin-induced acute kidney injury. J. Cell. Mol. Med. 2018, 22, 2908–2921. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; She, H.; Zhang, T.; Xu, H.; Cheng, L.; Yepes, M.; Zhao, Y.; Mao, Z. p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J. Cell Biol. 2018, 217, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Bryant, K.L.; Stalnecker, C.A.; Zeitouni, D.; Klomp, J.E.; Peng, S.; Tikunov, A.P.; Gunda, V.; Pierobon, M.; Waters, A.M.; George, S.D.; et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019, 25, 628–640. [Google Scholar] [CrossRef]
- Liu, G.Y.; Jiang, X.X.; Zhu, X.; He, W.Y.; Kuang, Y.L.; Ren, K.; Lin, Y.; Gou, X. ROS activates JNK-mediated autophagy to counteract apoptosis in mouse mesenchymal stem cells in vitro. Acta Pharmacol. Sin. 2015, 36, 1473–1479. [Google Scholar] [CrossRef]
- Suda, K.; Woo, J.T.; Takami, M.; Sexton, P.M.; Nagai, K. Lipopolysaccharide supports survival and fusion of preosteoclasts independent of TNF-alpha, IL-1, and RANKL. J. Cell. Physiol. 2002, 190, 101–108. [Google Scholar] [CrossRef]
- Wu, H.; Hu, B.; Zhou, X.; Zhou, C.; Meng, J.; Yang, Y.; Zhao, X.; Shi, Z.; Yan, S. Artemether attenuates LPS-induced inflammatory bone loss by inhibiting osteoclastogenesis and bone resorption via suppression of MAPK signaling pathway. Cell Death Dis. 2018, 9, 498. [Google Scholar] [CrossRef]



| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| MMP9 | CAAAGACCTGAAAACCTCCAAC | GACTGCTTCTCTCCCATCATC |
| CTSK | GCTTGGCATCTTTCCAGTTTTA | CAACACTGCATGGTTCACATTA |
| Atg5 | AGTCAAGTGATCAACGAAATGC | TATTCCATGAGTTTCCGGTTGA |
| Atg12 | GCCTCGGAACAGTTGTTTATTT | CAGTTTACCATCACTGCCAAAA |
| Beclin1 | TAATAGCTTCACTCTGATCGGG | CAAACAGCGTTTGTAGTTCTGA |
| LC3 | CTGTCCTGGATAAGACCAAGTT | GTCTTCATCCTTCTCCTGTTCA |
| GAPDH | GAPDH primer F and R were purchased from Sangon Biotech (Shanghai, China) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, T.; Fu, X.; Liu, Y.; Ji, Y.; Shang, Z. Sulforaphane Inhibits Osteoclastogenesis via Suppression of the Autophagic Pathway. Molecules 2021, 26, 347. https://doi.org/10.3390/molecules26020347
Luo T, Fu X, Liu Y, Ji Y, Shang Z. Sulforaphane Inhibits Osteoclastogenesis via Suppression of the Autophagic Pathway. Molecules. 2021; 26(2):347. https://doi.org/10.3390/molecules26020347
Chicago/Turabian StyleLuo, Tingting, Xiazhou Fu, Yaoli Liu, Yaoting Ji, and Zhengjun Shang. 2021. "Sulforaphane Inhibits Osteoclastogenesis via Suppression of the Autophagic Pathway" Molecules 26, no. 2: 347. https://doi.org/10.3390/molecules26020347
APA StyleLuo, T., Fu, X., Liu, Y., Ji, Y., & Shang, Z. (2021). Sulforaphane Inhibits Osteoclastogenesis via Suppression of the Autophagic Pathway. Molecules, 26(2), 347. https://doi.org/10.3390/molecules26020347






