Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants
Abstract
:1. Introduction
2. Results
2.1. Cloning and Expression of UGT93Y1 and UGT93Y2
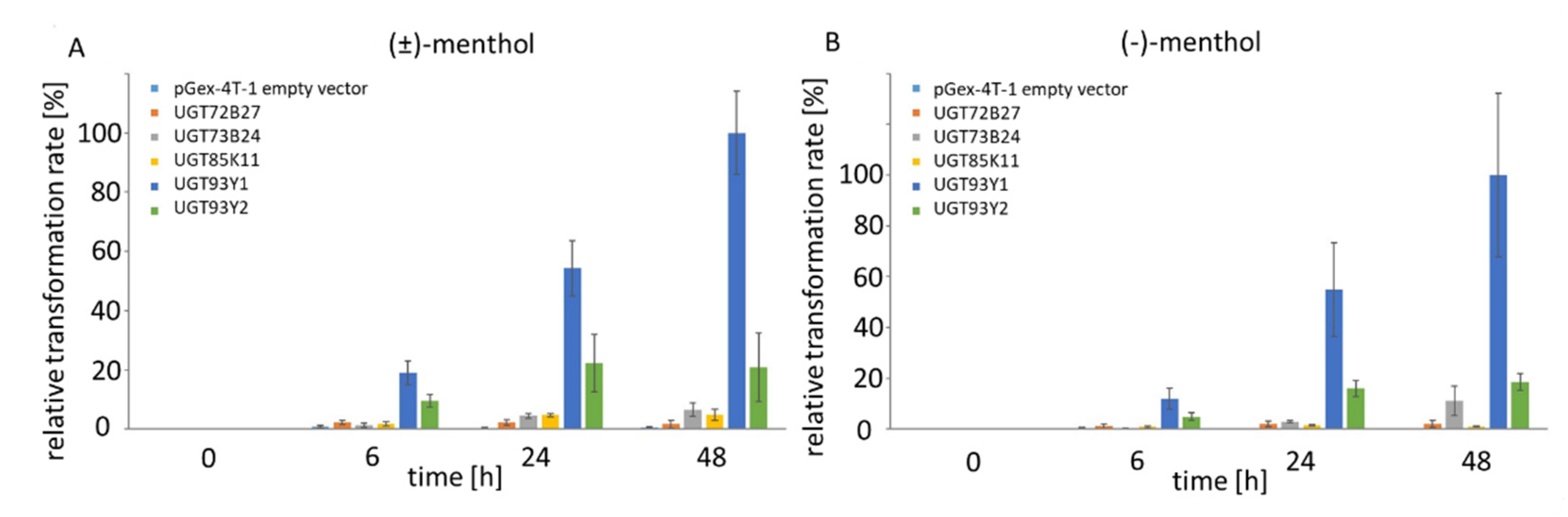
2.2. In Vivo Substrate Screening Using E. coli Waksman
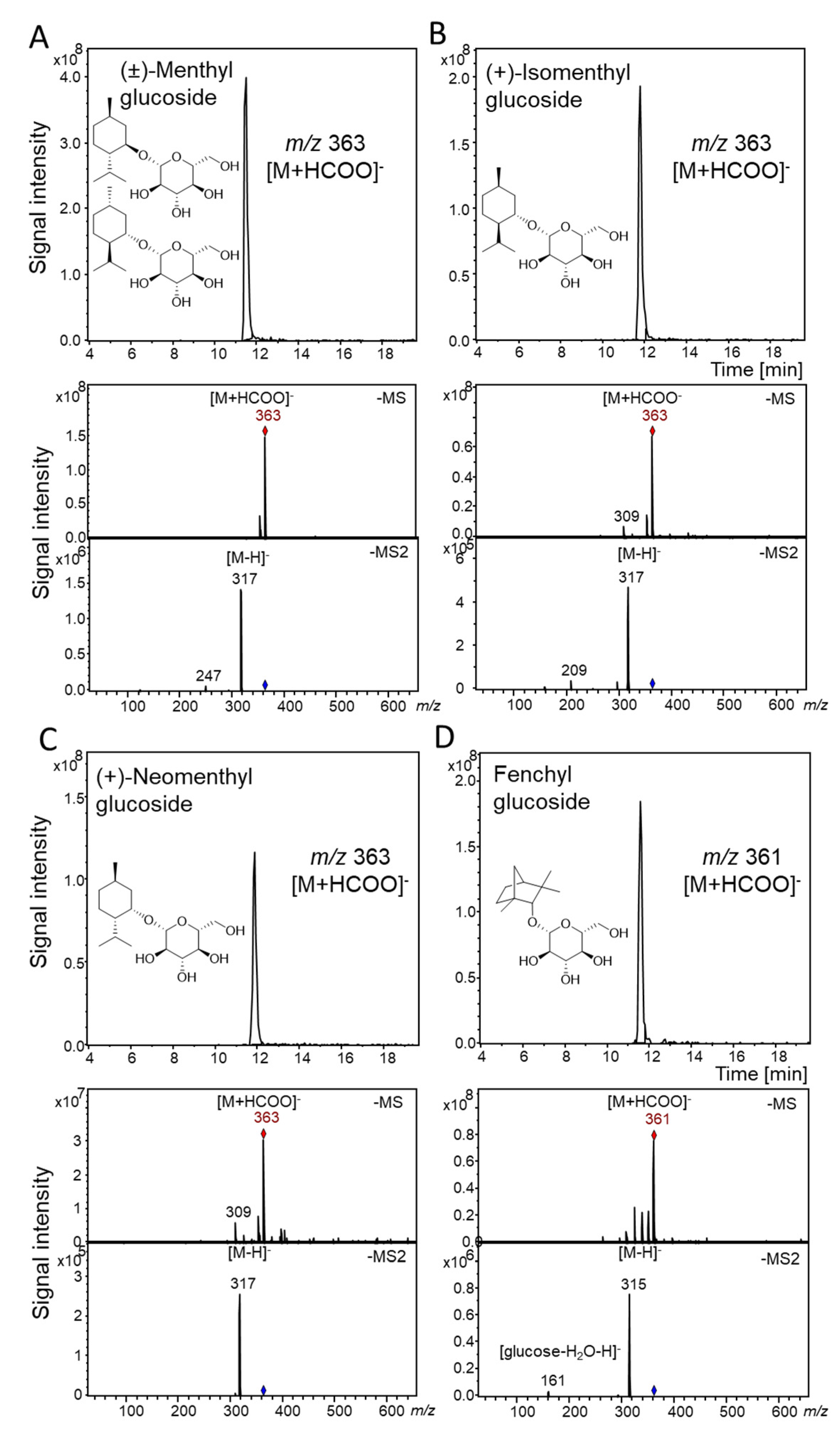
2.3. Qualitative Substrate Screening of UGT93Y1 and UGT93Y2 by LC-MS
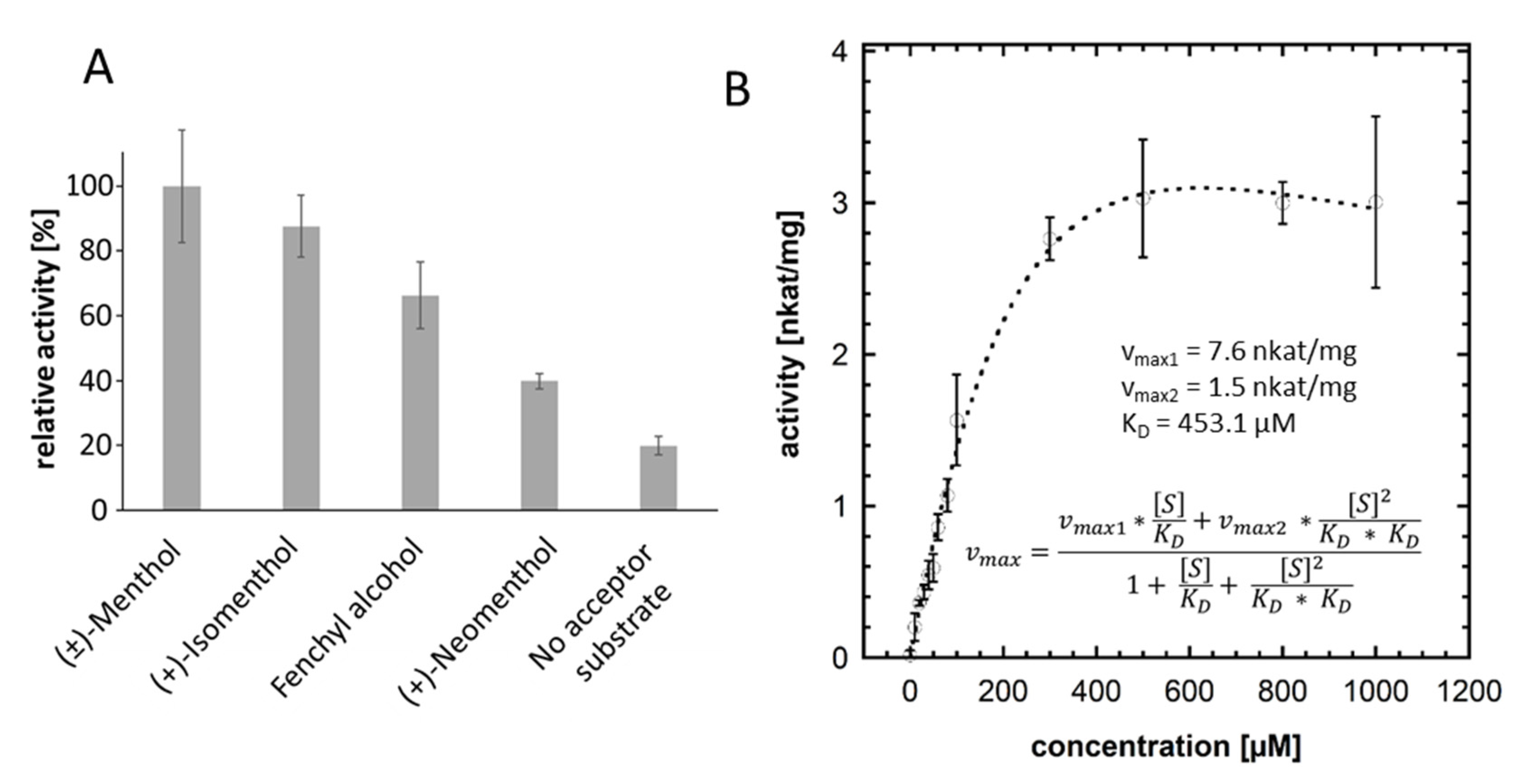
2.4. UDP Glo™ Glycosyltransferase Assay
2.5. Characterization of Menthyl Glucoside by NMR
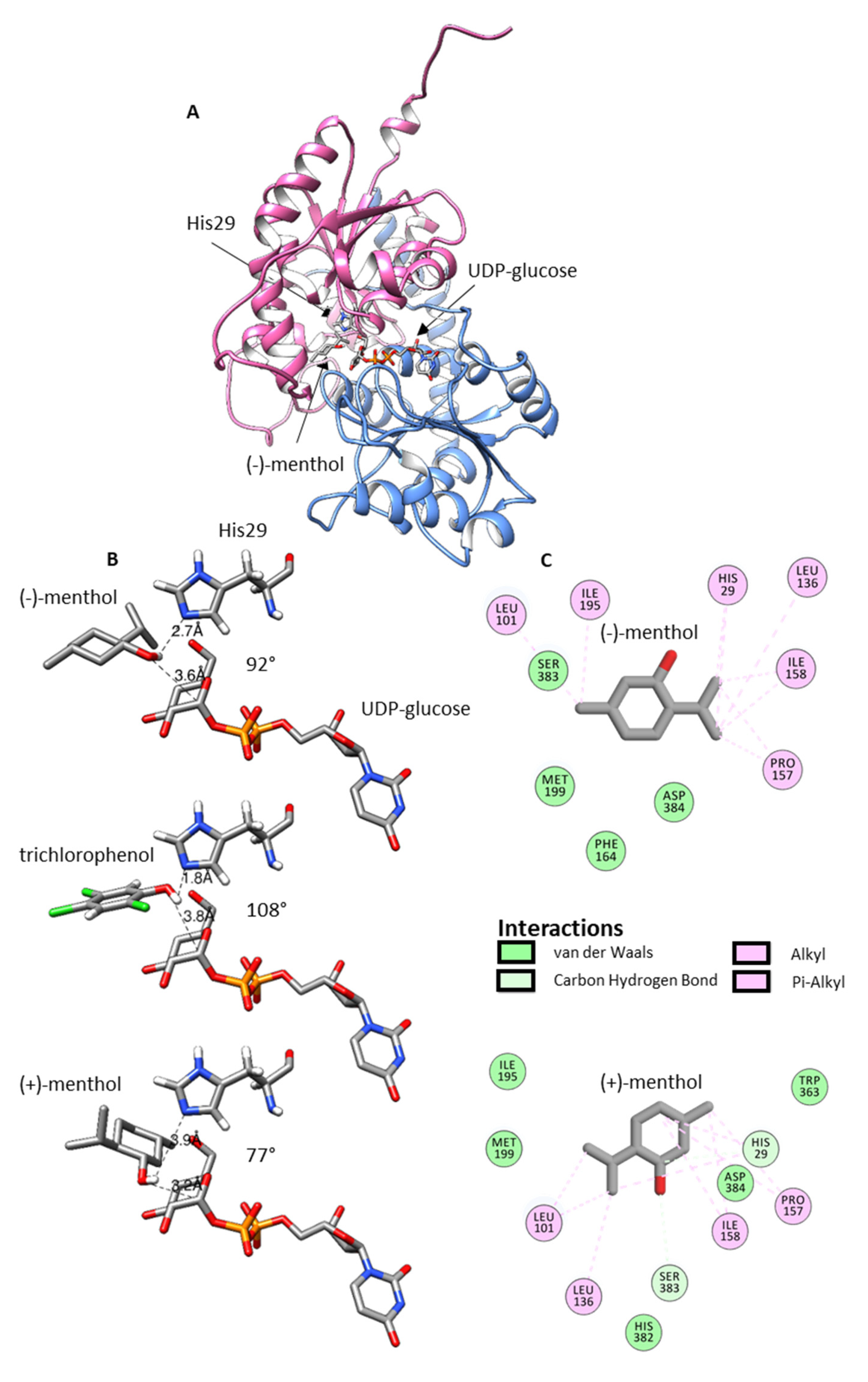
2.6. Homology Modeling and Ligand Docking
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. RNA Isolation, cDNA Synthesis and Cloning of UGT93Y1 and UGT93Y2
4.3. Recombinant Protein Production and Protein Purification
4.4. Qualitative Enzyme Assay Using LC-MS Screening
4.5. Quantitative UDP Glo™ Glycosyltransferase Screening
4.6. Screening of UGT-Library for Whole-Cell Biotransformation
4.7. Production and Purification of Menthyl Glucoside from Large Scale Biotransformation
4.8. NMR Analysis
4.9. Homology Modelling of UGT93Y1 and Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Härtner, J.; Reinscheid, U.M. Conformational analysis of menthol diastereomers by NMR and DFT computation. J. Mol. Struct. 2008, 872, 145–149. [Google Scholar] [CrossRef]
- Kamatou, G.P.P.; Vermaak, I.; Viljoen, A.M.; Lawrence, B.M. Menthol: A simple monoterpene with remarkable biological properties. Phytochemistry 2013, 96, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Oz, M.; El Nebrisi, E.G.; Yang, K.-H.S.; Howarth, F.C.; Al Kury, L.T. Cellular and molecular targets of menthol actions. Front. Pharmacol. 2017, 8, 472. [Google Scholar] [CrossRef] [Green Version]
- Fahlbusch, K.-G.; Hammerschmidt, F.-J.; Panten, J.; Pickenhagen, W.; Schatkowski, D.; Bauer, K.; Garbe, D.; Surburg, H. Flavors and Fragrances. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley Online Library: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Jerković, I.; Mastelić, J. Composition of free and glycosidically bound volatiles of Mentha aquatica L. Croat. Chem. Acta 2001, 74, 431–439. [Google Scholar]
- Mastelić, J.; Mladen, M.; Danica, K. Free and glycosidically bound volatiles of Mentha citrata Ehrh. Croat. Chem. Acta 2000, 73, 781–794. [Google Scholar]
- Croteau, R.B.; Davis, E.M.; Ringer, K.L.; Wildung, M.R. (−)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 2005, 92, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.Q.; Qamar, N.; Yadav, P.; Kulkarni, P.; Kumar, A.; Shasany, A.K. Comparative glandular trichome transcriptome-based gene characterization reveals reasons for differential (−)-menthol biosynthesis in Mentha species. Physiol. Plant. 2017, 160, 128–141. [Google Scholar] [CrossRef]
- Nilo, M.C.S.; Riachi, L.G.; Simas, D.L.R.; Coleho, G.C.; da Silva, A.J.R.; Costa, D.C.M.; Alviano, D.S.; Alviano, C.S.; Maria, C.A.B. Chemical composition and antioxidant and antifungal properties of Mentha x piperita L. (peppermint) and Mentha arvensis L. (cornmint) samples. Food Res. 2017, 1, 147–156. [Google Scholar] [CrossRef]
- Sgorbini, B.; Cagliero, C.; Pagani, A.; Sganzerla, M.; Boggia, L.; Bicchi, C.; Rubiolo, P. Determination of free and glucosidically-bound volatiles in plants. Two case studies: L-menthol in peppermint (Mentha x piperita L.) and eugenol in clove (Syzygium aromaticum (L.) Merr. & L.M.Perry). Phytochemistry 2015, 117, 296–305. [Google Scholar] [CrossRef]
- Stengele, M.; Stahl-Biskup, E. Influencing the level of glycosidically bound volatiles by feeding experiments with a Mentha × piperita l. cultivar. Flavour Fragr. J. 1994, 9, 261–263. [Google Scholar] [CrossRef]
- Sutour, S.; Tomi, F.; Bradesi, P.; Casanova, J. Chemical composition of the essential oil from corsican Mentha aquatica—Combined analysis by GC(RI), GC-MS and 13C NMR spectroscopy. Nat. Prod. Commun. 2011, 1479–1482. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, B.M. Mint: The Genus Mentha; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar] [CrossRef]
- Choi, H.-Y.; Kim, B.-M.; Morgan, A.M.A.; Kim, J.S.; Kim, W.-G. Improvement of the pharmacological activity of menthol via enzymatic β-anomer-selective glycosylation. AMB Express 2017, 7, 167. [Google Scholar] [CrossRef]
- Yosipovitch, G.; Szolar, C.; Hui, X.Y.; Maibach, H. Effect of topically applied menthol on thermal, pain and itch sensations and biophysical properties of the skin. Arch. Dermatol. Res. 1996, 288, 245–248. [Google Scholar] [CrossRef]
- Bautista, D.M.; Siemens, J.; Glazer, J.M.; Tsuruda, P.R.; Basbaum, A.I.; Stucky, C.L.; Jordt, S.-E.; Julius, D. The menthol receptor TRPM8 is the principal detector of environmental cold. Nature 2007, 448, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Bharate, S.S.; Bharate, S.B. Modulation of thermoreceptor TRPM8 by cooling compounds. ACS Chem. Neurosci. 2012, 3, 248–267. [Google Scholar] [CrossRef] [Green Version]
- Toogood, H.S.; Ní Cheallaigh, A.; Tait, S.; Mansell, D.J.; Jervis, A.; Lygidakis, A.; Humphreys, L.; Takano, E.; Gardiner, J.M.; Scrutton, N.S. Enzymatic menthol production: One-pot approach using engineered Escherichia coli. ACS Synth. Biol. 2015, 4, 1112–1123. [Google Scholar] [CrossRef]
- Schwab, W.; Fischer, T.; Wüst, M. Terpene glucoside production: Improved biocatalytic processes using glycosyltransferases. Eng. Life Sci. 2015, 15, 376–386. [Google Scholar] [CrossRef]
- Weber, J.; Schwarz, M.; Schiefer, A.; Hametner, C.; Häubl, G.; Fröhlich, J.; Mikula, H. Chemical glucosylation of labile natural products using a (2-nitrophenyl)acetyl-protected glucosyl acetimidate donor. Eur. J. Org. Chem. 2018, 2018, 2701–2706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakata, I.; Iwamura, H. Synthesis and properties of menthyl glycosides. Agric. Biol. Chem. 1979, 43, 307–312. [Google Scholar] [CrossRef]
- Do, H.; Sato, T.; Kirimura, K.; Kino, K.; Usami, S. Enzymatic synthesis of l-menthyl alpha-maltoside and l-menthyl alpha-maltooligosides from l-menthyl alpha-glucoside by cyclodextrin glucanotransferase. J. Biosci. Bioeng. 2002, 94, 119–123. [Google Scholar] [CrossRef]
- Noguchi, K.; Nakagawa, H.; Yoshiyama, M.; Shimura, S.; Kirimura, K.; Usami, S. Anomer-selective glucosylation of l-menthol using lyophilized cells of Saccharomyces cerevisiae. J. Ferment. Bioeng. 1998, 85, 436–438. [Google Scholar] [CrossRef]
- Nakagawa, H.; Yoshiyama, M.; Shimura, S.; Kirimura, K.; Usami, S. Anomer-selective glucosylation of l-menthol by yeast alpha-glucosidase. Biotechnol. Biochem. 1998, 62, 1332–1336. [Google Scholar] [CrossRef]
- Nakagawa, H.; Dobashi, Y.; Sato, T.; Yoshida, K.; Tsugane, T.; Shimura, S.E.A. Alpha-anomer-selective glucosylation of menthol with high yield through a crystal accumulation reaction using lyophilized cells of Xanthomonas campestris WU-9701. J. Biosci. Bioeng. 2000, 89, 138–144. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Y.; Lu, C.; Ma, Z.; Chen, H.; Zhu, L.; Lu, Y.; Chen, X. Efficient production of l-menthyl alpha-glucopyranoside from l-menthol via whole-cell biotransformation using recombinant Escherichia coli. Biotechnol. Lett. 2021, 43, 1757–1764. [Google Scholar] [CrossRef]
- Shimoda, K.; Kubota, N.; Hamada, H. Enantioselective glucosylation of (±)-secondary alcohols with plant glucosyltransferases. Tetrahedron Asymmetry 2004, 15, 2319–2321. [Google Scholar] [CrossRef]
- Green, M.D.; Clarke, D.J.; Oturu, E.M.; Styczynski, P.B.; Jackson, M.R.; Burchell, B.; Tephly, T.R. Cloning and expression of a rat liver phenobarbital-inducible UDP-glucuronosyltransferase (2B12) with specificity for monoterpenoid alcohols. Arch. Biochem. Biophys. 1995, 322, 460–468. [Google Scholar] [CrossRef]
- Mestrom, L.; Przypis, M.; Kowalczykiewicz, D.; Pollender, A.; Kumpf, A.; Marsden, S.R.; Bento, I.; Jarzębski, A.B.; Szymańska, K.; Chruściel, A.; et al. Leloir Glycosyltransferases in applied biocatalysis: A multidisciplinary approach. Int. J. Mol. Sci. 2019, 20, 5263. [Google Scholar] [CrossRef] [Green Version]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwab, W.; Fischer, T.C.; Giri, A.; Wüst, M. Potential applications of glucosyltransferases in terpene glucoside production: Impacts on the use of aroma and fragrance. Appl. Microbiol. Biotechnol. 2015, 99, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Shibata, H.; Maejima, S. A New Monoterpene Glucoside ℓ-Menthyl 6′-O-acetyl-β-d-glucoside. J. Essent. Oil Res. 1990, 2, 21–24. [Google Scholar] [CrossRef]
- Nunes, I.S.; Faria, J.M.S.; Figueiredo, A.C.; Pedro, L.G.; Trindade, H.; Barroso, J.G. Menthol and geraniol biotransformation and glycosylation capacity of Levisticum officinale hairy roots. Planta Med. 2009, 75, 387–391. [Google Scholar] [CrossRef] [Green Version]
- Furuya, T.; Orihara, Y.; Miyatake, H. Biotransformation of (−)-menthol by Eucalyptus perriniana cultured cells. J. Chem. Soc. Perkin Trans. 1989, 10, 1711–1719. [Google Scholar] [CrossRef]
- Berger, R.G.; Drawert, F. Glycosylation of terpenols and aromatic alcohols by cell suspension cultures of peppermint (Mentha piperita L.). Z. für Nat. C 1988, 43, 485–490. [Google Scholar] [CrossRef]
- Sakata, I.; Mitsui, T. Isolation and identification of l-menthyl-β-d-glucoside from Shubi. Agric. Biol. Chem. 1975, 39, 1329–1330. [Google Scholar] [CrossRef]
- Effenberger, I.; Hoffmann, T.; Jonczyk, R.; Schwab, W. Novel biotechnological glucosylation of high-impact aroma chemicals, 3(2H)- and 2(5H)-furanones. Sci. Rep. 2019, 9, 10943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G. Tarling Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.-C.; Giri, A.; Daniilidis, M.; Sun, G.; Härtl, K.; Hoffmann, T.; Schwab, W. Structural and functional analysis of UGT92G6 suggests an evolutionary link between mono- and disaccharide glycoside-forming transferases. Plant Cell Physiol. 2018, 59, 857–870. [Google Scholar] [CrossRef]
- McGraphery, K.; Schwab, W. Comparative analysis of high-throughput assays of family-1 plant glycosyltransferases. Int. J. Mol. Sci. 2020, 21, 2208. [Google Scholar] [CrossRef] [Green Version]
- Yano, D.; Suzuki, T. Kinetic analyses of the substrate inhibition of paramecium arginine kinase. Protein J. 2018, 37, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Tomita, Y.; Tori, K.; Yoshimura, Y. Determination of the absolute configuration of a secondary hydroxy group in a chiral secondary alcohol using glycosidation shifts in carbon-13 nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 1978, 100, 3331–3339. [Google Scholar] [CrossRef]
- Kasai, R.; Okihara, M.; Asakawa, J.; Mizutani, K.; Tanaka, O. 13C NMR Study of α- and β-anomeric pairs of d-mannopyranosides and L-rhamnopyranosides. Tetrahedron 1979, 35, 1427–1432. [Google Scholar] [CrossRef]
- Kasai, R.; Suzuo, M.; Asakawa, J.; Tanaka, O. Carbon-13 chemical shifts of isoprenoid-β-d-glucopyranosides and -β-d-mannopyranosides. Stereochemical influences of aglycone alcohols. Tetrahedron Lett. 1977, 18, 175–178. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Adiyaman, R.; Maghrabi, A.H.A.; Shuid, A.N.; Brackenridge, D.A.; Nealon, J.O.; Philomina, L.S. IntFOLD: An integrated web resource for high performance protein structure and function prediction. Nucleic Acids Res. 2019, 47, W408–W413. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.; Sun, Y.; Zhang, P.; Wang, Y. Structural insights into the catalytic mechanism of a plant diterpene glycosyltransferase SrUGT76G1. Plant Commun. 2020, 1, 100004. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Lim, E.-K.; Bowles, D.J. Discovery of new biocatalysts for the glycosylation of terpenoid scaffolds. Chemistry 2008, 14, 6656–6662. [Google Scholar] [CrossRef]
- Song, C.; Härtl, K.; McGraphery, K.; Hoffmann, T.; Schwab, W. Attractive but toxic: Emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation. Mol. Plant 2018, 11, 1225–1236. [Google Scholar] [CrossRef] [Green Version]
- Ciesla, W.P.; Bobak, D.A. Clostridium difficile toxins A and B are cation-dependent UDP-glucose hydrolases with differing catalytic activities. J. Biol. Chem. 1998, 273, 16021–16026. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, M.O.; Halmo, S.M.; Patel, S.; Middleton, D.; Takeuchi, H.; Schafer, C.M.; West, C.M.; Haltiwanger, R.S.; Avci, F.Y.; Moremen, K.W.; et al. Rapid screening of sugar-nucleotide donor specificities of putative glycosyltransferases. Glycobiology 2017, 27, 206–212. [Google Scholar] [CrossRef]
- Levanova, N.; Mattheis, C.; Carson, D.; To, K.-N.; Jank, T.; Frankel, G.; Aktories, K.; Schroeder, G.N. The Legionella effector LtpM is a new type of phosphoinositide-activated glucosyltransferase. J. Biol. Chem. 2019, 294, 2862–2879. [Google Scholar] [CrossRef] [Green Version]
- Weng, J.; Chen, L.; Cheng, Y.; Li, Y.; Jia, H.; Zhou, H.; Wei, P. Expression, characterization, and site-directed mutagenesis of UDP-glycosyltransferase UGT88A1 from Arabidopsis thaliana. Bioengineered 2019, 10, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Rüdiger, J.; Schwab, W. Improvement of an Escherichia coli whole-cell biocatalyst for geranyl glucoside production using directed evolution. Eng. Rep. 2021, e12440, Early view. [Google Scholar] [CrossRef]
- Wu, B. Substrate inhibition kinetics in drug metabolism reactions. Drug Metab. Rev. 2011, 43, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Yang, T.; Whitaker, B.D.; Shangguan, L.; Fang, J. Calcium/calmodulin alleviates substrate inhibition in a strawberry UDP-glucosyltransferase involved in fruit anthocyanin biosynthesis. BMC Plant Biol. 2016, 16, 197. [Google Scholar] [CrossRef] [Green Version]
- Dong, D.; Wu, B. What are the real causes of substrate inhibition in the glucuronidation reaction? Single Cell Biol. 2012, 1, e105. [Google Scholar] [CrossRef]
- Hewitt, C.O.; Eszes, C.M.; Sessions, R.B.; Moreton, K.M.; Dafforn, T.R.; Takei, J.; Dempsey, C.E.; Clarke, A.R.; Holbrook, J.J. A general method for relieving substrate inhibition in lactate dehydrogenases. Protein Eng. 1999, 12, 491–496. [Google Scholar] [CrossRef] [Green Version]
- Orihara, Y.; Miyatake, H.; Furuya, T. Triglucosylation on the biotransformation of (+)-menthol by cultured cells of Eucalyptus perriniana. Phytochemistry 1991, 30, 1843–1845. [Google Scholar] [CrossRef]
- Parajuli, P.; Pandey, R.P.; Trang, N.T.H.; Chaudhary, A.K.; Sohng, J.K. Synthetic sugar cassettes for the efficient production of flavonol glycosides in Escherichia coli. Microb. Cell Fact. 2015, 14, 76. [Google Scholar] [CrossRef] [Green Version]
- Scheibenzuber, S.; Hoffmann, T.; Effenberger, I.; Schwab, W.; Asam, S.; Rychlik, M. Enzymatic synthesis of modified Alternaria mycotoxins using a whole-cell biotransformation system. Toxins 2020, 12, 264. [Google Scholar] [CrossRef] [Green Version]
- Priebe, X.; Hoang, M.D.; Rüdiger, J.; Turgel, M.; Tröndle, J.; Schwab, W.; Weuster-Botz, D. Byproduct-free geraniol glycosylation by whole-cell biotransformation with recombinant Escherichia coli. Biotechnol. Lett. 2021, 43, 247–259. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, X.-L.; Ullah, N.; Tao, Y.-S. Aroma glycosides in grapes and wine. J. Food Sci. 2017, 82, 248–259. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Li, J.; Wang, S.; Yao, L.; Liang, W.; Wang, J.; Gao, W. Advances in ginsenoside biosynthesis and metabolic regulation. Biotechnol. Appl. Biochem. 2018, 65, 514–522. [Google Scholar] [CrossRef]
- Liao, Z.; Chen, M.; Guo, L.; Gong, Y.; Tang, F.; Sun, X.; Tang, K. Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep. Biochem. Biotechnol. 2004, 34, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Bönisch, F.; Frotscher, J.; Stanitzek, S.; Rühl, E.; Wüst, M.; Bitz, O.; Schwab, W. Activity-based profiling of a physiologic aglycone library reveals sugar acceptor promiscuity of family 1 UDP-glucosyltransferases from grape. Plant Physiol. 2014, 166, 23–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, G.; Strebl, M.; Merz, M.; Blamberg, R.; Huang, F.-C.; McGraphery, K.; Hoffmann, T.; Schwab, W. Glucosylation of the phytoalexin N-feruloyl tyramine modulates the levels of pathogen-responsive metabolites in Nicotiana benthamiana. Plant J. 2019, 100, 20–37. [Google Scholar] [CrossRef]
- Butt, S.S.; Badshah, Y.; Shabbir, M.; Rafiq, M. Molecular Docking Using Chimera and Autodock Vina Software for Nonbioinformaticians. JMIR Bioinform. Biotechnol. 2020, 1, e14232. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurze, E.; Ruß, V.; Syam, N.; Effenberger, I.; Jonczyk, R.; Liao, J.; Song, C.; Hoffmann, T.; Schwab, W. Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants. Molecules 2021, 26, 5511. https://doi.org/10.3390/molecules26185511
Kurze E, Ruß V, Syam N, Effenberger I, Jonczyk R, Liao J, Song C, Hoffmann T, Schwab W. Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants. Molecules. 2021; 26(18):5511. https://doi.org/10.3390/molecules26185511
Chicago/Turabian StyleKurze, Elisabeth, Victoria Ruß, Nadia Syam, Isabelle Effenberger, Rafal Jonczyk, Jieren Liao, Chuankui Song, Thomas Hoffmann, and Wilfried Schwab. 2021. "Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants" Molecules 26, no. 18: 5511. https://doi.org/10.3390/molecules26185511
APA StyleKurze, E., Ruß, V., Syam, N., Effenberger, I., Jonczyk, R., Liao, J., Song, C., Hoffmann, T., & Schwab, W. (2021). Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants. Molecules, 26(18), 5511. https://doi.org/10.3390/molecules26185511