Structural Characterization of Act c 10.0101 and Pun g 1.0101—Allergens from the Non-Specific Lipid Transfer Protein Family
Abstract
1. Introduction
2. Results
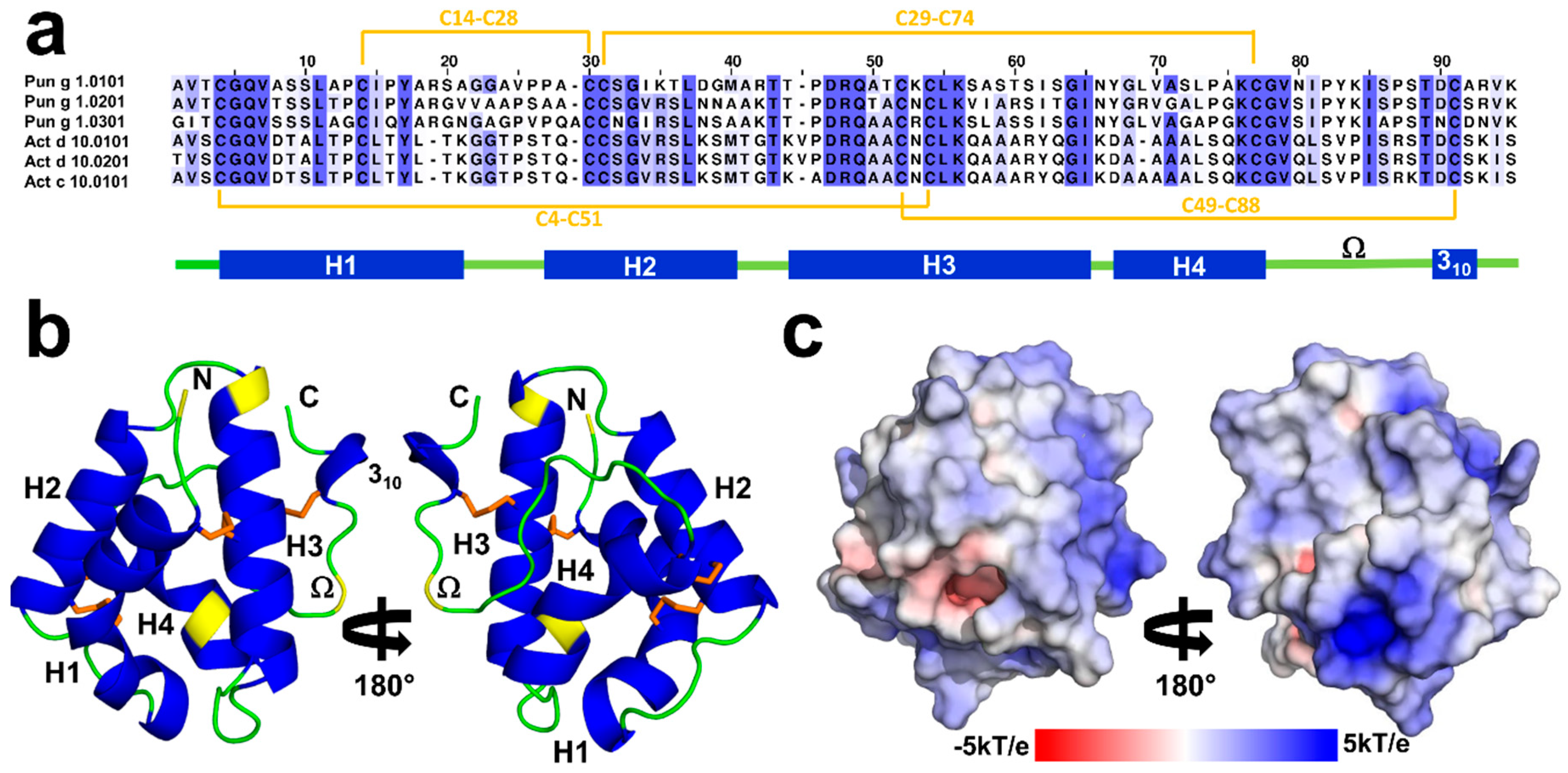
2.1. Overall Structures of Act c 10.0101
2.2. Overall Structures of Pun g 1.0101
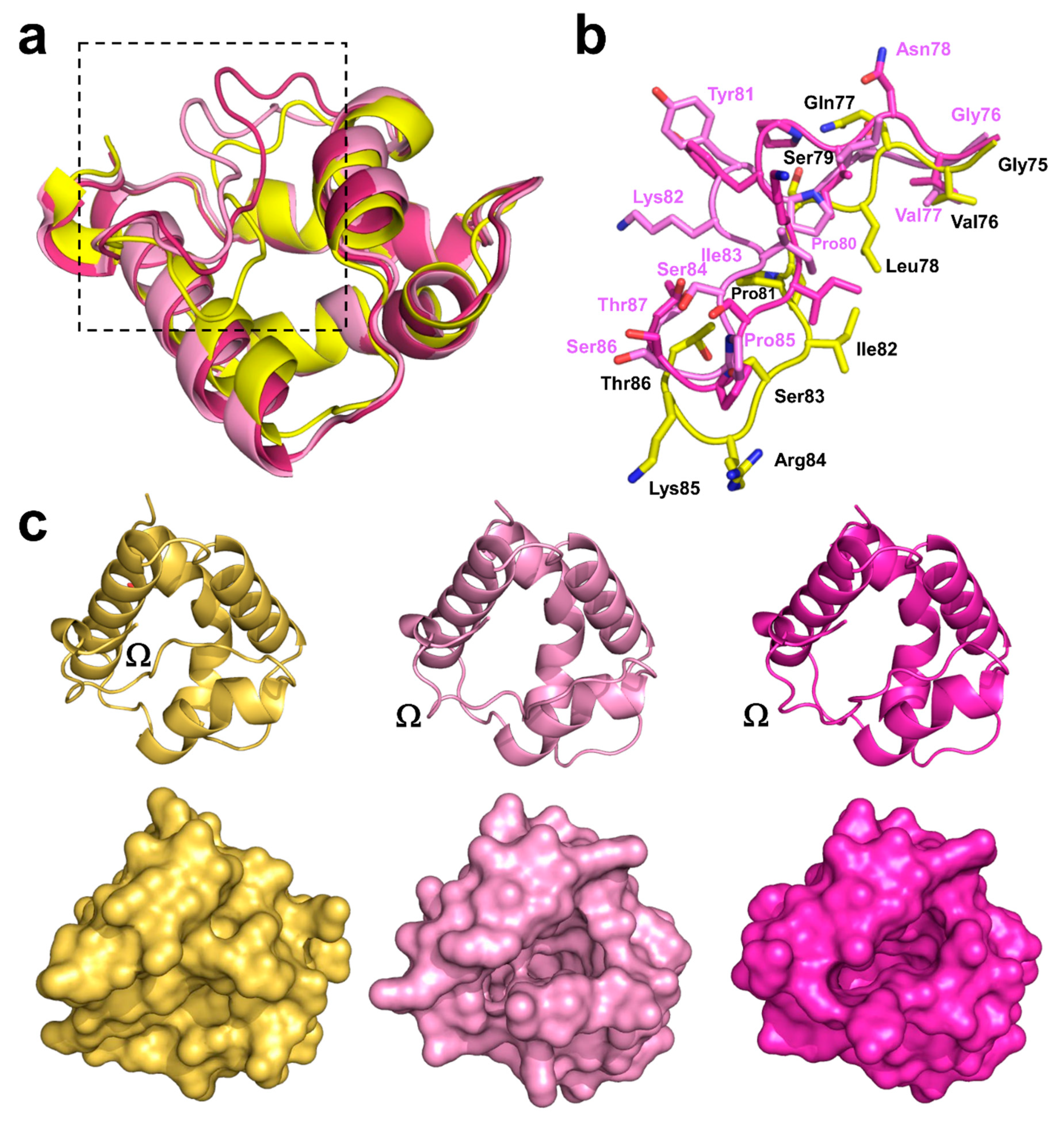
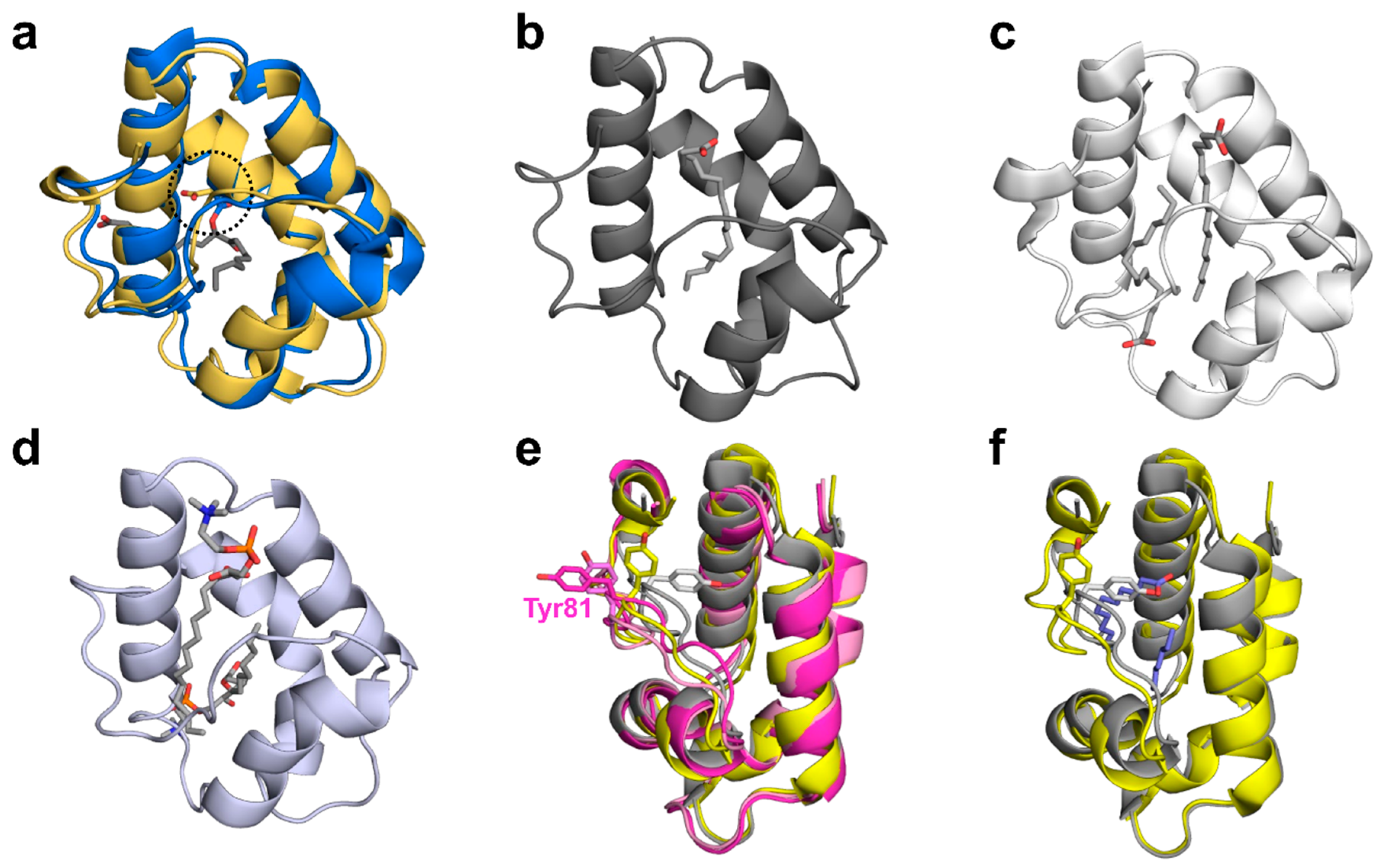
2.3. Structural Comparison with Other nsLTPs
2.3.1. Overall Protein Structure
2.3.2. Ligand Binding
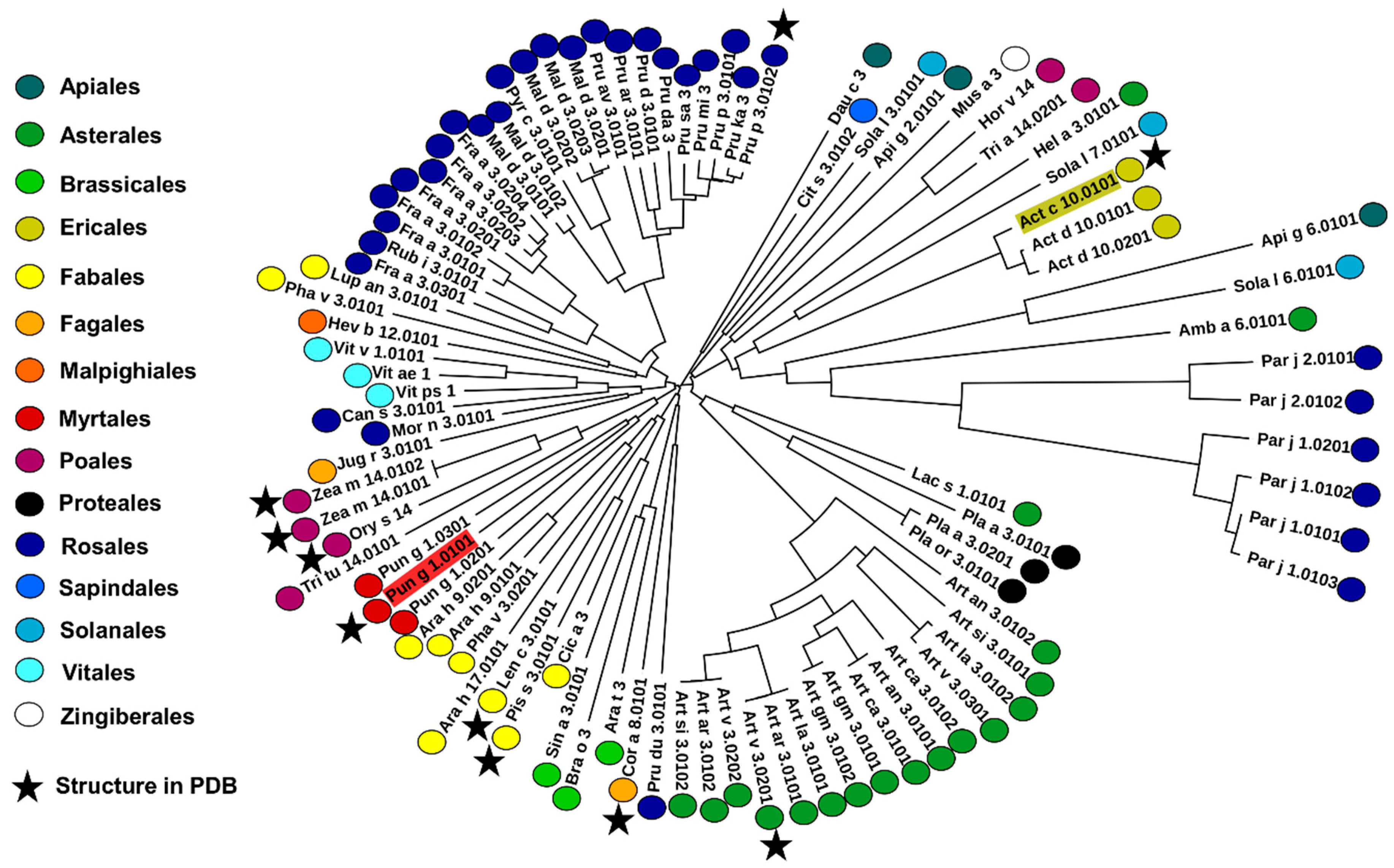
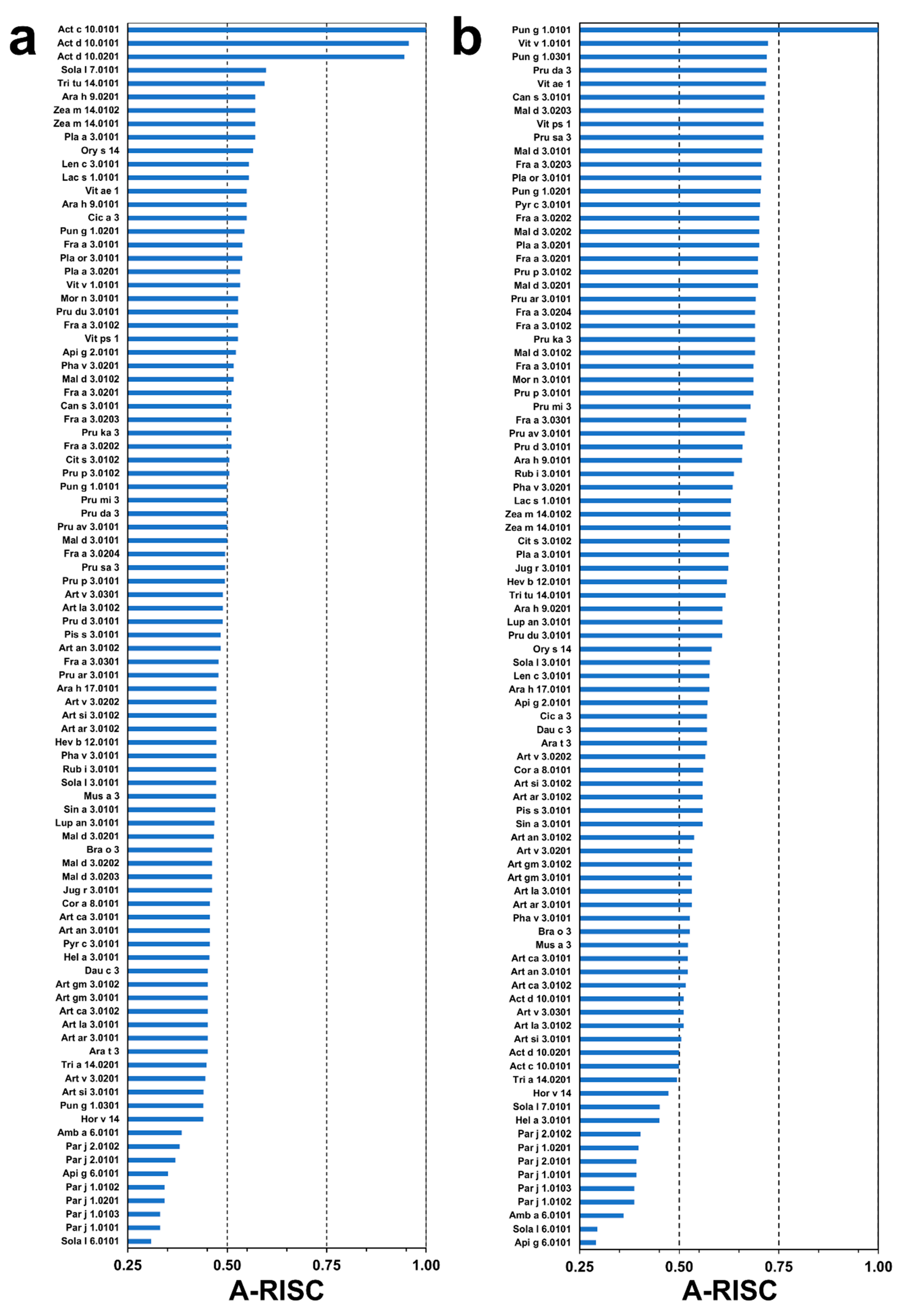
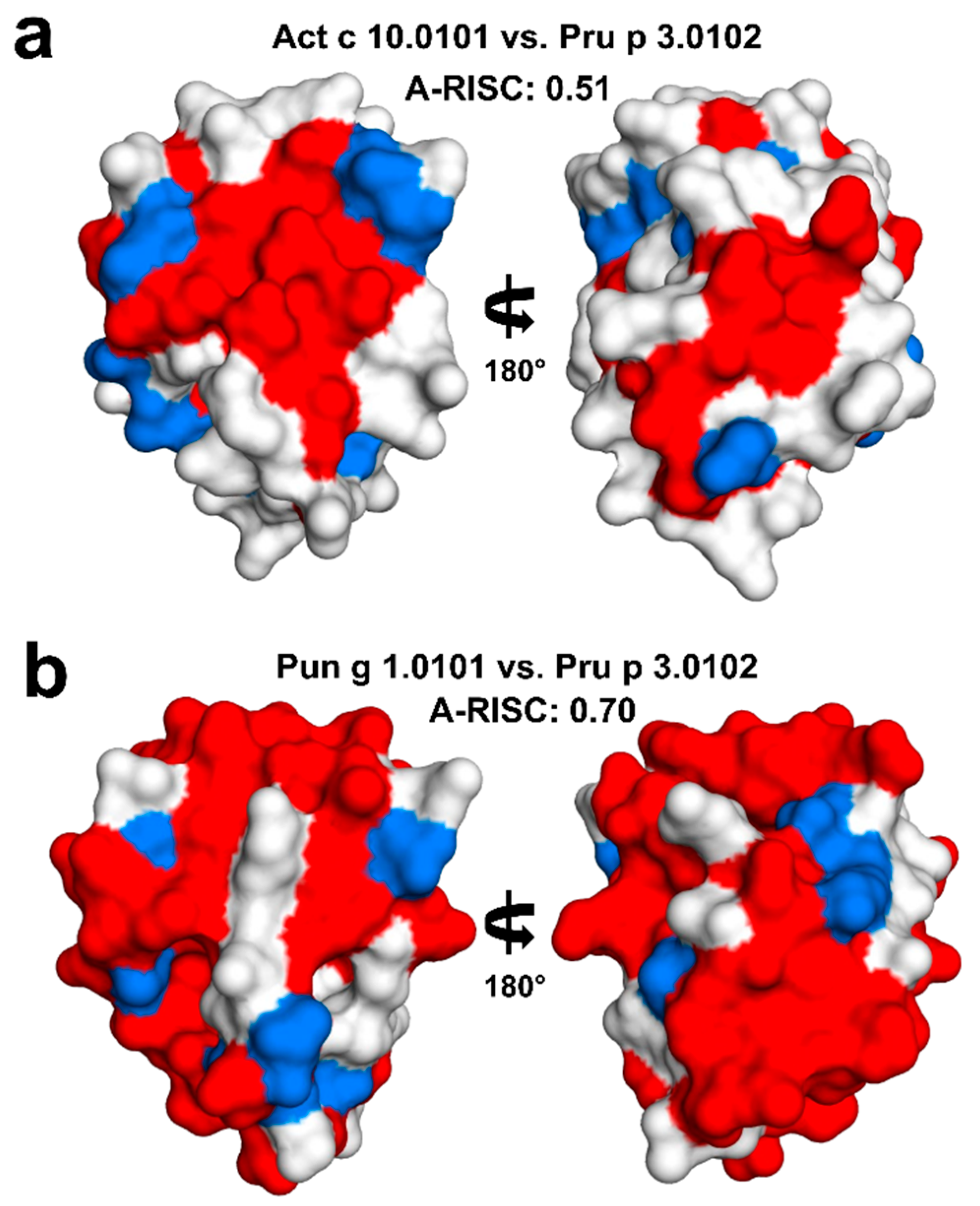
2.4. Analysis of Potential Cross-Reactivity between Act c 10.0101, Pun g 1.0101, and Other nsLTPs
3. Discussion
4. Materials and Methods
4.1. Protein Purification
4.2. Size-Exclusion Chromatography
4.3. Mass Spectrometric Analysis
4.4. Protein Crystallization and Data Collection
4.5. Structure Determination, Refinement, and Validation
4.6. Various Computational Approaches
4.7. Calculations of A-RISC Indexes
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gomar, J.; Petit, M.C.; Sodano, P.; Sy, D.; Marion, D.; Kader, J.C.; Vovelle, F.; Ptak, M. Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds. Protein Sci. 1996, 5, 565–577. [Google Scholar] [CrossRef]
- Lerche, M.H.; Poulsen, F.M. Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer proteins. Protein Sci. 1998, 7, 2490–2498. [Google Scholar] [CrossRef] [PubMed]
- Pasquato, N.; Berni, R.; Folli, C.; Folloni, S.; Cianci, M.; Pantano, S.; Helliwell, J.R.; Zanotti, G. Crystal structure of peach Pru p 3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens. J. Mol. Biol. 2006, 356, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Tassin-Moindrot, S.; Caille, A.; Douliez, J.P.; Marion, D.; Vovelle, F. The wide binding properties of a wheat nonspecific lipid transfer protein-Solution structure of a complex with prostaglandin B-2. Eur. J. Biochem. 2000, 267, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, I.V.; Shenkarev, Z.O.; Finkina, E.I.; Melnikova, D.N.; Rumynskiy, E.I.; Arseniev, A.S.; Ovchinnikova, T.V. A novel lipid transfer protein from the pea Pisum sativum: Isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties. BMC Plant Biol. 2016, 16, 107. [Google Scholar] [CrossRef]
- Melnikova, D.N.; Mineev, K.S.; Finkina, E.I.; Arseniev, A.S.; Ovchinnikova, T.V. A novel lipid transfer protein from the dill Anethum graveolens L.: Isolation, structure, heterologous expression, and functional characteristics. J. Pept. Sci. 2016, 22, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Carvalho Ade, O.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 2007, 28, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 2008, 17, 191–198. [Google Scholar] [CrossRef]
- Giangrieco, I.; Alessandri, C.; Rafaiani, C.; Santoro, M.; Zuzzi, S.; Tuppo, L.; Tamburrini, M.; D’Avino, R.; Ciardiello, M.A.; Mari, A. Structural features, IgE binding and preliminary clinical findings of the 7kDa Lipid Transfer Protein from tomato seeds. Mol. Immunol. 2015, 66, 154–163. [Google Scholar] [CrossRef]
- Salcedo, G.; Sanchez-Monge, R.; Diaz-Perales, A.; Garcia-Casado, G.; Barber, D. Plant non-specific lipid transfer proteins as food and pollen allergens. Clin. Exp. Allergy 2004, 34, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Vejvar, E.; Himly, M.; Briza, P.; Eichhorn, S.; Ebner, C.; Hemmer, W.; Ferreira, F.; Gadermaier, G. Allergenic relevance of nonspecific lipid transfer proteins 2: Identification and characterization of Api g 6 from celery tuber as representative of a novel IgE-binding protein family. Mol. Nutr. Food Res. 2013, 57, 2061–2070. [Google Scholar] [CrossRef] [PubMed]
- Edstam, M.M.; Viitanen, L.; Salminen, T.A.; Edqvist, J. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 2011, 4, 947–964. [Google Scholar] [CrossRef] [PubMed]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Plant Pathogenesis-Related Proteins PR-10 and PR-14 as Components of Innate Immunity System and Ubiquitous Allergens. Curr. Med. Chem. 2017, 24, 1772–1787. [Google Scholar] [CrossRef] [PubMed]
- Samuel, D.; Liu, Y.J.; Cheng, C.S.; Lyu, P.C. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa). J. Biol. Chem. 2002, 277, 35267–35273. [Google Scholar] [CrossRef]
- Ballmer-Weber, B.K. Lipid transfer protein as a potential panallergen? Allergy 2002, 57, 873–875. [Google Scholar] [CrossRef]
- Hauser, M.; Roulias, A.; Ferreira, F.; Egger, M. Panallergens and their impact on the allergic patient. Allergy Asthma Clin. Immunol. 2010, 6, 1. [Google Scholar] [CrossRef]
- Asero, R.; Piantanida, M.; Pinter, E.; Pravettoni, V. The clinical relevance of lipid transfer protein. Clin. Exp. Allergy 2018, 48, 6–12. [Google Scholar] [CrossRef]
- Romano, A.; Scala, E.; Rumi, G.; Gaeta, F.; Caruso, C.; Alonzi, C.; Maggioletti, M.; Ferrara, R.; Palazzo, P.; Palmieri, V.; et al. Lipid transfer proteins: The most frequent sensitizer in Italian subjects with food-dependent exercise-induced anaphylaxis. Clin. Exp. Allergy 2012, 42, 1643–1653. [Google Scholar] [CrossRef]
- Garcia-Casado, G.; Pacios, L.F.; Diaz-Perales, A.; Sanchez-Monge, R.; Lombardero, M.; Garcia-Selles, F.J.; Polo, F.; Barber, D.; Salcedo, G. Identification of IgE-binding epitopes of the major peach allergen Pru p 3. J. Allergy Clin. Immunol. 2003, 112, 599–605. [Google Scholar] [CrossRef]
- Zoccatelli, G.; Dalla Pellegrina, C.; Consolini, M.; Fusi, M.; Sforza, S.; Aquino, G.; Dossena, A.; Chignola, R.; Peruffo, A.; Olivieri, M.; et al. Isolation and identification of two lipid transfer proteins in pomegranate (Punica granatum). J. Agric. Food Chem. 2007, 55, 11057–11062. [Google Scholar] [CrossRef] [PubMed]
- Martin-Pedraza, L.; Gonzalez, M.; Gomez, F.; Blanca-Lopez, N.; Garrido-Arandia, M.; Rodriguez, R.; Torres, M.J.; Blanca, M.; Villalba, M.; Mayorga, C. Two nonspecific lipid transfer proteins (nsLTPs) from tomato seeds are associated to severe symptoms of tomato-allergic patients. Mol. Nutr. Food Res. 2016, 60, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Scala, E.; Till, S.J.; Asero, R.; Abeni, D.; Guerra, E.C.; Pirrotta, L.; Paganelli, R.; Pomponi, D.; Giani, M.; De Pita, O.; et al. Lipid transfer protein sensitization: Reactivity profiles and clinical risk assessment in an Italian cohort. Allergy 2015, 70, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Radauer, C.; Nandy, A.; Ferreira, F.; Goodman, R.E.; Larsen, J.N.; Lidholm, J.; Pomes, A.; Raulf-Heimsoth, M.; Rozynek, P.; Thomas, W.R.; et al. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy 2014, 69, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, M.L.; Giangrieco, I.; Camardella, L.; Ferrara, R.; Palazzo, P.; Panico, M.R.; Crescenzo, R.; Carratore, V.; Zennaro, D.; Liso, M.; et al. Allergenic lipid transfer proteins from plant-derived foods do not immunologically and clinically behave homogeneously: The kiwifruit LTP as a model. PLoS ONE 2011, 6, e27856. [Google Scholar] [CrossRef]
- Bublin, M.; Radauer, C.; Knulst, A.; Wagner, S.; Scheiner, O.; Mackie, A.R.; Mills, E.N.; Breiteneder, H. Effects of gastrointestinal digestion and heating on the allergenicity of the kiwi allergens Act d 1, actinidin, and Act d 2, a thaumatin-like protein. Mol. Nutr. Food Res. 2008, 52, 1130–1139. [Google Scholar] [CrossRef]
- Ciardiello, M.A.; Giangrieco, I.; Tuppo, L.; Tamburrini, M.; Buccheri, M.; Palazzo, P.; Bernardi, M.L.; Ferrara, R.; Mari, A. Influence of the natural ripening stage, cold storage, and ethylene treatment on the protein and IgE-binding profiles of green and gold kiwi fruit extracts. J. Agric. Food Chem. 2009, 57, 1565–1571. [Google Scholar] [CrossRef]
- Le, T.M.; Bublin, M.; Breiteneder, H.; Fernandez-Rivas, M.; Asero, R.; Ballmer-Weber, B.; Barreales, L.; Bures, P.; Belohlavkova, S.; de Blay, F.; et al. Kiwifruit allergy across Europe: Clinical manifestation and IgE recognition patterns to kiwifruit allergens. J. Allergy Clin. Immunol. 2013, 131, 164–171. [Google Scholar] [CrossRef]
- Lucas, J.S.; Lewis, S.A.; Trewin, J.B.; Grimshaw, K.E.; Warner, J.O.; Hourihane, J.O. Comparison of the allergenicity of Actinidia deliciosa (kiwi fruit) and Actinidia chinensis (gold kiwi). Pediatr. Allergy Immunol. 2005, 16, 647–654. [Google Scholar] [CrossRef]
- Palacin, A.; Rodriguez, J.; Blanco, C.; Lopez-Torrejon, G.; Sanchez-Monge, R.; Varela, J.; Jimenez, M.A.; Cumplido, J.; Carrillo, T.; Crespo, J.F.; et al. Immunoglobulin E recognition patterns to purified Kiwifruit (Actinidinia deliciosa) allergens in patients sensitized to Kiwi with different clinical symptoms. Clin. Exp. Allergy 2008, 38, 1220–1228. [Google Scholar] [CrossRef]
- Chruszcz, M.; Ciardiello, M.A.; Osinski, T.; Majorek, K.A.; Giangrieco, I.; Font, J.; Breiteneder, H.; Thalassinos, K.; Minor, W. Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins. Mol. Immunol. 2013, 56, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Offermann, L.R.; Giangrieco, I.; Perdue, M.L.; Zuzzi, S.; Santoro, M.; Tamburrini, M.; Cosgrove, D.J.; Mari, A.; Ciardiello, M.A.; Chruszcz, M. Elusive Structural, Functional, and Immunological Features of Act d 5, the Green Kiwifruit Kiwellin. J. Agric. Food Chem. 2015, 63, 6567–6576. [Google Scholar] [CrossRef] [PubMed]
- Hamiaux, C.; Maddumage, R.; Middleditch, M.J.; Prakash, R.; Brummell, D.A.; Baker, E.N.; Atkinson, R.G. Crystal structure of kiwellin, a major cell-wall protein from kiwifruit. J. Struct. Biol. 2014, 187, 276–281. [Google Scholar] [CrossRef]
- Baker, E.N. Structure of actinidin, after refinement at 1.7 A resolution. J. Mol. Biol. 1980, 141, 441–484. [Google Scholar] [CrossRef]
- Tuppo, L.; Alessandri, C.; Pasquariello, M.S.; Petriccione, M.; Giangrieco, I.; Tamburrini, M.; Mari, A.; Ciardiello, M.A. Pomegranate Cultivars: Identification of the New IgE-Binding Protein Pommaclein and Analysis of Antioxidant Variability. J. Agric. Food Chem. 2017, 65, 2702–2710. [Google Scholar] [CrossRef]
- Tuppo, L.; Giangrieco, I.; Alessandri, C.; Ricciardi, T.; Rafaiani, C.; Ciancamerla, M.; Ferrara, R.; Zennaro, D.; Bernardi, M.L.; Tamburrini, M.; et al. Pomegranate chitinase III: Identification of a new allergen and analysis of sensitization patterns to chitinases. Mol. Immunol. 2018, 103, 89–95. [Google Scholar] [CrossRef]
- Masuda, T.; Zhao, G.; Mikami, B. Crystal structure of class III chitinase from pomegranate provides the insight into its metal storage capacity. Biosci. Biotech. Biochem. 2015, 79, 45–50. [Google Scholar] [CrossRef]
- Fine, A.J. Hypersensitivity reaction to kiwi fruit (Chinese gooseberry, Actinidia chinensis). J. Allergy Clin. Immunol. 1981, 68, 235–237. [Google Scholar] [CrossRef]
- Gaig, P.; Bartolome, B.; Lleonart, R.; Garcia-Ortega, P.; Palacios, R.; Richart, C. Allergy to pomegranate (Punica granatum). Allergy 1999, 54, 287–288. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Ponstingl, H.; Henrick, K.; Thornton, J.M. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins 2000, 41, 47–57. [Google Scholar] [CrossRef]
- Cheng, H.C.; Cheng, P.T.; Peng, P.; Lyu, P.C.; Sun, Y.J. Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa. Protein Sci. 2004, 13, 2304–2315. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Melnikova, D.N.; Finkina, E.I.; Sukhanov, S.V.; Boldyrev, I.A.; Gizatullina, A.K.; Mineev, K.S.; Arseniev, A.S.; Ovchinnikova, T.V. Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake. Biochemistry 2017, 56, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Gizatullina, A.K.; Finkina, E.I.; Mineev, K.S.; Melnikova, D.N.; Bogdanov, I.V.; Telezhinskaya, I.N.; Balandin, S.V.; Shenkarev, Z.O.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris. Biochem. Biophys. Res. Commun. 2013, 439, 427–432. [Google Scholar] [CrossRef]
- Masthoff, L.J.; Hoff, R.; Verhoeckx, K.C.; van Os-Medendorp, H.; Michelsen-Huisman, A.; Baumert, J.L.; Pasmans, S.G.; Meijer, Y.; Knulst, A.C. A systematic review of the effect of thermal processing on the allergenicity of tree nuts. Allergy 2013, 68, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Offermann, L.R.; Bublin, M.; Perdue, M.L.; Pfeifer, S.; Dubiela, P.; Borowski, T.; Chruszcz, M.; Hoffmann-Sommergruber, K. Structural and Functional Characterization of the Hazelnut Allergen Cor a 8. J. Agric. Food Chem. 2015, 63, 9150–9158. [Google Scholar] [CrossRef] [PubMed]
- Scheurer, S.; Lauer, I.; Foetisch, K.; San Miguel Moncin, M.; Retzek, M.; Hartz, C.; Enrique, E.; Lidholm, J.; Cistero-Bahima, A.; Vieths, S. Strong allergenicity of Pru av 3, the lipid transfer protein from cherry, is related to high stability against thermal processing and digestion. J. Allergy Clin. Immunol. 2004, 114, 900–907. [Google Scholar] [CrossRef]
- Gomez, F.; Bogas, G.; Gonzalez, M.; Campo, P.; Salas, M.; Diaz-Perales, A.; Rodriguez, M.J.; Prieto, A.; Barber, D.; Blanca, M.; et al. The clinical and immunological effects of Pru p 3 sublingual immunotherapy on peach and peanut allergy in patients with systemic reactions. Clin. Exp. Allergy 2017, 47, 339–350. [Google Scholar] [CrossRef]
- Gonzalez-Mancebo, E.; Gonzalez-de-Olano, D.; Trujillo, M.J.; Santos, S.; Gandolfo-Cano, M.; Melendez, A.; Juarez, R.; Morales, P.; Calso, A.; Mazuela, O.; et al. Prevalence of sensitization to lipid transfer proteins and profilins in a population of 430 patients in the south of Madrid. J. Investig. Allergol. Clin. Immunol. 2011, 21, 278–282. [Google Scholar]
- Schulten, V.; Nagl, B.; Scala, E.; Bernardi, M.L.; Mari, A.; Ciardiello, M.A.; Lauer, I.; Scheurer, S.; Briza, P.; Jurets, A.; et al. Pru p 3, the nonspecific lipid transfer protein from peach, dominates the immune response to its homolog in hazelnut. Allergy 2011, 66, 1005–1013. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Han, G.W.; Lee, J.Y.; Song, H.K.; Chang, C.; Min, K.; Moon, J.; Shin, D.H.; Kopka, M.L.; Sawaya, M.R.; Yuan, H.S.; et al. Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 2001, 308, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Charvolin, D.; Douliez, J.P.; Marion, D.; Cohen-Addad, C.; Pebay-Peyroula, E. The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 A resolution. Eur. J. Biochem. 1999, 264, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Deboever, E.; Deleu, M.; Mongrand, S.; Lins, L.; Fauconnier, M.L. Plant-Pathogen Interactions: Underestimated Roles of Phyto-oxylipins. Trends Plant Sci. 2020, 25, 22–34. [Google Scholar] [CrossRef]
- Signini, E.F.; Nieman, D.C.; Silva, C.D.; Sakaguchi, C.A.; Catai, A.M. Oxylipin Response to Acute and Chronic Exercise: A Systematic Review. Metabolites 2020, 10, 264. [Google Scholar] [CrossRef]
- Gangemi, S.; Mistrello, G.; Roncarolo, D.; Amato, S.; Minciullo, P.L. Pomegranate-dependent exercise-induced anaphylaxis. J. Investig. Allergol. Clin. Immunol. 2008, 18, 491–492. [Google Scholar]
- Borges, J.P.; Barre, A.; Culerrier, R.; Granier, C.; Didier, A.; Rouge, P. Lipid transfer proteins from Rosaceae fruits share consensus epitopes responsible for their IgE-binding cross-reactivity. Biochem. Biophys. Res. Commun. 2008, 365, 685–690. [Google Scholar] [CrossRef]
- Pacios, L.F.; Tordesillas, L.; Cuesta-Herranz, J.; Compes, E.; Sanchez-Monge, R.; Palacin, A.; Salcedo, G.; Diaz-Perales, A. Mimotope mapping as a complementary strategy to define allergen IgE-epitopes: Peach Pru p 3 allergen as a model. Mol. Immunol. 2008, 45, 2269–2276. [Google Scholar] [CrossRef]
- Tordesillas, L.; Cuesta-Herranz, J.; Gonzalez-Munoz, M.; Pacios, L.F.; Compes, E.; Garcia-Carrasco, B.; Sanchez-Monge, R.; Salcedo, G.; Diaz-Perales, A. T-cell epitopes of the major peach allergen, Pru p 3: Identification and differential T-cell response of peach-allergic and non-allergic subjects. Mol. Immunol. 2009, 46, 722–728. [Google Scholar] [CrossRef]
- Almeida, E.M.; Bartolome, B.; Faria, E.G.; Sousa, N.G.; Luis, A.S. Pomegranate anaphylaxis due to cross-reactivity with Peach LTP (Pru p 3). Allergol. Immunopathol. 2015, 43, 104–106. [Google Scholar] [CrossRef]
- Bublin, M.; Eiwegger, T.; Breiteneder, H. Do lipids influence the allergic sensitization process? J. Allergy Clin. Immunol. 2014, 134, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Dubiela, P.; Aina, R.; Polak, D.; Geiselhart, S.; Humeniuk, P.; Bohle, B.; Alessandri, S.; Del Conte, R.; Cantini, F.; Borowski, T.; et al. Enhanced Pru p 3 IgE-binding activity by selective free fatty acid-interaction. J. Allergy Clin. Immunol. 2017, 140, 1728–1731. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, M.A.; Palazzo, P.; Bernardi, M.L.; Carratore, V.; Giangrieco, I.; Longo, V.; Melis, M.; Tamburrini, M.; Zennaro, D.; Mari, A.; et al. Biochemical, immunological and clinical characterization of a cross-reactive nonspecific lipid transfer protein 1 from mulberry. Allergy 2010, 65, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. Macromol. Crystallogr. Part A 1997, 276, 307–326. [Google Scholar]
- Vagin, A.; Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Minor, W.; Cymborowski, M.; Otwinowski, Z.; Chruszcz, M. HKL-3000: The integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. 2006, 62, 859–866. [Google Scholar] [CrossRef]
- Langer, G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef]
- Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. 2006, 62, 1002–1011. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Painter, J.; Merritt, E.A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 2006, 39, 109–111. [Google Scholar] [CrossRef]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., 3rd; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Rosenstrom, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; Version 2.3.5; Schrödinger, LLC: New York, NY, USA, 2020.
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Chruszcz, M.; Kapingidza, A.B.; Dolamore, C.; Kowal, K. A robust method for the estimation and visualization of IgE cross-reactivity likelihood between allergens belonging to the same protein family. PLoS ONE 2018, 13, e0208276. [Google Scholar] [CrossRef] [PubMed]
- Mari, A.; Rasi, C.; Palazzo, P.; Scala, E. Allergen databases: Current status and perspectives. Curr. Allergy Asthma Rep. 2009, 9, 376–383. [Google Scholar] [CrossRef] [PubMed]

| Protein | Act c 10.0101 | Pun g 1.0101 |
|---|---|---|
| PDB accession code | 7KSB | 7KSC |
| Data collection | ||
| Diffraction source | APS, 22ID | APS, 22ID |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Space group | P21 | P21 |
| a, b, c, β (Å, °) | 38.8, 38.1, 48.4, 98.9 | 29.5, 89.4, 58.9, 102.8 |
| Resolution range (Å) | 40.00–1.95 (1.98–1.95) | 50.00–2.40 (2.44–2.40) |
| No. of unique reflections | 10,217 (497) | 10,593 (548) |
| Completeness (%) | 98.7 (98.2) | 90.6 (90.4) |
| Redundancy | 4.0 (3.9) | 2.8 (2.0) |
| <I/σ(I)> | 25.1 (4.3) | 6.6 (2.3) |
| Rmeas | 0.113 (0.483) | 0.220 (0.511) |
| Rp.i.m | 0.057 (0.240) | 0.124 (0.316) |
| CC1/2 | (0.904) | (0.751) |
| Refinement | ||
| Resolution range (Å) | 40.00–1.95 (2.00–1.95) | 40.00–2.40 (2.46–2.40) |
| Completeness (%) | 98.4 (92.7) | 90.1 (85.0) |
| No. of reflections, working set | 9707 | 10014 |
| No. of reflections, test set | 503 | 541 |
| Final Rcryst | 0.199 (0.223) | 0.204 (0.233) |
| Final Rfree | 0.257 (0.380) | 0.252 (0.348) |
| RMSD Bonds (Å) | 0.013 | 0.010 |
| RMSD Angles (°) | 1.8 | 1.5 |
| Ramachandran Plot | ||
| Allowed regions (%) | 100.0 | 100.0 |
| Favored regions (%) | 98.4 | 99.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Malley, A.; Pote, S.; Giangrieco, I.; Tuppo, L.; Gawlicka-Chruszcz, A.; Kowal, K.; Ciardiello, M.A.; Chruszcz, M. Structural Characterization of Act c 10.0101 and Pun g 1.0101—Allergens from the Non-Specific Lipid Transfer Protein Family. Molecules 2021, 26, 256. https://doi.org/10.3390/molecules26020256
O’Malley A, Pote S, Giangrieco I, Tuppo L, Gawlicka-Chruszcz A, Kowal K, Ciardiello MA, Chruszcz M. Structural Characterization of Act c 10.0101 and Pun g 1.0101—Allergens from the Non-Specific Lipid Transfer Protein Family. Molecules. 2021; 26(2):256. https://doi.org/10.3390/molecules26020256
Chicago/Turabian StyleO’Malley, Andrea, Swanandi Pote, Ivana Giangrieco, Lisa Tuppo, Anna Gawlicka-Chruszcz, Krzysztof Kowal, Maria Antonietta Ciardiello, and Maksymilian Chruszcz. 2021. "Structural Characterization of Act c 10.0101 and Pun g 1.0101—Allergens from the Non-Specific Lipid Transfer Protein Family" Molecules 26, no. 2: 256. https://doi.org/10.3390/molecules26020256
APA StyleO’Malley, A., Pote, S., Giangrieco, I., Tuppo, L., Gawlicka-Chruszcz, A., Kowal, K., Ciardiello, M. A., & Chruszcz, M. (2021). Structural Characterization of Act c 10.0101 and Pun g 1.0101—Allergens from the Non-Specific Lipid Transfer Protein Family. Molecules, 26(2), 256. https://doi.org/10.3390/molecules26020256







