Conditional Cooperativity in DNA Minor-Groove Recognition by Oligopeptides
Abstract
1. Introduction
2. Results and Discussion
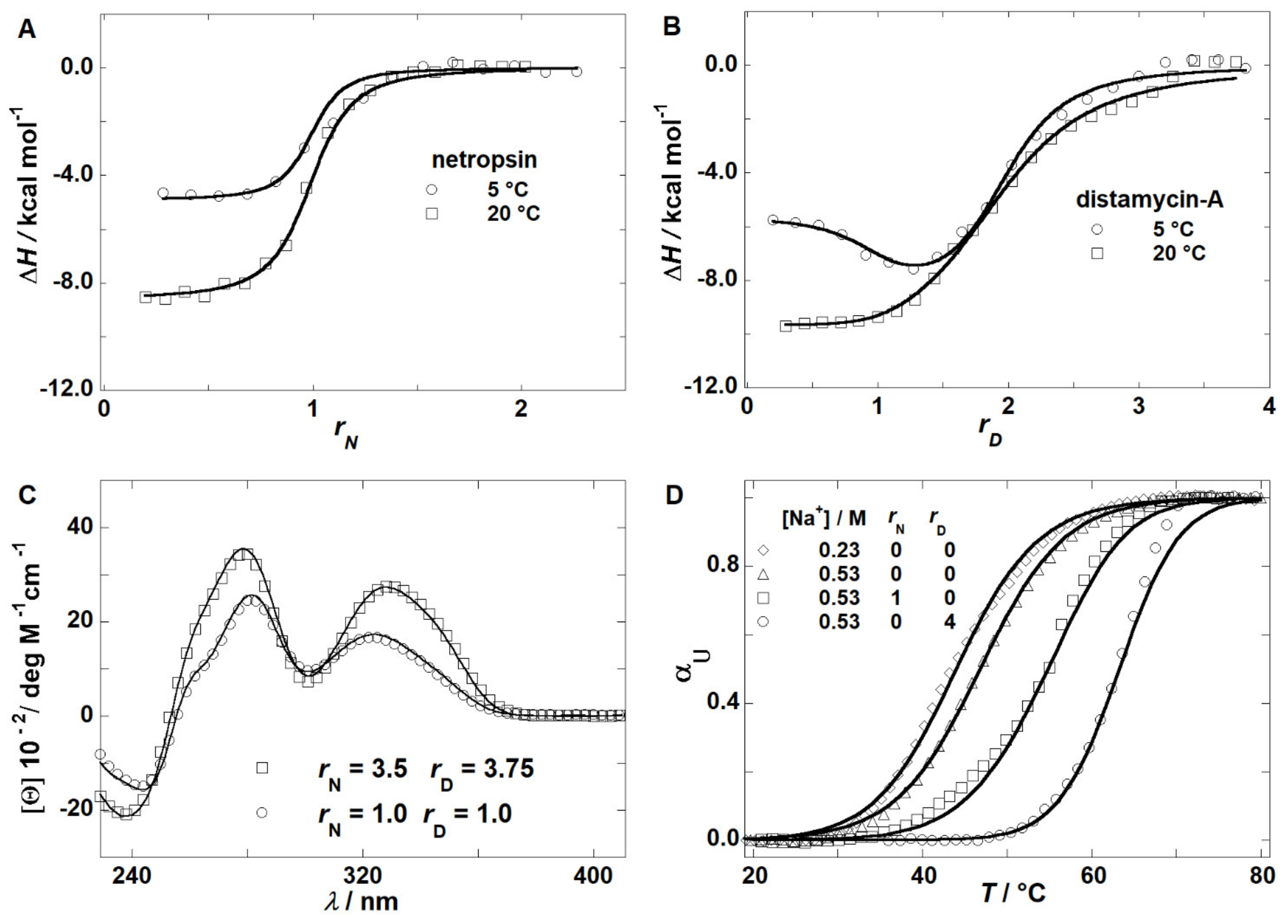
2.1. Thermodynamics of Binding
2.2. Thermodynamics of (un)Folding
2.3. Thermodynamic Parameters of Binding and Folding Define the Species Populations
2.4. Model Predicted Species Populations Are in Accordance with Experimental Observations
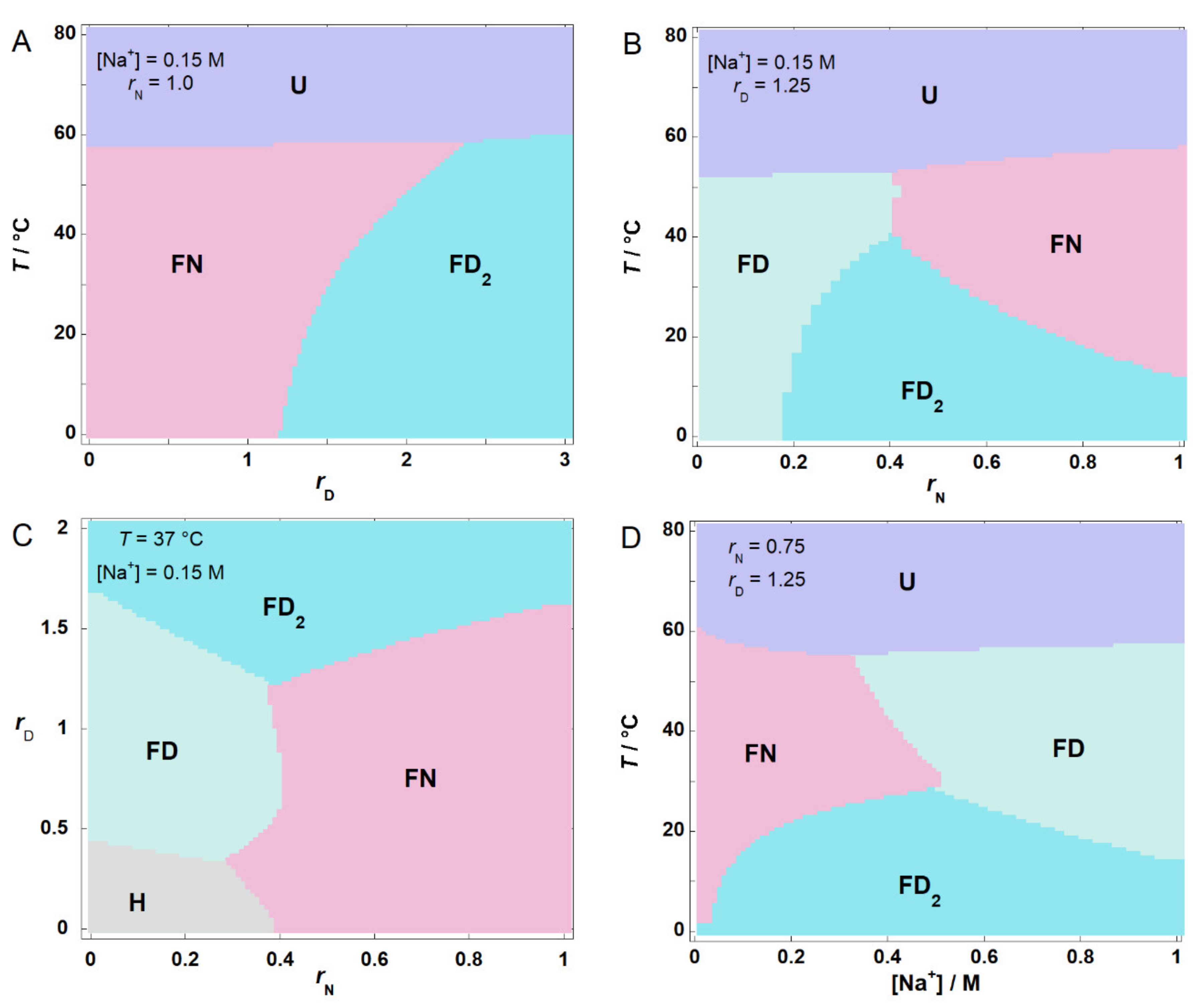
2.5. Phase Diagrams Illustrate the Nature of Conditional Cooperativity
3. Conclusions
4. Materials and Methods
4.1. DNA and Ligand Preparation
4.2. Isothermal Titration Calorimetry (ITC)
4.3. UV-Absorption Spectroscopy (UV)
4.4. Circular Dichroism Spectroscopy (CD)
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Overgaard, M.; Borch, J.; Jørgensen, M.G.; Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by condi-tional cooperativity. Mol. Microbiol. 2008, 69, 841. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pino, A.; Balasubramanian, S.; Wyns, L.; Gazit, E.; De Greve, H.; Magnuson, R.D.; Charlier, D.; van Nuland, N.A.; Loris, R. Allostery and Intrinsic Disorder Mediate Transcription Regulation by Conditional Cooperativity. Cell 2010, 142, 101–111. [Google Scholar] [CrossRef]
- Garcia-Pino, A.; De Gieter, S.S.; Talavera, A.A.; De Greve, H.; Efremov, R.; Loris, R. An intrinsically disordered entropic switch determines allostery in Phd–Doc regulation. Nat. Chem. Biol. 2016, 12, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Vandervelde, A.; Drobnak, I.; Hadži, S.; Sterckx, Y.; Welte, T.; De Greve, H.; Charlier, D.; Efremov, R.; Loris, R.; Lah, J. Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon. Nucleic Acids Res. 2017, 45, 2937–2950. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, P.; Girardin, Y.; Loris, R. Prokaryote toxin–antitoxin modules: Complex regulation of an unclear function. Protein Sci. 2021, 30, 1103–1113. [Google Scholar] [CrossRef]
- Chaires, J.B. Drug-DNA interactions. Curr. Opin. Struct. Biol. 1998, 8, 314. [Google Scholar] [CrossRef]
- Smidlehner, T.; Badovinac, M.; Piantanida, I. Pyrene-cyanine conjugates as multipurpose fluorescent probes for non-covalent recognition of ds-DNA, RNA and proteins. New J. Chem. 2018, 42, 6655. [Google Scholar] [CrossRef]
- Ban, Ž.; Žinić, B.; Vianello, R.; Schmuck, C.; Piantanida, I. Nucleobase–Guanidiniocarbonyl-Pyrrole Conjugates as Novel Fluo-rimetric Sensors for Single Stranded RNA. Molecules 2017, 22, 2213. [Google Scholar] [CrossRef]
- Radić-Stojković, M.; Piotrowski, P.; Schmuck, C.; Piantanida, I. Short, rigid linker between pyrene and guanidiniocarbon-yl-pyrrole induced new set of spectroscopic responses to ds-DNA secondary structure. Org. Biomol. Chem. 2015, 13, 1629. [Google Scholar] [CrossRef]
- White, S.K.; Szewczyk, J.W.; Turner, J.M.; Baird, E.E.; Dervan, P.B. Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature 1998, 391, 468–471. [Google Scholar] [CrossRef]
- Geierstanger, B.H.; Wemmer, D.E. Complexes of the minor groove of DNA. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 463–493. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Nucleic Acid Structure and Recognition; Oxford University Press: Oxford, UK, 2002; pp. 89–137. [Google Scholar]
- Kopka, M.L.; Yoon, C.; Goodsell, D.; Pjura, P.; Dickerson, R.E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. USA 1985, 82, 1376–1380. [Google Scholar] [CrossRef]
- Marky, L.A.; Breslauer, K.J. Origins of netropsin binding affinity and specificity: Correlations of thermodynamic and structural data. Proc. Natl. Acad. Sci. USA 1987, 84, 4359–4363. [Google Scholar] [CrossRef] [PubMed]
- Rentzeperis, D.; Marky, L.A. Netropsin binding as a thermodynamic probe of the grooves of parallel DNA. J. Am. Chem. Soc. 1993, 115, 1645–1650. [Google Scholar] [CrossRef]
- Pelton, J.G.; Wemmer, D.E. Binding modes of distamycin A with d(CGCAAATTTGCG)2 determined by two-dimensional NMR. J. Am. Chem. Soc. 1990, 112, 1393–1399. [Google Scholar] [CrossRef]
- Capobianco, M.L.; Colonna, F.P.; Forni, A.; Gabresi, A.; Lotti, S.; Moretti, I.; Samori, B.; Tondelli, L. Interactions of nucleic acids with distamycins. Binding of Dst-3 to d(CGTTTAAACG)2 and d(CGTACGTACG). Nucleic Acids Res. 1991, 19, 1695. [Google Scholar] [CrossRef][Green Version]
- Pelton, J.G.; Wemmer, D.E. Structuralcharacterization of a 2:1 distamycin Ad(CGCAAATTGGC) complex by two-dimensional NMR. Proc. Natl. Acad. Sci. USA 1989, 86, 5723. [Google Scholar] [CrossRef]
- Rentzeperis, D.; Marky, L.A.; Dwyer, T.J.; Geierstanger, B.H.; Pelton, J.G.; Wemmer, D.E. Interaction of minor groove ligands to an AAATT/AATTT site: Correlation of thermodynamic characterization and solution structure. Biochemistry 1995, 34, 2937–2945. [Google Scholar] [CrossRef]
- Chen, F.-M.; Sha, F. Circular Dichroic and Kinetic Differentiation of DNA Binding Modes of Distamycin. Biochemistry 1998, 37, 11143–11151. [Google Scholar] [CrossRef]
- Lah, J.; Vesnaver, G. Binding of Distamycin A and Netropsin to the 12mer DNA Duplexes Containing Mixed AT·GC Sequences with At Most Five or Three Successive AT Base Pairs. Biochemistry 2000, 39, 9317–9326. [Google Scholar] [CrossRef]
- Lah, J.; Drobnak, I.; Dolinar, M.; Vesnaver, G. What drives the binding of minor groove-directed ligands to DNA hairpins? Nucleic Acids Res. 2008, 36, 897. [Google Scholar] [CrossRef]
- Lah, J.; Vesnaver, G. Energetic Diversity of DNA Minor-groove Recognition by Small Molecules Displayed Through Some Model Ligand-DNA Systems. J. Mol. Biol. 2004, 342, 73–89. [Google Scholar] [CrossRef]
- Drobnak, I.; Vesnaver, G.; Lah, J. Model-Based Thermodynamic Analysis of Reversible Unfolding Processes. J. Phys. Chem. B 2010, 114, 8713. [Google Scholar] [CrossRef] [PubMed]
- Santa Lucia, J., Jr.; Hicks, D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 415. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L.; Crane-Robinson, C. Forces maintaining the DNA double helix and its complexes with transcription factors. Prog. Biophys. Mol. Biol. 2018, 135, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Hadži, S.; Lah, J. Origin of heat capacity increment in DNA folding: The hydration effect. BBA Gen. Subj. 2021, 1865, 1297742. [Google Scholar] [CrossRef]
- Lah, J.; Seručnik, M.; Vesnaver, G. Influence of a Hairpin Loop on the Thermodynamic Stability of a DNA Oligomer. J. Nucleic Acids 2011, 2011, 1–9. [Google Scholar] [CrossRef]
- Cantor, C.R.; Warshaw, M.M.; Shapiro, H. Oligonucleotide interactions. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 1970, 9, 1059. [Google Scholar] [CrossRef]

| t | U → F | F + N → FN | F + D → FD | FD + D →FD2 | F + 2D → FD2 |
|---|---|---|---|---|---|
| / kcal mol−1 | −3.1 ± 0.2 | −9.9 ± 0.1 | −10.2 ± 0.1 | −8.3 ± 0.1 | −18.5 ± 0.2 |
| / kcal mol−1 | −34.1 ± 0.2 | −8.6 ± 0.3 | −9.7 ± 0.4 | −10.9 ± 0.6 | −20.6 ± 1.0 |
| / kcal mol−1 | −31.0 ± 0.2 | 1.3 ± 0.4 | 0.5 ± 0.5 | −2.6 ± 0.7 | −2.1 ± 1.2 |
| / kcal mol−1K−1 | −0.26 ± 0.05 | −0.25 ± 0.02 | −0.26 ± 0.04 | −0.13 ± 0.5 | −0.39 ± 0.09 |
| nNa,j | −1.18 ± 0.2 | 1.95 ± 0.5 a | 0.62 ± 0.25 a | 0.63 ± 0.25 a | 1.25 ± 0.5 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lah, J.; Hadži, S. Conditional Cooperativity in DNA Minor-Groove Recognition by Oligopeptides. Molecules 2021, 26, 5188. https://doi.org/10.3390/molecules26175188
Lah J, Hadži S. Conditional Cooperativity in DNA Minor-Groove Recognition by Oligopeptides. Molecules. 2021; 26(17):5188. https://doi.org/10.3390/molecules26175188
Chicago/Turabian StyleLah, Jurij, and San Hadži. 2021. "Conditional Cooperativity in DNA Minor-Groove Recognition by Oligopeptides" Molecules 26, no. 17: 5188. https://doi.org/10.3390/molecules26175188
APA StyleLah, J., & Hadži, S. (2021). Conditional Cooperativity in DNA Minor-Groove Recognition by Oligopeptides. Molecules, 26(17), 5188. https://doi.org/10.3390/molecules26175188






