Effects of Light Intensity and Spectral Composition on the Transcriptome Profiles of Leaves in Shade Grown Tea Plants (Camellia sinensis L.) and Regulatory Network of Flavonoid Biosynthesis
Abstract
:1. Introduction
2. Results
2.1. Effects of Light Intensity and Light Spectral Composition on the Flavonoid Compositions of Tea Leaves
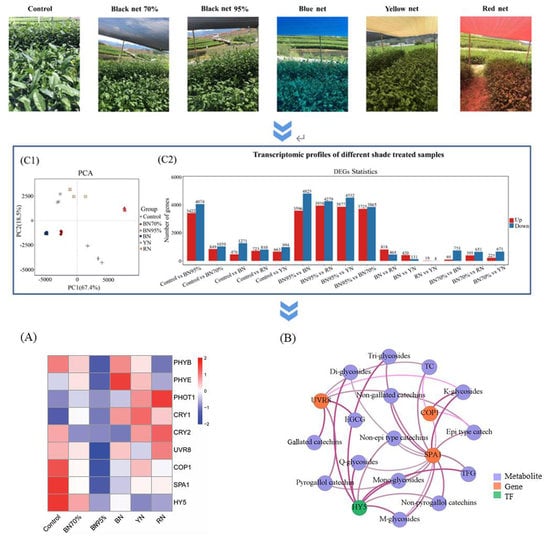
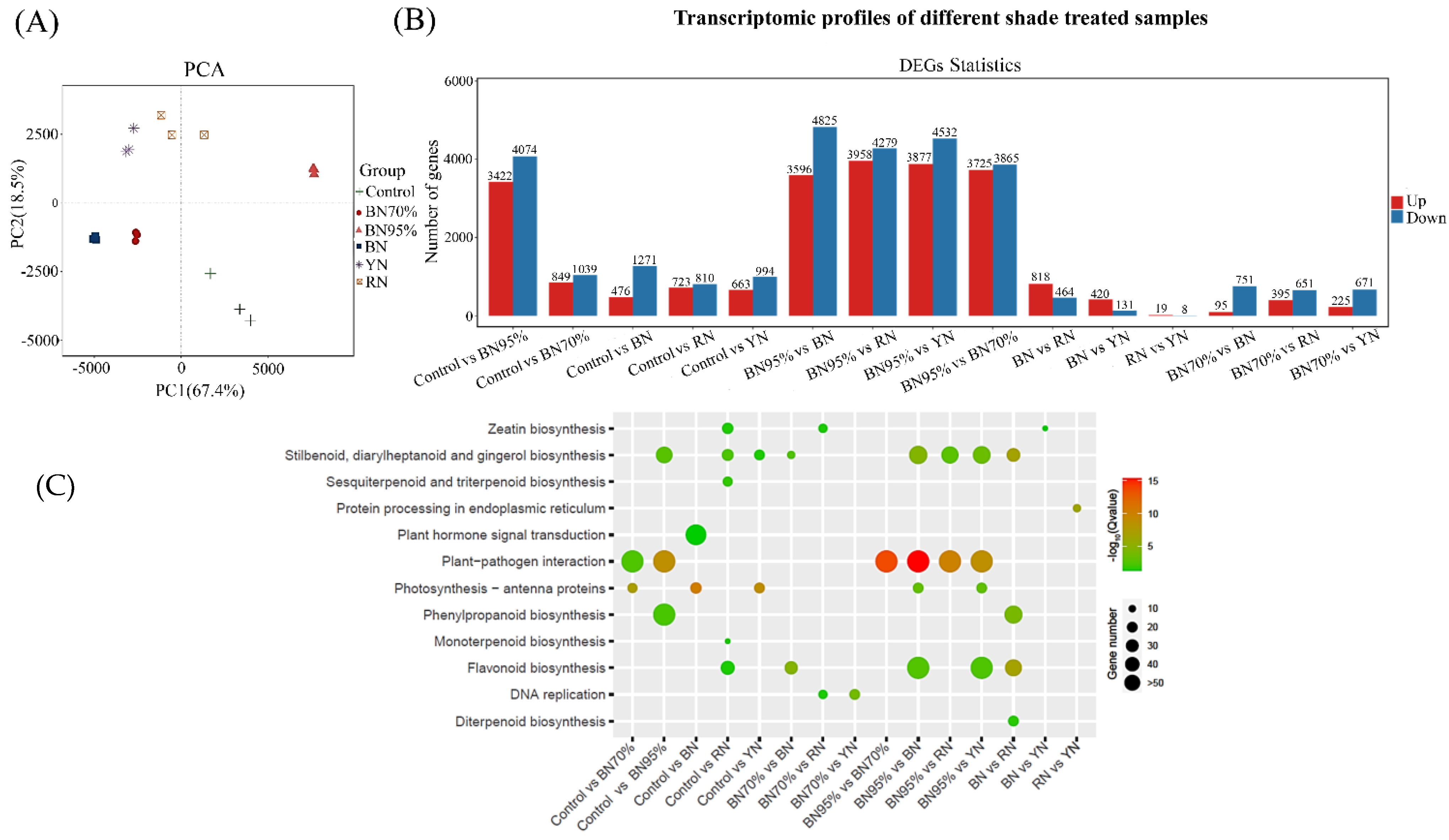
2.2. The Transcriptome Profiles of Tea Leaves under Different Shade Treatments
2.3. KEGG Functional Enrichment Analyses of DEGs
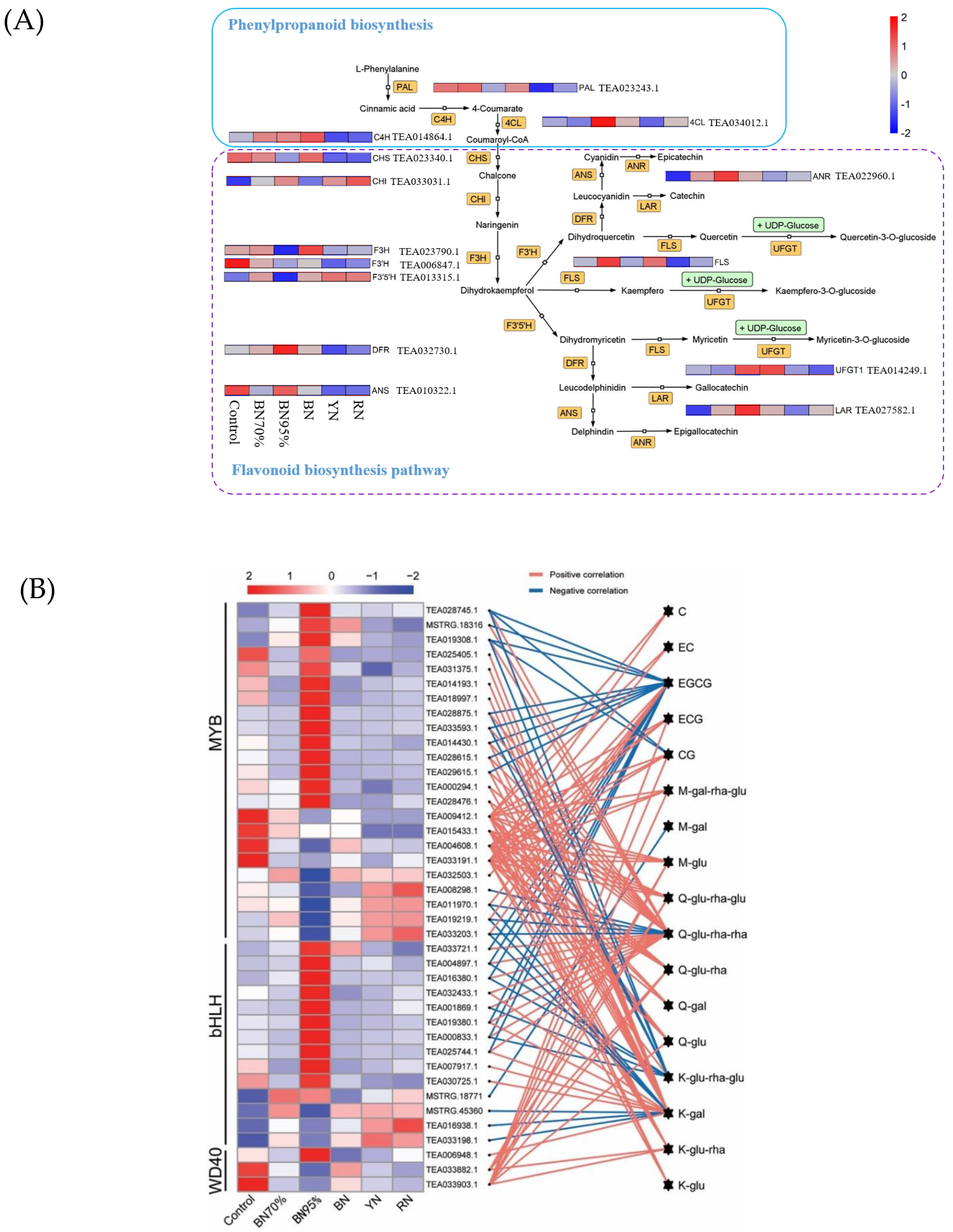
2.4. Regulatory Effects of Shade Treatments on the Biosynthetic Pathway of Flavonoids
2.5. TFs Related to Flavonoid Biosynthesis
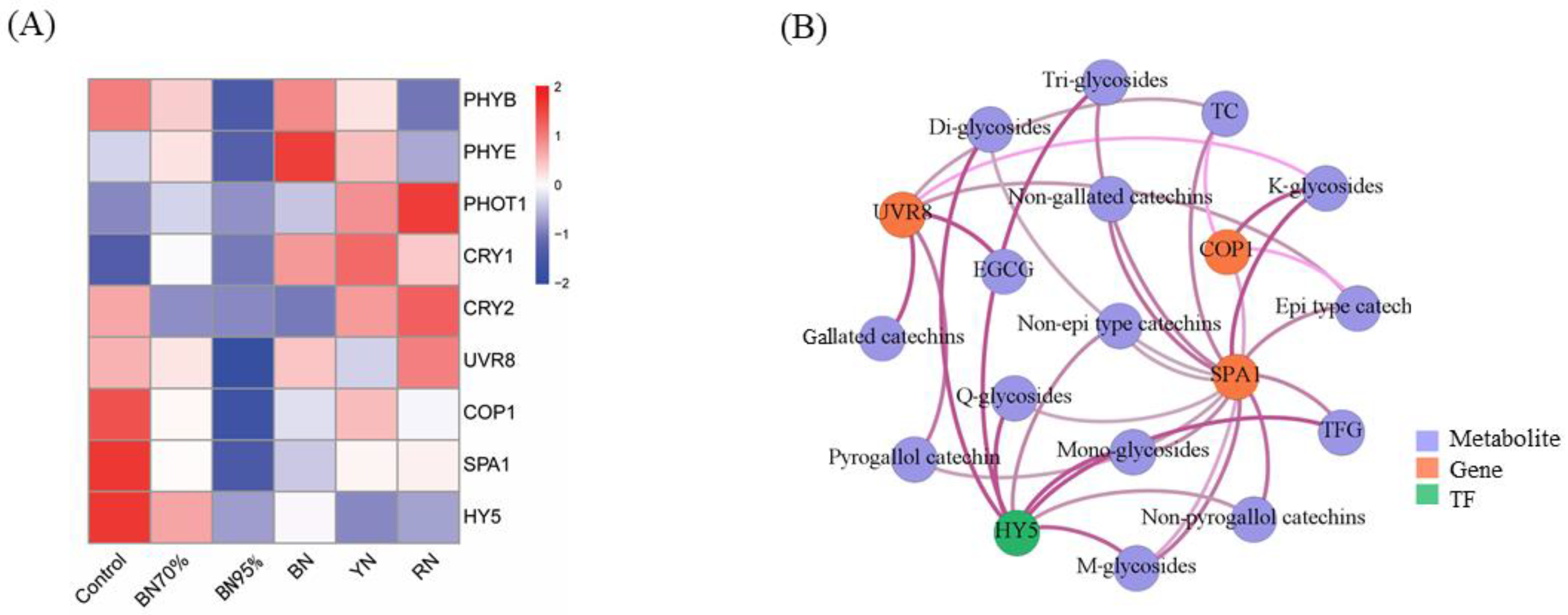
2.6. Expressions of Light Signal Perception and Signaling Genes and the Association with Flavonoids
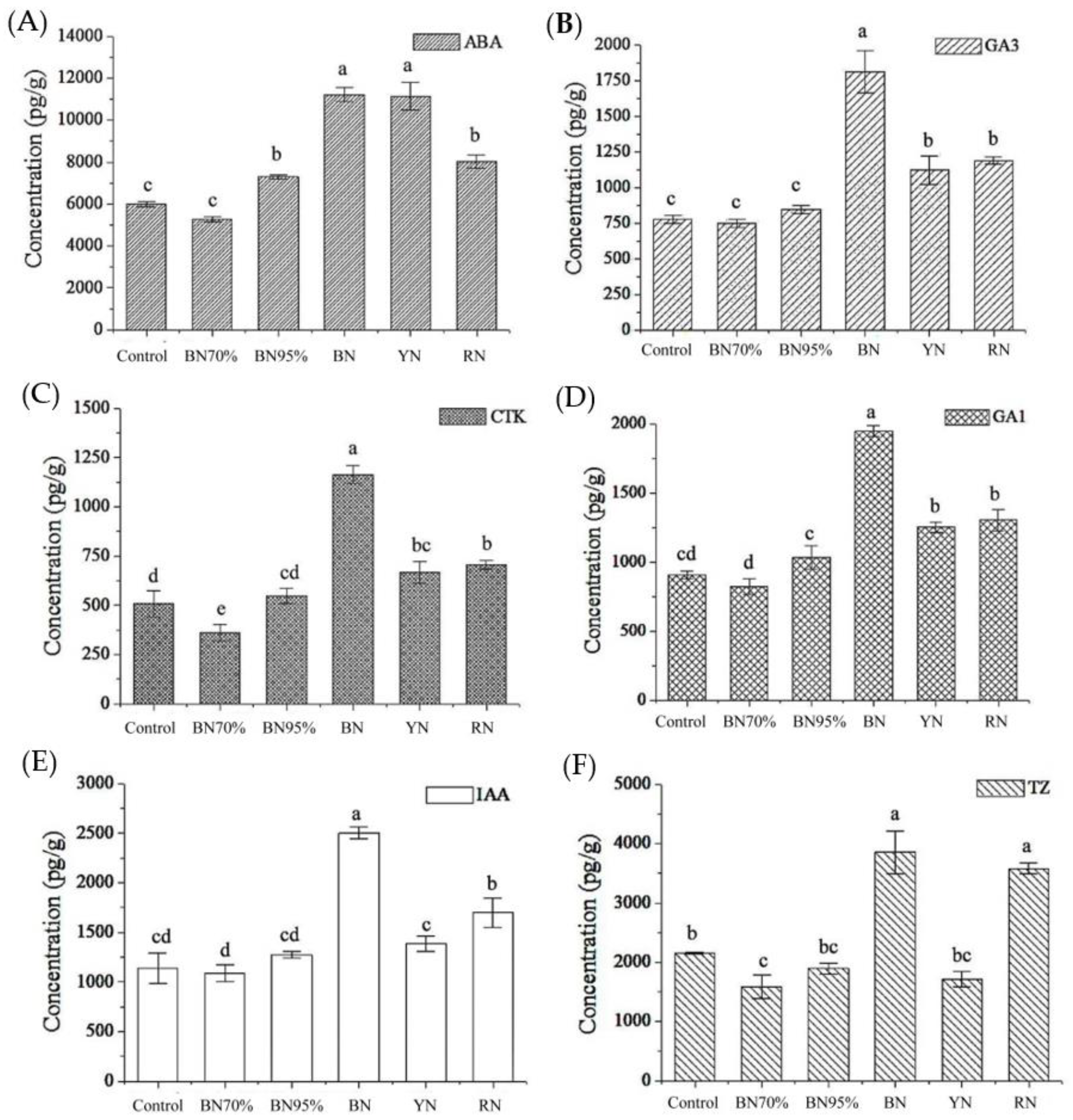
2.7. Phytohormone Biosynthesis in Shade Grown Tea Leaves and the Association with Flavonoids
3. Discussion
4. Materials and Methods
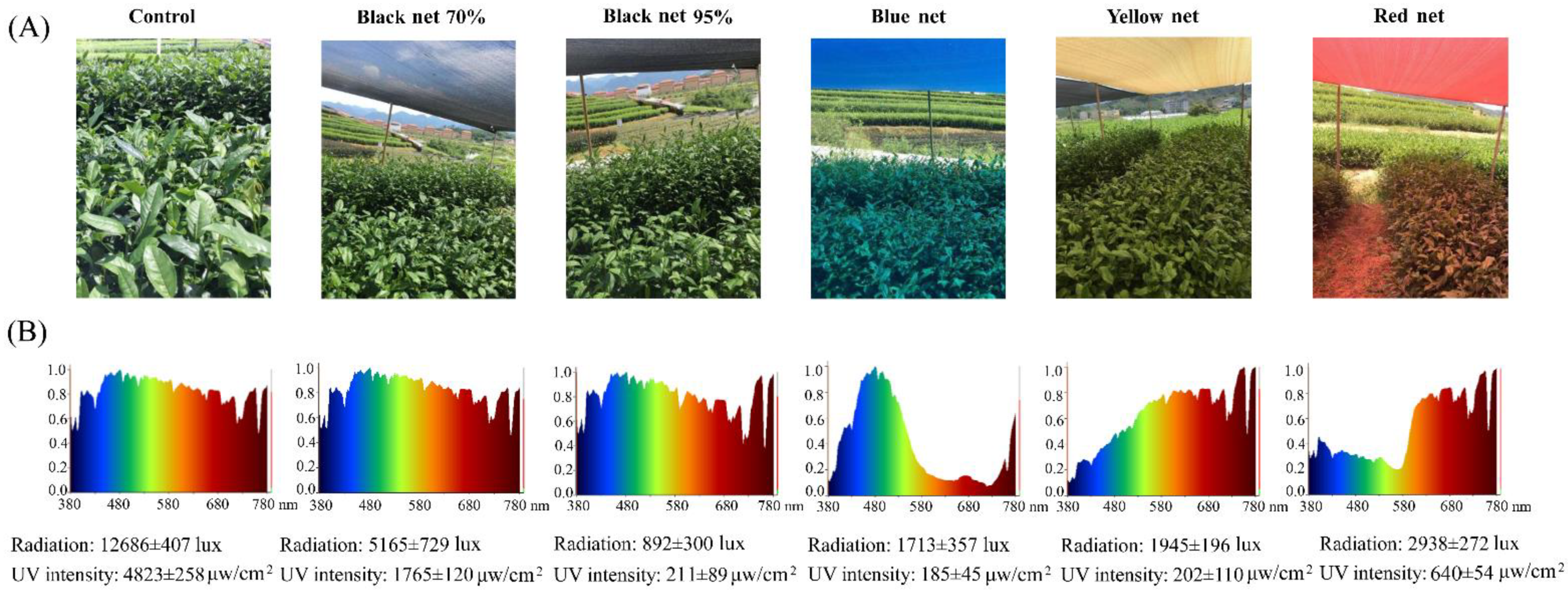
4.1. Plant Materials, Shading Treatments, and Sampling
4.2. Determination of Flavonoids in Tea Leaves
4.3. Transcriptomic and Bioinformatic Analyses
4.4. LC–MS/MS Analysis of Phytohormones
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Khan, N.; Mukhtar, H. Tea polyphenols in promotion of human health. Nutrients 2019, 11, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.D.; Xin, H.H.; Wang, M.L.; Ma, Q.P.; Wang, L.; Kaleri, N.A.; Wang, Y.H.; Li, X.H. Transcriptomic analysis reveals the molecular mechanisms of drought-stress-induced decreases in Camellia sinensis leaf quality. Front. Plant Sci. 2016, 7, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Liu, M.; Ruan, J. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Wang, W.; Xia, T.; Gao, L.; Shen, G.; Pang, Y. Characterization of a heat responsive UDP: Flavonoid glucosyltransferase gene in tea plant (Camellia sinensis). PLoS ONE 2018, 13, e0207212. [Google Scholar] [CrossRef] [PubMed]
- Mamati, G.E.; Liang, Y.R.; Lu, J.L. Expression of basic genes involved in tea polyphenol synthesis in relation to accumulation of catechins and total tea polyphenols. J. Sci. Food Agric. 2006, 86, 459–464. [Google Scholar] [CrossRef]
- Scharbert, S.; Hofmann, T. Molecular definition of black tea taste by means of quantitative studies, taste reconstitution, and omission experiments. J. Agric. Food Chem. 2005, 53, 5377–5384. [Google Scholar] [CrossRef]
- Wu, L.Y.; Fang, Z.T.; Lin, J.K.; Sun, Y.; Du, Z.Z.; Guo, Y.L.; Liu, J.H.; Liang, Y.R.; Ye, J.H. Complementary iTRAQ proteomic and transcriptomic analyses of leaves in tea plant (Camellia sinensis L.) with different maturity and regulatory network of flavonoid biosynthesis. J. Proteome Res. 2019, 18, 252–264. [Google Scholar]
- Mouradov, A.; Spangenberg, G. Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef]
- Zoratti, L.; Karppinen, K.; Escobar, A.L.; Haggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef]
- Wagner, J.R.; Brunzelle, J.S.; Forest, K.T.; Vierstra, R.D. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 2005, 438, 325–331. [Google Scholar] [CrossRef]
- Park, Y.; Runkle, E.S. Spectral effects of light-emitting diodes on plant growth, visual color quality, and photosynthetic photon efficacy: White versus blue plus red radiation. PLoS ONE 2018, 13, e0202386. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Ma, J.Q.; Ma, C.L.; Shen, S.Y.; Liu, Y.F.; Chen, L. Regulation of growth and flavonoid formation of tea plants (Camellia sinensis) by blue and green light. J. Agric. Food Chem. 2019, 67, 2408–2419. [Google Scholar] [CrossRef]
- Miao, L.X.; Zhang, Y.C.; Yang, X.F.; Xiao, J.P.; Zhang, H.Q.; Zhang, Z.F.; Wang, Y.Z.; Jiang, G.H. Colored light-quality selective plastic films affect anthocyanin content, enzyme activities, and the expression of flavonoid genes in strawberry (Fragaria × ananassa) fruit. Food Chem. 2016, 207, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Li, S.T.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef]
- Bhatia, C.; Gaddam, S.R.; Pandey, A.; Trivedi, P.K. COP1 mediates light-dependent regulation of flavonol biosynthesis through HY5 in Arabidopsis. Plant Sci. 2021, 303, 110760. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Li, Y.; She, G.; Zhang, X.; Jordan, B.; Chen, Q.; Zhao, J.; Wan, X. Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading. BMC Plant Biol. 2018, 18, 233. [Google Scholar] [CrossRef]
- Latowski, D.; Kruk, J.; Burda, K.; Skrzynecka-Jaskier, M.; Kostecka-Gugala, A.; Strzalka, K. Kinetics of violaxanthin de-epoxidation by violaxanthin de-epoxidase, a xanthophyll cycle enzyme, is regulated by membrane fluidity in model lipid bilayers. Eur. J. Biochem. 2002, 269, 4656–4665. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Chen, X.-R.; Wang, J.-P.; Cui, W.-Q.; Xing, X.-X.; Chen, X.-Y.; Ding, W.-Y.; God’Spower, B.-O.; Eliphaz, N.; Sun, M.-Q.; et al. Transcriptomic analysis reveals flavonoid biosynthesis of Syringa oblata Lindl. in response to different light intensity. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gao, L.; Shan, Y.; Liu, Y.; Tian, Y.; Xia, T. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2012, 141, 7–16. [Google Scholar] [CrossRef]
- Sano, S.; Takemoto, T.; Ogihara, A.; Suzuki, K.; Masumura, T.; Satoh, S.; Takano, K.; Mimura, Y.; Morita, S. Stress Responses of Shade-Treated Tea Leaves to High Light Exposure after Removal of Shading. Plants 2020, 9, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.Y.; Wang, L.T.; Ma, J.H.; Ma, E.D.; Li, J.Y.; Gong, M. Effects of light quality on growth and development, photosynthetic characteristics and content of carbohydrates in tobacco (Nicotiana tabacum L.) plants. Photosynthetica 2017, 55, 467–477. [Google Scholar] [CrossRef]
- Jenkins, G.I. UV and blue light signal transduction in the regulation of flavonoid biosynthesis gene expression in Arabidopsis. Cell. Integr. Signal. Pathw. Plant Dev. 1998, 104, 71–82. [Google Scholar] [CrossRef]
- Kadomura-Ishikawa, Y.; Miyawaki, K.; Noji, S.; Takahashi, A. Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria x ananassa fruits. J. Plant Res. 2013, 126, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Neugart, S.; Krumbein, A.; Zrenner, R. Influence of Light and Temperature on Gene Expression Leading to Accumulation of Specific Flavonol Glycosides and Hydroxycinnamic Acid Derivatives in Kale (Brassica oleracea var. sabellica). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-S.; Xu, Y.-J.; Gao, L.-P.; Yu, O.; Wang, X.-Z.; He, X.-J.; Jiang, X.-L.; Liu, Y.-J.; Xia, T. Functional analysis of Flavonoid 3′,5′-hydroxylase from Tea plant (Camellia sinensis): Critical role in the accumulation of catechins. BMC Plant Biol. 2014, 14, 347. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Chen, S.; Gu, M.; Chen, X.; Chen, X.; Yang, J.; Zhao, F.; Ye, N. Exploration of the Effects of Different Blue LED Light Intensities on Flavonoid and Lipid Metabolism in Tea Plants via Transcriptomics and Metabolomics. Int. J. Mol. Sci. 2020, 21, 4606. [Google Scholar] [CrossRef]
- Xie, L.; Cao, Y.; Zhao, Z.; Ren, C.; Xing, M.; Wu, B.; Zhang, B.; Xu, C.; Chen, K.; Li, X. Involvement of MdUGT75B1 and MdUGT71B1 in flavonol galactoside/glucoside biosynthesis in apple fruit. Food Chem. 2020, 312, 126124. [Google Scholar] [CrossRef]
- Siipola, S.M.; Kotilainen, T.; Sipari, N.; Morales, L.O.; Lindfors, A.V.; Robson, T.M.; Aphalo, P.J. Epidermal UV-A absorbance and whole-leaf flavonoid composition in pea respond more to solar blue light than to solar UV radiation. Plant Cell Environ. 2015, 38, 941–952. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Kitayama, M.; Lu, N.; Takagaki, M. Improving secondary metabolite accumulation, mineral content, and growth of coriander (Coriandrum sativum L.) by regulating light quality in a plant factory. J. Hortic. Sci. Biotechnol. 2020, 95, 356–363. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, S.; Yang, W.; Shang, X.; Fu, X. Light quality affects flavonoid production and related gene expression in Cyclocarya paliurus. J. Photochem. Photobiol. B Biol. 2018, 179, 66–73. [Google Scholar] [CrossRef]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef]
- Yao, P.; Huang, Y.; Dong, Q.; Wan, M.; Wang, A.; Chen, Y.; Li, C.; Wu, Q.; Chen, H.; Zhao, H. FtMYB6, a Light-Induced SG7 R2R3-MYB Transcription Factor, Promotes Flavonol Biosynthesis in Tartary Buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 2020, 68, 13685–13696. [Google Scholar] [CrossRef] [PubMed]
- Premathilake, A.T.; Ni, J.; Bai, S.; Tao, R.; Ahmad, M.; Teng, Y. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 2020, 252, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Liu, X.; Zhu, W.; Cheng, X.; Wang, X.; Wan, X.; Liu, L. Ambient Ultraviolet B Signal Modulates Tea Flavor Characteristics via Shifting a Metabolic Flux in Flavonoid Biosynthesis. J. Agric. Food Chem. 2021, 69, 3401–3414. [Google Scholar] [CrossRef] [PubMed]
- Van Gelderen, K.; Kang, C.; Pierik, R. Light Signaling, Root Development, and Plasticity. Plant Physiol. 2018, 176, 1049–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetti, C.; Sebastiani, F.; Tattini, M. Review: ABA, flavonols, and the evolvability of land plants. Plant Sci. 2019, 280, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, W.; Dou, Y.; Zhang, J.; Jiang, G.; Miao, L.; Han, G.; Liu, Y.; Li, H.; Zhang, Z. Transcript Quantification by RNA-Seq Reveals Differentially Expressed Genes in the Red and Yellow Fruits of Fragaria vesca. PLoS ONE 2015, 10, e0144356. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Mou, W.; Xia, R.; Li, L.; Zawora, C.; Ying, T.; Mao, L.; Liu, Z.; Luo, Z. Integrated analysis of high-throughput sequencing data shows abscisic acid-responsive genes and miRNAs in strawberry receptacle fruit ripening. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bovy, A.; de Vos, R.; Kemper, M.; Schijlen, E.; Pertejo, M.A.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.M.; Park, H.J.; Yim, S.D.; Baek, N.I.; Lee, C.H.; An, G.H.; Woo, Y.M. Transgenic rice lines expressing maize C1 and R-S regulatory genes produce various flavonoids in the endosperm. Plant Biotechnol. J. 2006, 4, 303–315. [Google Scholar] [CrossRef]
- Li, M.Z.; Li, Y.Z.; Guo, L.L.; Gong, N.D.; Pang, Y.Z.; Jiang, W.B.; Liu, Y.J.; Jiang, X.L.; Zhao, L.; Wang, Y.S.; et al. Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant. J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Mitra, M.; Agarwal, P.; Kundu, A.; Banerjee, V.; Roy, S. Investigation of the effect of UV-B light on Arabidopsis MYB4 (AtMYB4) transcription factor stability and detection of a putative MYB4-binding motif in the promoter proximal region of AtMYB4. PLoS ONE 2019, 14, e0220123. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Lai, L.; Li, L.; Liu, L.; Jakada, B.; Huang, Y.; He, Q.; Chai, M.; Niu, X.; Qin, Y. AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling. Int. J. Mol. Sci. 2020, 21, 5727. [Google Scholar] [CrossRef]
- Emiliani, J.; D’Andrea, L.; Ferreyra, M.L.F.; Maulion, E.; Rodriguez, E.; Rodriguez-Concepcion, M.; Casati, P. A role for β,β-xanthophylls in Arabidopsis UV-B photoprotection. J. Exp. Bot. 2018, 69, 4921–4933. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zeng, L.; Liu, J.; Xing, D. Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to Sclerotinia sclerotiorum. PLoS Pathog. 2015, 11, e1004878. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.F.; Zhang, W.H.; Zhao, Y.; Gong, X.M.; Guo, L.; Zhu, G.L.; Wang, X.C.; Gong, Z.Z.; Schumaker, K.S.; Guo, Y. SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 2007, 19, 3805–3818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verslues, P.E.; Guo, Y.; Dong, C.H.; Ma, W.; Zhu, J.K. Mutation of SAD2, an importin β-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant. J. 2006, 47, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Mentzen, W.I.; Wurtele, E.S. Regulon organization of Arabidopsis. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.R.; Ramirez, M.V.; Miller, N.D.; Vallabhaneni, P.; Ray, W.K.; Helm, R.F.; Winkel, B.S.J.; Muday, G.K. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 2011, 156, 144–164. [Google Scholar] [CrossRef] [Green Version]
- Mou, W.; Li, D.; Bu, J.; Jiang, Y.; Khan, Z.U.; Luo, Z.; Mao, L.; Ying, T. Comprehensive Analysis of ABA Effects on Ethylene Biosynthesis and Signaling during Tomato Fruit Ripening. PLoS ONE 2016, 11, e0154072. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Dong, L.X.; Jiang, Z.F.; Yang, X.L.; Sun, Z.H.; Li, J.X.; Wu, Y.H.; Xu, M.J. Salicylic acid-induced flavonoid accumulation in Ginkgo biloba leaves is dependent on red and far-red light. Ind. Crop. Prod. 2018, 118, 102–110. [Google Scholar] [CrossRef]
- Moreira-Rodríguez, M.; Nair, V.; Benavides, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. UVA, UVB Light, and Methyl Jasmonate, Alone or Combined, Redirect the Biosynthesis of Glucosinolates, Phenolics, Carotenoids, and Chlorophylls in Broccoli Sprouts. Int. J. Mol. Sci. 2017, 18, 2330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berli, F.J.; Fanzone, M.; Piccoli, P.; Bottini, R. Solar UV-B and ABA are involved in phenol metabolism of Vitis vinifera L. increasing biosynthesis of berry skin polyphenols. J. Agric. Food Chem. 2011, 59, 4874–4884. [Google Scholar] [CrossRef] [PubMed]
- Gai, Z.; Wang, Y.; Ding, Y.; Qian, W.; Qiu, C.; Xie, H.; Sun, L.; Jiang, Z.; Ma, Q.; Wang, L.; et al. Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [Green Version]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPL Module during the Red Peel Coloration of Bagging-Treated Chinese Sand Pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.Q.; Nie, Y.; Gao, Y.; Huang, B.; Ye, J.H.; Lu, J.L.; Liang, Y.R. Screening the cultivar and processing factors based on the flavonoid profiles of dry teas using principal component analysis. J. Food Compos. Anal. 2018, 67, 29–37. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
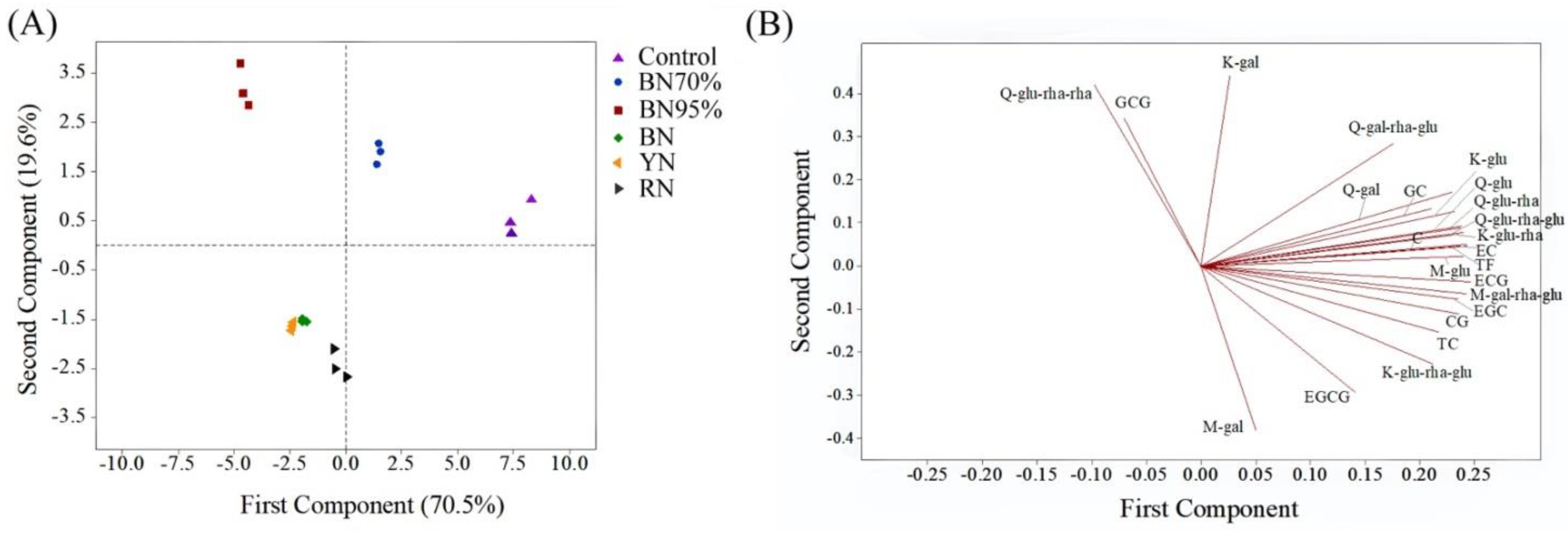
| Compounds a | Control | BN70% | BN95% | BN | YN | RN |
|---|---|---|---|---|---|---|
| Catechins (mg/g Dry Weight) | ||||||
| GC b | 1.5 ± 0.1a | 1.2 ± 0.1b | 0.9 ± 0.0c | 0.6 ± 0.1d | 0.6 ± 0.1d | 1.1 ± 0.1b |
| EGC b | 31.2 ± 0.4a | 21.6 ± 0.4b | 9.2 ± 0.4d | 13.5 ± 0.2c | 14.9 ± 0.4c | 22.9 ± 1.7b |
| C b | 0.9 ± 0.1a | 0.7 ± 0.0b | 0.4 ± 0.0d | 0.4 ± 0.0d | 0.4 ± 0.0d | 0.5 ± 0.0c |
| EC b | 12.0 ± 0.5a | 8.5 ± 0.1b | 4.6 ± 0.1c | 4.5 ± 0.2c | 5.3 ± 0.1c | 7.5 ± 1.0b |
| EGCG b | 85.3 ± 0.8b | 86.0 ± 0.5b | 64.9 ± 0.3d | 81.2 ± 0.3c | 79.4 ± 1.5c | 92.4 ± 0.9a |
| GCG b | 0.5 ± 0.1c | 0.8 ± 0.0a | 0.8 ± 0.1ab | 0.6 ± 0.1bc | 0.6 ± 0.0c | 0.5 ± 0.1c |
| ECG b | 13.4 ± 0.1a | 11.1 ± 0.1b | 8.6 ± 0.0d | 9.9 ± 0.0c | 9.7 ± 0.1c | 10.7 ± 0.4b |
| CG b | 2.1 ± 0.0a | 1.4 ± 0.1b | 0.9 ± 0.1c | 1.2 ± 0.0b | 1.3 ± 0.1b | 1.4 ± 0.2b |
| TC d | 146.9 ± 1.0a (100%) | 131.2 ± 0.7c (89.3%) | 90.1 ± 0.4e (61.3%) | 112.0 ± 0.3d (76.2%) | 112.2 ± 1.7d (76.4%) | 137.0 ± 3.7b (93.3%) |
| Flavonol Glycosides (µg/g Dry Weight) | ||||||
| M-gal-rha-glu c | 199.0 ± 4.8a | 126.5 ± 1.6b | 78.5 ± 8.8d | 112.0 ± 2.5bc | 102.8 ± 3.6c | 129.6 ± 15.4b |
| M-gal c | 131.2 ± 0.6b | 56.8 ± 2.5c | 66.1 ± 2.0c | 157.0 ± 4.8a | 117.8 ± 2.4b | 129.9 ± 11.6b |
| M-glu c | 1202.1 ± 8.1a | 613.2 ± 14.4b | 116.9 ± 1.6e | 478.0 ± 3.2c | 297.9 ± 3.9d | 305.5 ± 17.8d |
| K-glu-rha-gal c | 1775.2 ± 26.8a | 1379.5 ± 28.6bc | 788.2 ± 12.0d | 1397.3 ± 7.7b | 1327.5 ± 2.4c | 1425.3 ± 25.3b |
| K-gal c | 6.6 ± 0.7b | 5.3 ± 1.0b | 9.4 ± 0.7a | 1.9 ± 0.6c | 1.7 ± 0.1c | 1.4 ± 0.2c |
| K-glu-rha c | 17.0 ± 2.5a | 7.5 ± 0.4b | 3.5 ± 0.7c | 5.5 ± 0.6bc | 2.7 ± 0.1c | 5.2 ± 0.9bc |
| K-glu c | 204.0 ± 46.1a | 119.2 ± 6.6b | 42.5 ± 3.7c | 53.6 ± 4.2c | 47.7 ± 5.1c | 48.3 ± 4.5c |
| Q-gal-rha-glu c | 133.8 ± 3.1b | 150.8 ± 7.9a | 50.5 ± 2.8c | 22.6 ± 2.6d | 11.9 ± 1.1d | 15.3 ± 3.0d |
| Q-glu-rha c | 289.9 ± 0.6a | 133.7 ± 9.7b | 30.5 ± 1.7d | 77.5 ± 12.7c | 41.8 ± 6.5d | 45.5 ± 3.9d |
| Q-glu-rha-rha c | 30.5 ± 0.5c | 73.9 ± 4.5b | 143.8 ± 8.8a | 36.2 ± 5.1c | 14.4 ± 0.6d | 15.7 ± 0.7d |
| Q-glu-rha-glu c | 4436.9 ± 52.4a | 2036.1 ± 21.0b | 444.5 ± 5.4e | 1058.8 ± 3.2c | 814.7 ± 6.9d | 776.2 ± 15.5d |
| Q-gal c | 185.2 ± 14.0a | 104.1 ± 2.4b | 40.2 ± 2.7c | 28.3 ± 7.2c | 27.6 ± 0.7c | 31.7 ± 7.2c |
| Q-glu c | 605.2 ± 8.0a | 270.6 ± 5.0b | 65.6 ± 2.6e | 123.5 ± 4.4c | 96.9 ± 2.1d | 99.9 ± 10.3d |
| TFG d | 9216.5 ± 140.1a (100%) | 5077.1 ± 68.0b (55.1%) | 1880.2 ± 14.6e (20.4%) | 3552.0 ± 12.5c (38.5%) | 2905.5 ± 10.6d (31.5%) | 3029.4 ± 54.7d (32.9%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, J.-H.; Lv, Y.-Q.; Liu, S.-R.; Jin, J.; Wang, Y.-F.; Wei, C.-L.; Zhao, S.-Q. Effects of Light Intensity and Spectral Composition on the Transcriptome Profiles of Leaves in Shade Grown Tea Plants (Camellia sinensis L.) and Regulatory Network of Flavonoid Biosynthesis. Molecules 2021, 26, 5836. https://doi.org/10.3390/molecules26195836
Ye J-H, Lv Y-Q, Liu S-R, Jin J, Wang Y-F, Wei C-L, Zhao S-Q. Effects of Light Intensity and Spectral Composition on the Transcriptome Profiles of Leaves in Shade Grown Tea Plants (Camellia sinensis L.) and Regulatory Network of Flavonoid Biosynthesis. Molecules. 2021; 26(19):5836. https://doi.org/10.3390/molecules26195836
Chicago/Turabian StyleYe, Jian-Hui, Yi-Qing Lv, Sheng-Rui Liu, Jing Jin, Yue-Fei Wang, Chao-Ling Wei, and Shi-Qi Zhao. 2021. "Effects of Light Intensity and Spectral Composition on the Transcriptome Profiles of Leaves in Shade Grown Tea Plants (Camellia sinensis L.) and Regulatory Network of Flavonoid Biosynthesis" Molecules 26, no. 19: 5836. https://doi.org/10.3390/molecules26195836
APA StyleYe, J.-H., Lv, Y.-Q., Liu, S.-R., Jin, J., Wang, Y.-F., Wei, C.-L., & Zhao, S.-Q. (2021). Effects of Light Intensity and Spectral Composition on the Transcriptome Profiles of Leaves in Shade Grown Tea Plants (Camellia sinensis L.) and Regulatory Network of Flavonoid Biosynthesis. Molecules, 26(19), 5836. https://doi.org/10.3390/molecules26195836