Differential Protein Expression in Exponential and Stationary Growth Phases of Mycobacterium avium subsp. hominissuis 104
Abstract
1. Introduction
2. Results
3. Discussion
4. Methods
4.1. Bacterial Strains, Culture Media, and Growth Conditions
4.2. Bacterial Protein Preparation
4.3. SDS-PAGE
4.4. Isoelectric Focusing (IEF) and Second Dimension Gel Electrophoresis (2-DE)
4.5. Staining of the Gel and Visualization of the Spots
4.6. Processing of the Spot and Mass Identification (TripleTOF MS/MS)
4.7. Bioinformatic Analysis
4.8. Traditional PCR Reaction
4.9. RNA Extraction
4.10. Quantitative Reverse Transcription Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Park, S.C.; Kang, M.J.; Han, C.H.; Lee, S.M.; Kim, C.J.; Lee, J.M.; Kang, Y.A. Prevalence, incidence, and mortality of nontuberculous mycobacterial infection in Korea: A nationwide population-based study. BMC Pulm. Med. 2019, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, H.; Kurashima, A.; Morimoto, K.; Hoshino, Y.; Hasegawa, N.; Ato, M.; Mitarai, S. Epidemiology of Pulmonary Nontuberculous Mycobacterial Disease, Japan. Emerg. Infect. Dis. 2016, 22, 1116–1117. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Lezcano, O.M.; Gonzalez-Cortes, C.; Mirsaeidi, M. The unexplained increase of nontuberculous mycobacteriosis. Int. J. Mycobacteriol. 2019, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Kuronuma, K.; Saito, A.; Ikeda, K.; Ariki, S.; Saitou, A.; Otsuka, M.; Chiba, H.; Takahashi, S.; Takahashi, M.; et al. Insufficient serum L-ficolin is associated with disease presence and extent of pulmonary Mycobacterium avium complex disease. Respir. Res. 2019, 20, 224. [Google Scholar] [CrossRef] [PubMed]
- Sakatani, M. The non-tuberculous mycobacteriosis. Kekkaku [Tuberculosis] 2005, 80, 25–30. [Google Scholar]
- Uchiya, K.; Takahashi, H.; Yagi, T.; Moriyama, M.; Inagaki, T.; Ichikawa, K.; Nakagawa, T.; Nikai, T.; Ogawa, K. Comparative genome analysis of Mycobacterium avium revealed genetic diversity in strains that cause pulmonary and disseminated disease. PLoS ONE 2013, 8, e71831. [Google Scholar] [CrossRef]
- Horan, K.L.; Freeman, R.; Weigel, K.; Semret, M.; Pfaller, S.; Covert, T.C.; van Soolingen, D.; Leao, S.C.; Behr, M.A.; Cangelosi, G.A. Isolation of the genome sequence strain Mycobacterium avium 104 from multiple patients over a 17-year period. J. Clin. Microbiol. 2006, 44, 783–789. [Google Scholar] [CrossRef]
- Li, L.; Bannantine, J.P.; Zhang, Q.; Amonsin, A.; May, B.J.; Alt, D.; Banerji, N.; Kanjilal, S.; Kapur, V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 2005, 102, 12344–12349. [Google Scholar] [CrossRef]
- Ignatov, D.; Malakho, S.; Majorov, K.; Skvortsov, T.; Apt, A.; Azhikina, T. RNA-Seq analysis of Mycobacterium avium non-coding transcriptome. PLoS ONE 2013, 8, e74209. [Google Scholar] [CrossRef]
- Janagama, H.K.; Lamont, E.A.; George, S.; Bannantine, J.P.; Xu, W.W.; Tu, Z.J.; Wells, S.J.; Schefers, J.; Sreevatsan, S. Primary transcriptomes of Mycobacterium avium subsp. paratuberculosis reveal proprietary pathways in tissue and macrophages. BMC Genom. 2010, 11, 561. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Talaat, A.M. Genomic and transcriptomic studies in Mycobacterium avium subspecies paratuberculosis. Vet. Immunol. Immunopathol. 2010, 138, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Rojony, R.; Martin, M.; Campeau, A.; Wozniak, J.M.; Gonzalez, D.J.; Jaiswal, P.; Danelishvili, L.; Bermudez, L.E. Quantitative analysis of Mycobacterium avium subsp. hominissuis proteome in response to antibiotics and during exposure to different environmental conditions. Clin. Proteom. 2019, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Chinison, J.J.; Danelishvili, L.; Gupta, R.; Rose, S.J.; Babrak, L.M.; Bermudez, L.E. Identification of Mycobacterium avium subsp. hominissuis secreted proteins using an in vitro system mimicking the phagosomal environment. BMC Microbiol. 2016, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Radosevich, T.J.; Reinhardt, T.A.; Lippolis, J.D.; Bannantine, J.P.; Stabel, J.R. Proteome and differential expression analysis of membrane and cytosolic proteins from Mycobacterium avium subsp. paratuberculosis strains K-10 and 187. J. Bacteriol. 2007, 189, 1109–1117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- He, Z.; De Buck, J. Localization of proteins in the cell wall of Mycobacterium avium subsp. paratuberculosis K10 by proteomic analysis. Proteome Sci. 2010, 8, 21. [Google Scholar] [CrossRef]
- Lange, R.; Hengge-Aronis, R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 1991, 5, 49–59. [Google Scholar] [CrossRef]
- Phillips, Z.E.; Strauch, M.A. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. CMLS 2002, 59, 392–402. [Google Scholar] [CrossRef]
- Bernhardt, J.; Weibezahn, J.; Scharf, C.; Hecker, M. Bacillus subtilis during feast and famine: Visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 2003, 13, 224–237. [Google Scholar] [CrossRef]
- Ntolosi, B.A.; Betts, J.; Zappe, H.; Powles, R.; Steyn, L.M. Growth phase-associated changes in protein expression in Mycobacterium smegmatis identify a new low molecular weight heat shock protein. Tuberculosis 2001, 81, 279–289. [Google Scholar] [CrossRef]
- Yuan, Y.; Crane, D.D.; Barry, C.E., 3rd. Stationary phase-associated protein expression in Mycobacterium tuberculosis: Function of the mycobacterial alpha-crystallin homolog. J. Bacteriol. 1996, 178, 4484–4492. [Google Scholar] [CrossRef]
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.H.; Garrity, J.; Crosby, H.A.; Escalante-Semerena, J.C. In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Mol. Microbiol. 2011, 80, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Han, M.J.; Lee, S.Y.; Jeong, K.J.; Yoo, J.S. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 2003, 81, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.E.; Smalley, D.J.; Conway, T. Gene expression profiling of Escherichia coli growth transitions: An expanded stringent response model. Mol. Microbiol. 2002, 45, 289–306. [Google Scholar] [CrossRef]
- Mitchell, G.; Ge, L.; Huang, Q.; Chen, C.; Kianian, S.; Roberts, M.F.; Schekman, R.; Portnoy, D.A. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun. 2015, 83, 2175–2184. [Google Scholar] [CrossRef]
- Unnikrishnan, M.; Cohen, J.; Sriskandan, S. Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect. Immun. 1999, 67, 5495–5499. [Google Scholar] [CrossRef]
- Martinot, A.J.; Farrow, M.; Bai, L.; Layre, E.; Cheng, T.Y.; Tsai, J.H.; Iqbal, J.; Annand, J.W.; Sullivan, Z.A.; Hussain, M.M.; et al. Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis. PLoS Pathog. 2016, 12, e1005351. [Google Scholar] [CrossRef]
- Aguilar-Lopez, B.A.; Correa, F.; Moreno-Altamirano, M.M.B.; Espitia, C.; Hernandez-Longoria, R.; Oliva-Ramirez, J.; Padierna-Olivos, J.; Sanchez-Garcia, F.J. LprG and PE_PGRS33 Mycobacterium tuberculosis virulence factors induce differential mitochondrial dynamics in macrophages. Scand. J. Immunol. 2019, 89, e12728. [Google Scholar] [CrossRef]
- Viale, M.N.; Park, K.T.; Imperiale, B.; Gioffre, A.K.; Colombatti Olivieri, M.A.; Moyano, R.D.; Morcillo, N.; Santangelo, M.d.l.P.; Davis, W.; Romano, M.I. Characterization of a Mycobacterium avium subsp. Avium Operon Associated with Virulence and Drug Detoxification. BioMed Res. Int. 2014, 2014, 809585. [Google Scholar] [CrossRef] [PubMed]
- Lazdunski, A.M. Peptidases and proteases of Escherichia coli and Salmonella typhimurium. FEMS Microbiol. Rev. 1989, 5, 265–276. [Google Scholar] [CrossRef]
- Dragset, M.S.; Ioerger, T.R.; Loevenich, M.; Haug, M.; Sivakumar, N.; Marstad, A.; Cardona, P.J.; Klinkenberg, G.; Rubin, E.J.; Steigedal, M.; et al. Global Assessment of Mycobacterium avium subsp. hominissuis Genetic Requirement for Growth and Virulence. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Manca, C.; Paul, S.; Barry, C.E., 3rd; Freedman, V.H.; Kaplan, G. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 1999, 67, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Phong, W.Y.; Lin, W.; Rao, S.P.; Dick, T.; Alonso, S.; Pethe, K. Characterization of phosphofructokinase activity in Mycobacterium tuberculosis reveals that a functional glycolytic carbon flow is necessary to limit the accumulation of toxic metabolic intermediates under hypoxia. PLoS ONE 2013, 8, e56037. [Google Scholar] [CrossRef]
- Muthusamy, S.; Lundin, D.; Mamede Branca, R.M.; Baltar, F.; Gonzalez, J.M.; Lehtio, J.; Pinhassi, J. Comparative proteomics reveals signature metabolisms of exponentially growing and stationary phase marine bacteria. Environ. Microbiol. 2017, 19, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Bathke, J.; Konzer, A.; Remes, B.; McIntosh, M.; Klug, G. Comparative analyses of the variation of the transcriptome and proteome of Rhodobacter sphaeroides throughout growth. BMC Genom. 2019, 20, 358. [Google Scholar] [CrossRef]
- Lanigan, M.D.; Vaughan, J.A.; Shiell, B.J.; Beddome, G.J.; Michalski, W.P. Mycobacterial proteome extraction: Comparison of disruption methods. Proteomics 2004, 4, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Enany, S.; Yoshida, Y.; Yamamoto, T. Exploring extra-cellular proteins in methicillin susceptible and methicillin resistant Staphylococcus aureus by liquid chromatography-tandem mass spectrometry. World J. Microbiol. Biotechnol. 2014, 30, 1269–1283. [Google Scholar] [CrossRef]
- Magdeldin, S.; Enany, S.; Yoshida, Y.; Xu, B.; Zhang, Y.; Zureena, Z.; Lokamani, I.; Yaoita, E.; Yamamoto, T. Basics and recent advances of two dimensional- polyacrylamide gel electrophoresis. Clin. Proteom. 2014, 11, 16. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [PubMed]
- Enany, S.; Yoshida, Y.; Tateishi, Y.; Ozeki, Y.; Nishiyama, A.; Savitskaya, A.; Yamaguchi, T.; Ohara, Y.; Yamamoto, T.; Ato, M.; et al. Mycobacterial DNA-binding protein 1 is critical for long term survival of Mycobacterium smegmatis and simultaneously coordinates cellular functions. Sci. Rep. 2017, 7, 6810. [Google Scholar] [CrossRef] [PubMed]
- Arenas, N.E.; Salazar, L.M. Steps and Tools for PCR-Based Technique Design. In Biotechnology and Bioengineering; Jacob-Lopes, E., Zepka, L.Q., Eds.; IntechOpen: London, UK, 2019; pp. 1–16. [Google Scholar]

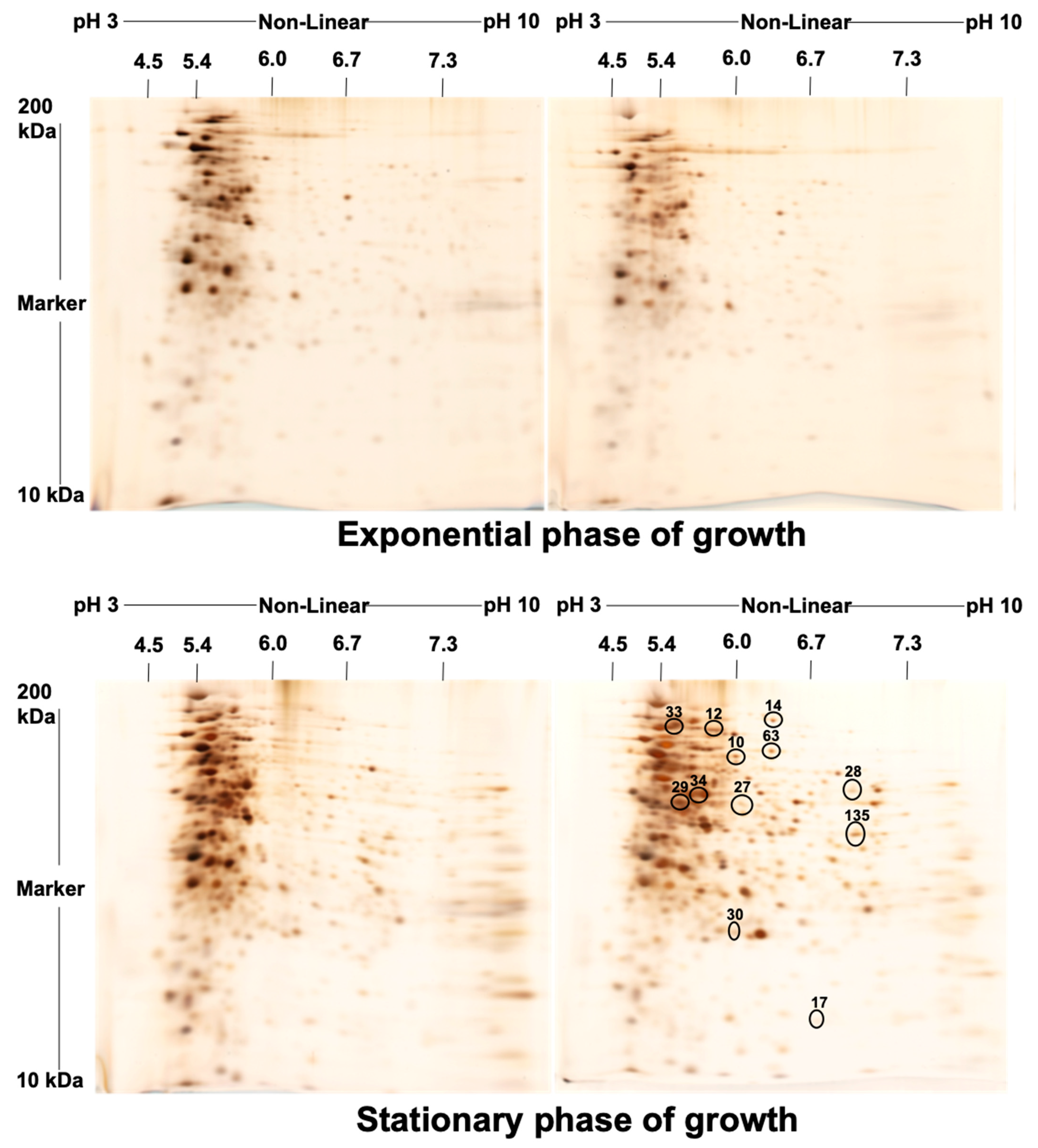

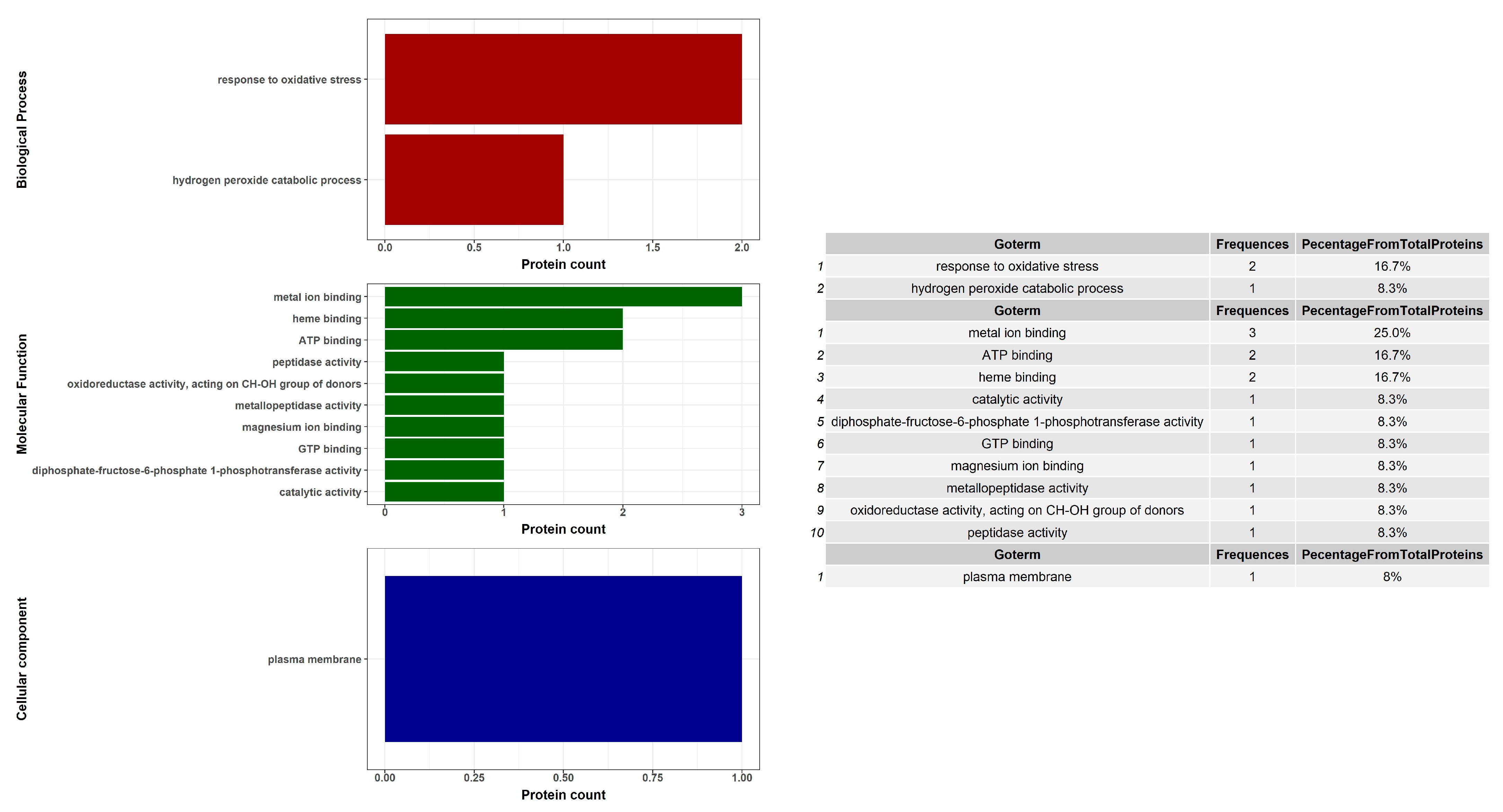

| Spot No. | Fold Change (+/−) * | ANOVA (p) | Average Normalized Volumes | Accession No. | Protein Name | Protein Score | No. Identified Peptides | Gene Name | No. (qRT-PCR) | Molecular Function | Regulation at Transcriptomic Level * | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exponential Growth Phase | Stationary Growth Phase | |||||||||||
| 10 | +4.4 | 0.018 | 11,896.756 | 52,584.334 | A0A0H2ZX69 | Pyruvate decarboxylase | 655 | 25 | MAV_4093 | 1 | Catalytic activity | + |
| 12 | +4.3 | 0.038 | 4334.274 | 18,578.945 | A0A0H2ZZB0 | Catalase | 2211 | 21 | katE | 2 | Catalase activity | + |
| 14 | +4 | 0.048 | 8140.139 | 32,853.588 | A0A0E2WBQ0 | ATP-dependent Clp protease ATP-binding protein | 1176 | 24 | MAVA5_02330 | 3 | ATP binding and Chaperone | − |
| 17 | −3.2 | 0.035 | 33,720.648 | 10,671.303 | A0A0H3A465 | Universal stress protein family protein | 348 | 24 | MAV_3137 | 4 | Response to stress | − |
| 27 | +2.2 | 0.003 | 7519.716 | 16,866.019 | A0A0H2ZYK5 | Dibenzothiophene desulfurization enzyme C | 353 | 21 | MAV_3806 | 5 | Oxidoreductase activity | − |
| 28 | +2.2 | 0.045 | 9994.532 | 21,965.532 | A0A0H2ZU78 | aspartyl aminopeptidase | 419 | 26 | apeB | 6 | Aminopeptidase, Hydrolase, Metalloprotease, and Aminopeptidase activity | + |
| 29 | −2.2 | 0.046 | 530,650.636 | 244,551.945 | A0A0H2ZT78 | Acetyl-CoA acetyltransferase | 1532 | 26 | MAV_1198 | 7 | Transferase activity | + |
| 30 | +2.1 | 0.029 | 19,961.352 | 42,368.898 | A0A0H3A4W6 | LprG protein | 323 | 26 | MAV_3367 | 8 | Phosphatidylinositol binding | + |
| 33 | −2 | 0.035 | 449,155.112 | 221,267.396 | A0QGA4 | Catalase-peroxidase | 2066 | 25 | katG | 9 | Catalase activity | − |
| 34 | −2 | 0.015 | 150,556.978 | 75,806.224 | A0QL35 | Elongation factor Tu | 2605 | 20 | tuf | 10 | GTPase activity + translation elongation factor activity | − |
| 63 | +5.1 | 0.050 | 6531.054 | 33,156.744 | A0A0H2ZRA7 | Glucose-methanol-choline | 1014 | 26 | MAV_4925 | 11 | Choline dehydrogenase activity | + |
| 135 | +2.2 | 0.050 | 35,082.949 | 77,486.389 | A0A0H3A360 | 6-phosphofructokinase 1 | 800 | 23 | pfkA | 12 | 6-phosphofructokinase activity | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enany, S.; Ato, M.; Matsumoto, S. Differential Protein Expression in Exponential and Stationary Growth Phases of Mycobacterium avium subsp. hominissuis 104. Molecules 2021, 26, 305. https://doi.org/10.3390/molecules26020305
Enany S, Ato M, Matsumoto S. Differential Protein Expression in Exponential and Stationary Growth Phases of Mycobacterium avium subsp. hominissuis 104. Molecules. 2021; 26(2):305. https://doi.org/10.3390/molecules26020305
Chicago/Turabian StyleEnany, Shymaa, Manabu Ato, and Sohkichi Matsumoto. 2021. "Differential Protein Expression in Exponential and Stationary Growth Phases of Mycobacterium avium subsp. hominissuis 104" Molecules 26, no. 2: 305. https://doi.org/10.3390/molecules26020305
APA StyleEnany, S., Ato, M., & Matsumoto, S. (2021). Differential Protein Expression in Exponential and Stationary Growth Phases of Mycobacterium avium subsp. hominissuis 104. Molecules, 26(2), 305. https://doi.org/10.3390/molecules26020305





