When UDG and hAPE1 Meet Cyclopurines. How (5′R) and (5′S) 5′,8-Cyclo-2′-deoxyadenosine and 5′,8-Cyclo-2′-deoxyguanosine Affect UDG and hAPE1 Activity?
Abstract
1. Introduction
2. Results and Discussion
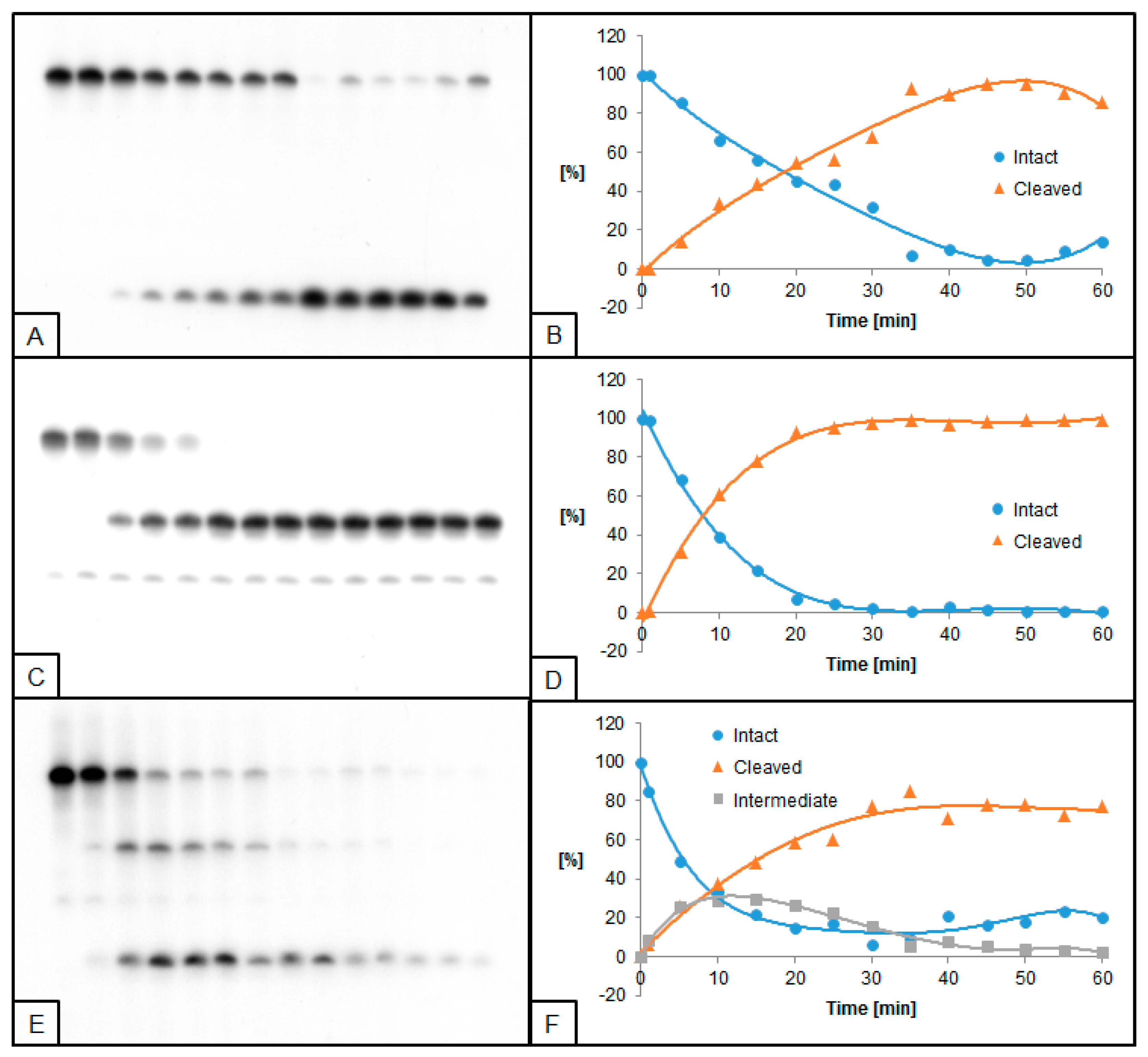
2.1. UDG and hAPE1 Action towards 2′-Deoxyuridine
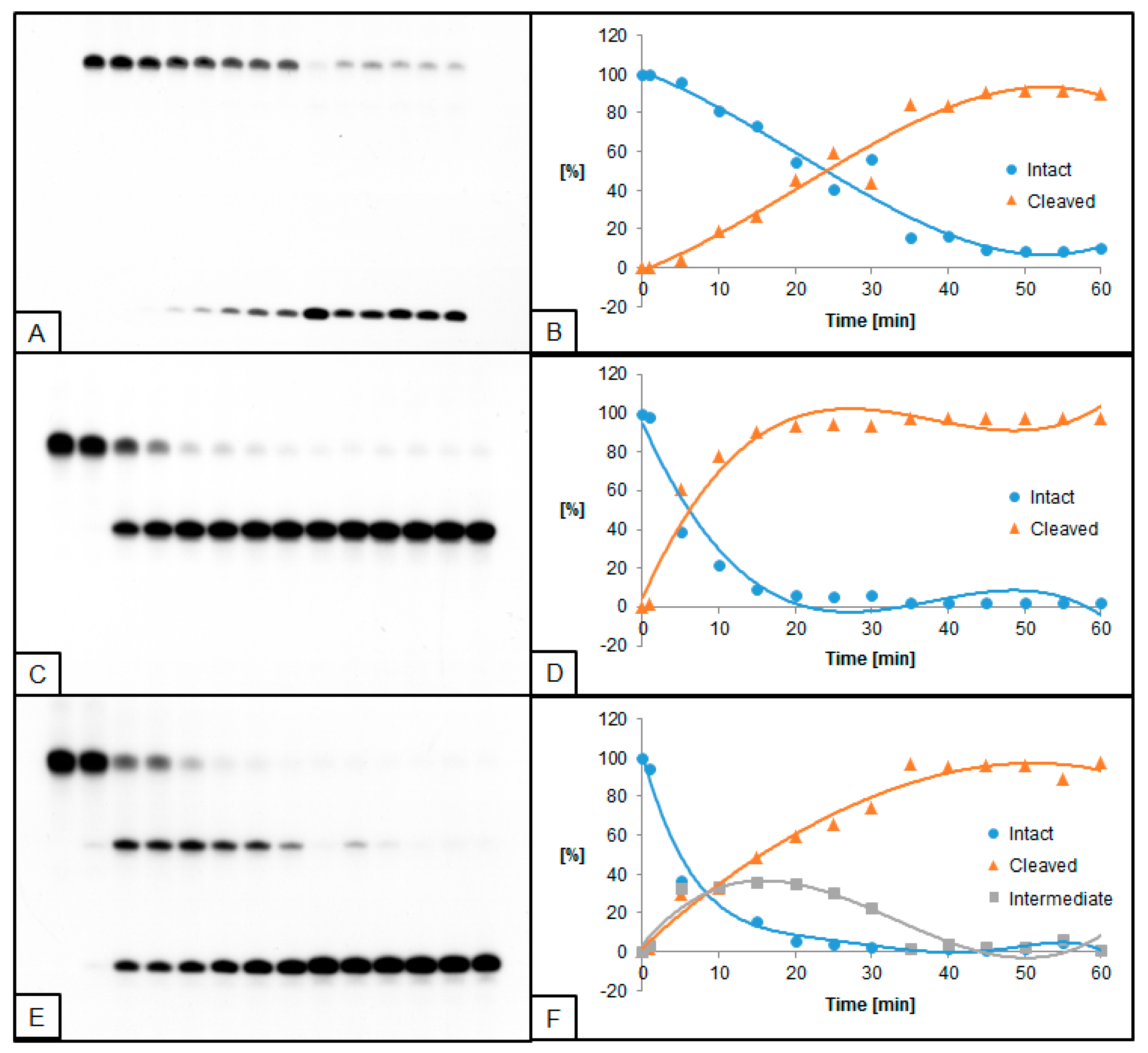
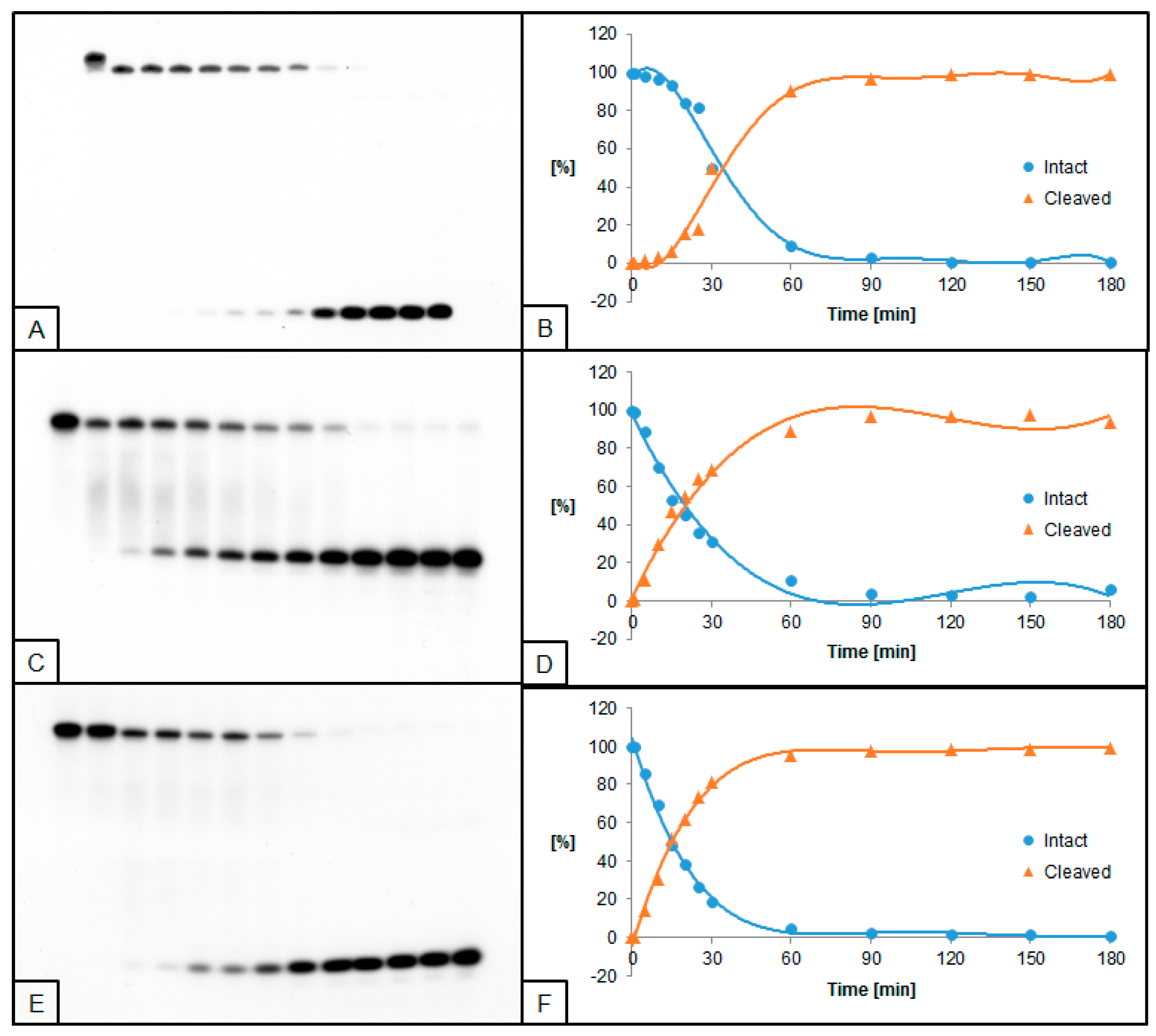
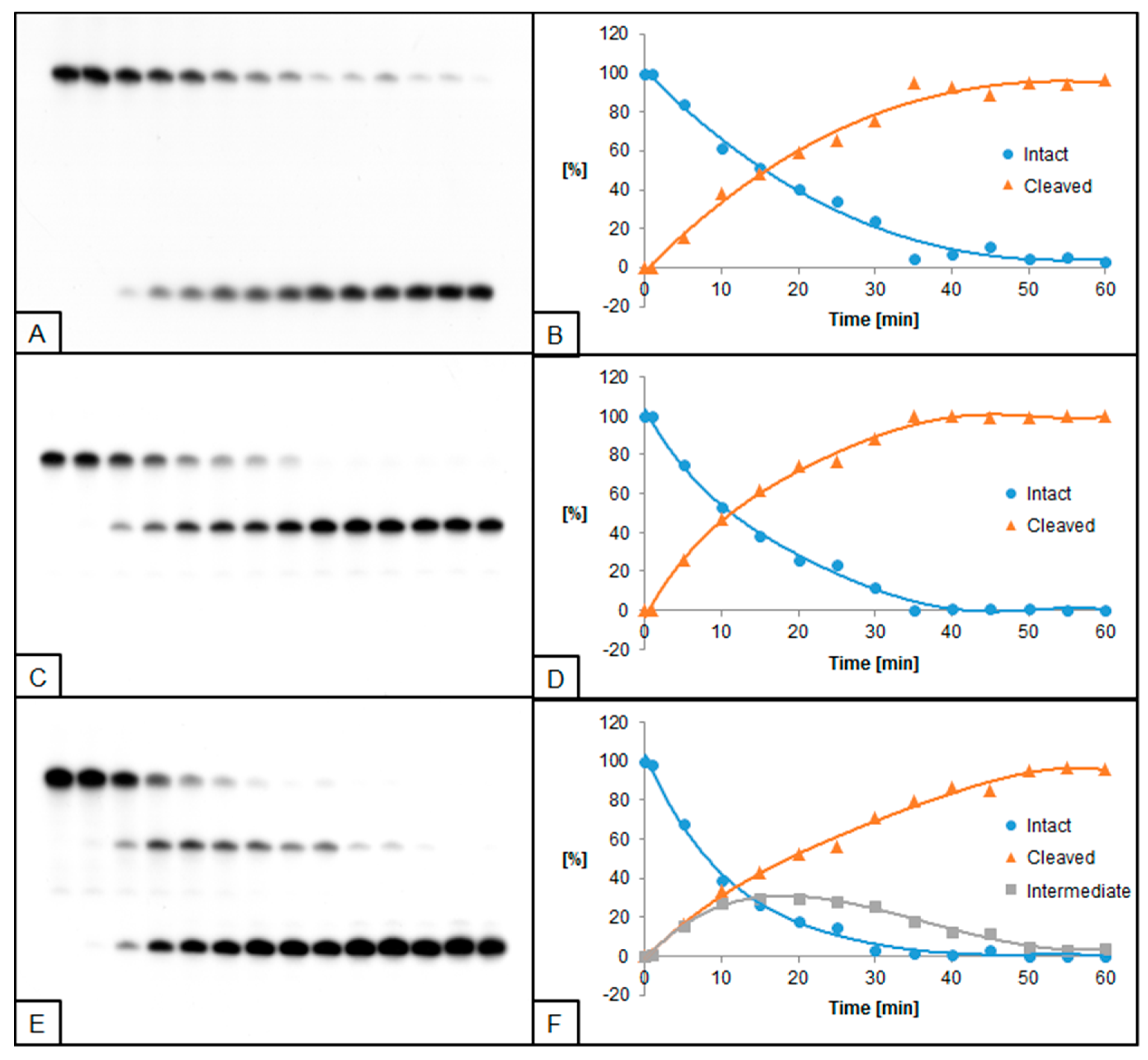
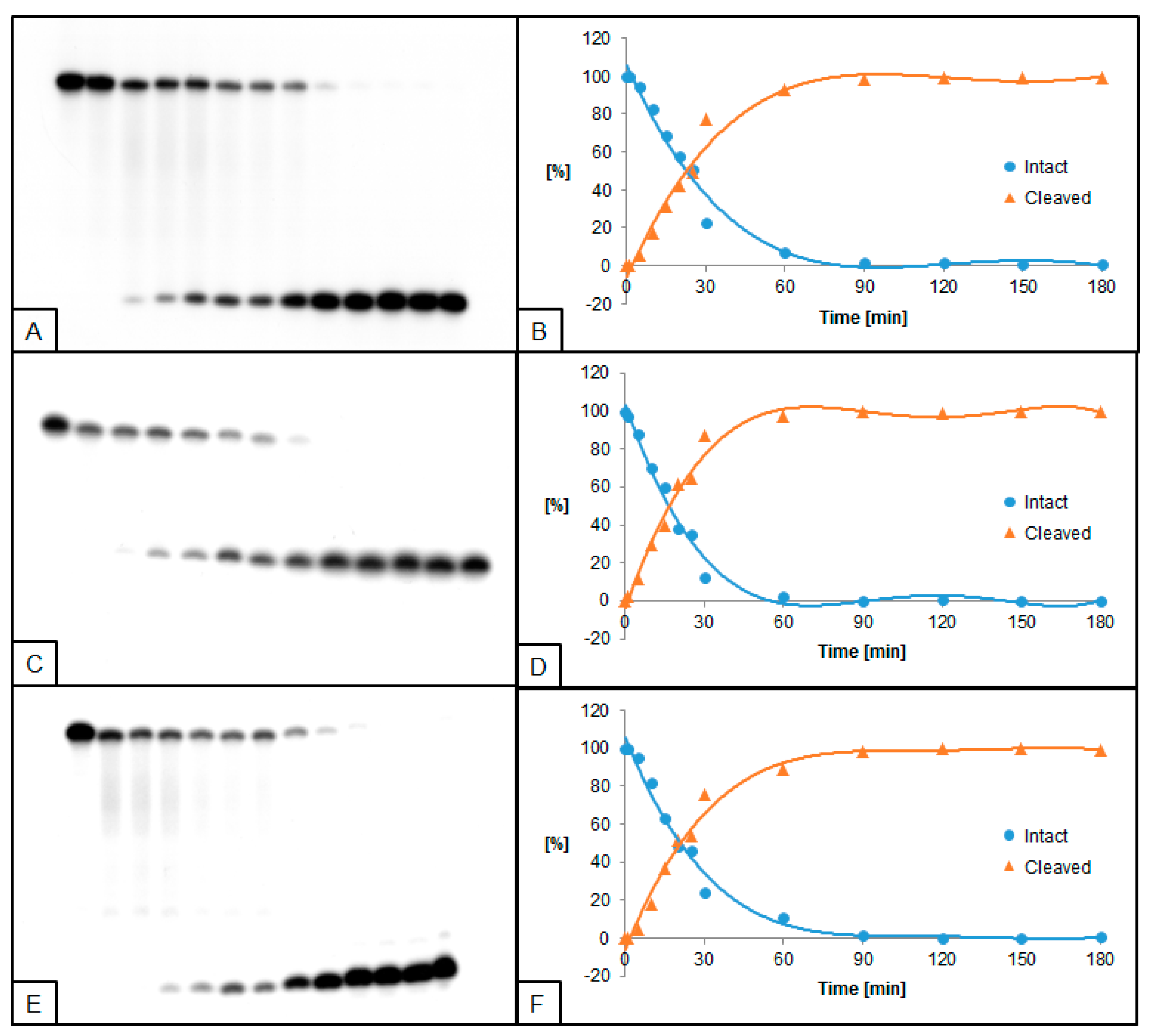
2.2. The Influence of ScdA on UDG and hAPE1 Activity
2.3. The Influence of RcdA on UDG and hAPE1 Activity
2.4. The Influence of ScdG on UDG and hAPE1 Activity
2.5. The Influence of RcdG on UDG and hAPE1 Activity
3. Materials and Methods
3.1. Substrate Oligonucleotides Synthesis and Purification
3.2. Oligonucleotides Concentration
3.3. Mass Spectroscopy of Oligonucleotides
3.4. Preparation of 5′-End-Labeled Oligonucleotides
3.5. Oligonucleotide Hybridization
3.6. UDG and hAPE1 Cleavage Assay
4. Conclusions
- For all substrate oligos, AP sites generated by UDG were suitable for subsequent hAPE1 action. This suggests that cdPus do not affect the structure of the AP site created after dU excision by UDG.
- dU present within CDL is excised more efficiently when located towards the 3′-end of cdPus (except for oligonucleotides containing RcdA and single dU lesion).
- The presence of ScdA within CDL (dU(−5)ScdA, dU(+5)ScdA and dU(−5)(+5)ScdA) forces the negative effect on dU excision by UDG in comparison with RcdA, however, the opposite trend for ScdG and RcdG was denoted.
- The cleavage of dU located in the +5 position within CDL containing two dU was higher for dU(−5)(+5)ScdA and dU(−5)(+5)RcdG than for dU(−5)(+5)RcdA and dU(−5)(+5)ScdG.
- The activity of hAPE1 increased for the majority of cdPus (except for dU(−5)(+5)RcdA and dU(−5)(+5) RcdG).
- The activity of UDG increased or did not change for the majority of cdPus, especially for oligonucleotides containing a single dU located towards 3′-end (except for dU(−5)ScdA and dU(−5)(+5)RcdA where enzyme activity was lower than the control).
- For oligos with a single dU (dU(−5) and dU(+5)), the hAPE1 activity increased for all substrates; the UDG activity was also elevated with the exception of dU(−5)ScdA and dU(−5)RcdG.
- The observed increase in enzymatic activities was higher for hAPE1 than for UDG.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Ambekar, S.S. DNA: Damage and Repair Mechanisms in Humans. Glob. J. Pharm. Pharm. Sci. 2017, 3, 58–66. [Google Scholar] [CrossRef][Green Version]
- Sage, E.; Shikazono, N. Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 2017, 107, 125–135. [Google Scholar] [CrossRef]
- Cadet, J.; Ravanat, J.-L.; TavernaPorro, M.; Menoni, H.; Angelov, D. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 2012, 327, 5–15. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef]
- Jaruga, P.; Dizdaroglu, M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: Mechanisms of formation, measurement, repair and biological effects. DNA Repair 2008, 7, 1413–1425. [Google Scholar] [CrossRef]
- Bukowska, B.; Karwowski, B.T. The Clustered DNA Lesions—Types, Pathways of Repair and Relevance to Human Health. Curr. Med. Chem. 2018, 25, 2722–2735. [Google Scholar] [CrossRef] [PubMed]
- Krokidis, M.G.; Terzidis, M.A.; Efthimiadou, E.; Zervou, S.-K.; Kordas, G.; Papadopoulos, K.; Hiskia, A.; Kletsas, D.; Chatgilialoglu, C. Purine 5′,8-Cyclo-2′-deoxynucleoside Lesions: Formation by Radical Stress and Repair in Human Breast Epithelial Cancer Cells. Free Radic. Res. 2017, 51, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J. The cyclopurine deoxynucleosides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Free Radic. Biol. Med. 2017, 107, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Suhasini, A.N.; Banerjee, T.; Sommers, J.A.; Kaplan, D.L.; Kuper, J.; Kisker, C.; Brosh, R.M. Impact of Age-Associated Cyclopurine Lesions on DNA Repair Helicases. PLoS ONE 2014, 9, e113293. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Krokidis, M.G.; Masi, A.; Barata-Vallejo, S.; Ferreri, C.; Terzidis, M.A.; Szreder, T.; Bobrowski, K. New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides. Molecules 2019, 24, 3860. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Eriksson, L.A.; Krokidis, M.G.; Masi, A.; Wang, S.; Zhang, R. Oxygen Dependent Purine Lesions in Double-Stranded Oligodeoxynucleotides: Kinetic and Computational Studies Highlight the Mechanism for 5′,8-Cyclopurine Formation. J. Am. Chem. Soc. 2020, 142, 5825–5833. [Google Scholar] [CrossRef]
- Huang, H.; Das, R.S.; Basu, A.K.; Stone, M.P. Structures of (5′S)-8,5′-Cyclo-2′-deoxyguanosine Mismatched with dA or dT. Chem. Res. Toxicol. 2012, 25, 478–490. [Google Scholar] [CrossRef]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of oxygen free-radical-induced 5’,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Dirksen, M.-L.; Blakely, W.F.; Holwitt, E.; Dizdaroglu, M. Effect of DNA Conformation on the Hydroxyl Radical-induced Formation of 8,5′-cyclopurine 2′-deoxyribonucleoside Residues in DNA. Int. J. Radiat. Biol. 1988, 54, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Kropachev, K.; Ding, S.; Terzidis, M.A.; Masi, A.; Liu, Z.; Cai, Y.; Kolbanovskiy, M.; Chatgilialoglu, C.; Broyde, S.; Geacintov, N.E.; et al. Structural basis for the recognition of diastereomeric 5′,8-cyclo-2′-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014, 42, 5020–5032. [Google Scholar] [CrossRef]
- Izumi, T.; Mellon, I. Base Excision Repair and Nucleotide Excision Repair. Genome Stability: From Virus to Human Application; Elsevier Inc.: London, UK, 2016. [Google Scholar]
- Gruber, C.C.; Walker, G.C. Incomplete base excision repair contributes to cell death from antibiotics and other stresses. DNA Repair 2018, 71, 108–117. [Google Scholar] [CrossRef]
- Carter, R.J.; Parsons, J.L. Base Excision Repair, a Pathway Regulated by Posttranslational Modifications. Mol. Cell. Biol. 2016, 36, 1426–1437. [Google Scholar] [CrossRef]
- Prakash, A.; Doublie, S. Base Excision Repair in the Mitochondria. J. Cell. Biochem. 2015, 116, 1490–1499. [Google Scholar] [CrossRef]
- Schormann, N.; Ricciardi, R.; Chattopadhyay, D. Uracil-DNA glycosylases-Structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014, 23, 1667–1685. [Google Scholar] [CrossRef] [PubMed]
- Zharkov, D.O. Base excision DNA repair. Cell. Mol. Life Sci. 2008, 65, 1544–1565. [Google Scholar] [CrossRef]
- Abbotts, R.; Madhusudan, S. Human AP endonuclease 1 (APE1): From mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010, 36, 425–435. [Google Scholar] [CrossRef]
- Maher, R.L.; Bloom, L.B. Pre-steady-state Kinetic Characterization of the AP Endonuclease Activity of Human AP Endonuclease 1. J. Biol. Chem. 2007, 282, 30577–30585. [Google Scholar] [CrossRef]
- Jaruga, P.; Rozalski, R.; Jawien, A.; Migdalski, A.; Olinski, R.; Dizdaroglu, M. DNA Damage Products (5′R)- and (5′S)-8,5′-Cyclo-2′- deoxyadenosines as Potential Biomarkers in Human Urine for Atherosclerosis. Biochemistry 2012, 51, 1822–1824. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.M.; Jaruga, P.; Ramsey, C.R.; Gilman, N.K.; Green, V.M.; Rostad, S.W.; Emerman, J.T.; Dizdaroglu, M.; Malins, D.C. Structural Alterations in Breast Stromal and Epithelial DNA: The Influence of 8,5-cyclo-2-Deoxyadenosine. Cell Cycle 2006, 5, 1240–1244. [Google Scholar] [CrossRef]
- Masi, A.; Fortini, P.; Krokidis, M.G.; Romeo, E.F.; Bascietto, C.; De Angelis, P.; Guglielmi, V.; Chatgilialoglu, C. Increased levels of 5′,8-Cyclopurine DNA lesions in inflammatory bowel diseases. Redox Biol. 2020, 34, 101562. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Geacintov, N.E.; Krokidis, M.G.; Liu, Y.; Masi, A.; Shafirovich, V.; Terzidis, M.A.; Tsegay, P.S. 5′,8-Cyclopurine Lesions in DNA Damage: Chemical, Analytical, Biological, and Diagnostic Significance. Cells 2019, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Robins, P.; Masutani, C.; Hanaoka, F.; Gasparutto, D.; Cadet, J.; Wood, R.D.; Lindahl, T. Oxygen free radical damage to DNA: Translesion synthesis by human DNA polymerase η and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J. Biol. Chem. 2001, 276, 49283–49288. [Google Scholar] [CrossRef]
- You, C.; Swanson, A.L.; Dai, X.; Yuan, B.; Wang, J.; Wang, Y. Translesion synthesis of 8,5’-cyclopurine-2’-deoxynucleosides by DNA polymerasesη, ℓ, and ζ. J. Biol. Chem. 2013, 288, 28548–28556. [Google Scholar] [CrossRef]
- Brooks, P.J.; Wise, D.S.; Berry, D.A.; Kosmoski, J.V.; Smerdon, M.J.; Somers, R.L.; Mackie, H.; Spoonde, A.Y.; Ackerman, E.J.; Coleman, K.; et al. The Oxidative DNA Lesion 8,5′-(S)-Cyclo-2′-deoxyadenosine Is Repaired by the Nucleotide Excision Repair Pathway and Blocks Gene Expression in Mammalian Cells. J. Biol. Chem. 2000, 275, 22355–22362. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T. The Influence of (5’R)- and (5’S)-5’,8-Cyclo-2’-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare. Cells 2019, 8, 1303. [Google Scholar] [CrossRef]
- Pande, P.; Das, R.S.; Sheppard, C.; Kow, Y.W.; Basu, A.K. Repair efficiency of (5’S)-8,5’-cyclo-2’-deoxyguanosine and (5’S)-8,5’-cyclo-2’-deoxyadenosine depends on the complementary base. DNA Repair. 2012, 11, 926–931. [Google Scholar] [CrossRef][Green Version]
- Boguszewska, K.; Szewczuk, M.; Kaźmierczak-Barańska, J.; Karwowski, B. How (5′S) and (5′R) 5′,8-Cyclo-2′-Deoxypurines Affect Base Excision Repair of Clustered DNA Damage in Nuclear Extracts of xrs5 Cells? A Biochemical Study. Cells 2021, 10, 725. [Google Scholar] [CrossRef] [PubMed]
- Romieu, A.; Gasparutto, D.; Cadet, J. Synthesis and characterization of oligonucleotides containing 5’,8-cyclopurine 2’-deoxyribonucleosides: (5’R)-5’,8-cyclo-2’-deoxyadenosine, (5’S)-5’,8-cyclo-2’-deoxyguanosine, and (5’R)-5’,8-cyclo-2’-deoxyguanosine. Chem. Res. Toxicol. 1999, 12, 412–421. [Google Scholar] [CrossRef]
- Szewczuk, M.; Boguszewska, K.; Kaźmierczak-Barańska, J.; Karwowski, B. The Influence of 5′R and 5′S cdA and cdG on the Activity of BsmAI and SspI Restriction Enzymes. Molecules 2021, 26, 3750. [Google Scholar] [CrossRef] [PubMed]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Sage, E.; Harrison, L. Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat. Res. Mol. Mech. Mutagen. 2011, 711, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P. The case for 8,5′-cyclopurine-2′-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience 2007, 145, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T.; Bellon, S.; O’Neill, P.; Lomax, M.E.; Cadet, J. Effects of (5′S)-5′,8-cyclo-2′-deoxyadenosine on the base excision repair of oxidatively generated clustered DNA damage. A biochemical and theoretical study. Org. Biomol. Chem. 2014, 12, 8671–8682. [Google Scholar] [CrossRef]



| End | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Matrix SA | 3′ | G | A | G | A | A | C | A | G | T | C | C | T | T | A | T | A | A | C | A | G | A | G | A | T | A | C | G | A | G | G | G | T | G | G | T | T | T | C | C | G |
| Matrix SA-A | 3′ | G | A | G | A | A | C | A | G | T | C | C | T | T | A | T | A | A | C | A | G | A | G | A | T | A | C | G | A | A | G | G | T | G | G | T | T | T | C | C | G |
| Matrix SG | 3′ | G | A | G | A | A | C | A | G | T | C | C | T | T | A | T | A | A | C | A | G | A | G | A | C | A | C | G | A | G | G | G | T | G | G | T | T | T | C | C | G |
| Matrix SG-A | 3′ | G | A | G | A | A | C | A | G | T | C | C | T | T | A | T | A | A | C | A | G | A | G | A | C | A | C | G | A | A | G | G | T | G | G | T | T | T | C | C | G |
| dU0 | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | U | C | T | A | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)(+5)dA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | A | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)ScdA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | SX | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C |
| dU(+5)ScdA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | SX | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)(+5)ScdA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | SX | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)RcdA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | RX | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C |
| dU(+5)RcdA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | RX | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)(+5)RcdA | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | RX | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)ScdG | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | SY | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C |
| dU(+5)ScdG | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | SY | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)(+5)ScdG | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | SY | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)RcdG | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | RY | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C |
| dU(+5)RcdG | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | RY | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| dU(−5)(+5)RcdG | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | RY | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C |
| Oligonucleotide | Quantity [OD] | Quantity [nmol] |
|---|---|---|
| Matrix SA | 27.2 | 62.56 |
| Matrix SA-A | 12.6 | 28.98 |
| Matrix SG | 30.0 | 69.00 |
| Matrix SG-A | 13.6 | 31.28 |
| dU0 | 26.6 | 61.18 |
| dU(−5)(+5)dA | 28.2 | 64.86 |
| dU(−5)ScdA | 24.5 | 56.35 |
| dU(+5)ScdA | 22.6 | 51.98 |
| dU(−5)(+5)ScdA | 22.5 | 51.75 |
| dU(−5)RcdA | 22.0 | 50.60 |
| dU(+5)RcdA | 25.0 | 57.50 |
| dU(−5)(+5)RcdA | 26.2 | 60.26 |
| dU(−5)ScdG | 18.6 | 42.78 |
| dU(+5)ScdG | 11.0 | 25.30 |
| dU(−5)(+5)ScdG | 9.6 | 22.08 |
| dU(−5)RcdG | 2.0 | 4.60 |
| dU(+5)RcdG | 1.3 | 2.99 |
| dU(−5)(+5)RcdG | 2.2 | 5.06 |
| Oligonucleotide | Calculated Mass | Found Mass |
|---|---|---|
| Matrix SA | 12,409.00 | 12,409.82 |
| dU0 | 12,158.83 | 12,168.25 |
| dU(−5)(+5)dA | 12,168.85 | 12,168.05 |
| dU(−5)ScdA | 12,166.84 | 12,166.50 |
| dU(+5)ScdA | 12,181.85 | 12,181.54 |
| dU(−5)(+5)ScdA | 12,167.85 | 12,167.20 |
| dU(−5)RcdA | 12,166.84 | 12,166.20 |
| dU(+5)RcdA | 12,181.85 | 12,181.10 |
| dU(−5)(+5)RcdA | 12,167.85 | 12,167.00 |
| dU(−5)ScdG | 12,182.84 | 12,182.10 |
| dU(+5)ScdG | 12,197.85 | 12,197.00 |
| dU(−5)(+5)ScdG | 12,183.85 | 12,182.90 |
| dU(−5)RcdG | 12,182.84 | 12,182.64 |
| dU(+5)RcdG | 12,197.85 | 12,195.75 |
| dU(−5)(+5)RcdG | 12,183.85 | 12,183.90 |
| cdPus | UDG Activity | hAPE1 Activity | ||||
|---|---|---|---|---|---|---|
| ScdA | - | + | ND | + | + | + |
| RcdA | + | + | - | + | + | - |
| ScdG | + | + | ND | + | + | + |
| RcdG | ND | + | + | + | + | ND |
| −5 | +5 | −5/+5 | −5 | +5 | −5/+5 | |
| The relative position of dU towards cdPus within CDL | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szewczuk, M.; Boguszewska, K.; Kaźmierczak-Barańska, J.; Karwowski, B.T. When UDG and hAPE1 Meet Cyclopurines. How (5′R) and (5′S) 5′,8-Cyclo-2′-deoxyadenosine and 5′,8-Cyclo-2′-deoxyguanosine Affect UDG and hAPE1 Activity? Molecules 2021, 26, 5177. https://doi.org/10.3390/molecules26175177
Szewczuk M, Boguszewska K, Kaźmierczak-Barańska J, Karwowski BT. When UDG and hAPE1 Meet Cyclopurines. How (5′R) and (5′S) 5′,8-Cyclo-2′-deoxyadenosine and 5′,8-Cyclo-2′-deoxyguanosine Affect UDG and hAPE1 Activity? Molecules. 2021; 26(17):5177. https://doi.org/10.3390/molecules26175177
Chicago/Turabian StyleSzewczuk, Michał, Karolina Boguszewska, Julia Kaźmierczak-Barańska, and Bolesław T. Karwowski. 2021. "When UDG and hAPE1 Meet Cyclopurines. How (5′R) and (5′S) 5′,8-Cyclo-2′-deoxyadenosine and 5′,8-Cyclo-2′-deoxyguanosine Affect UDG and hAPE1 Activity?" Molecules 26, no. 17: 5177. https://doi.org/10.3390/molecules26175177
APA StyleSzewczuk, M., Boguszewska, K., Kaźmierczak-Barańska, J., & Karwowski, B. T. (2021). When UDG and hAPE1 Meet Cyclopurines. How (5′R) and (5′S) 5′,8-Cyclo-2′-deoxyadenosine and 5′,8-Cyclo-2′-deoxyguanosine Affect UDG and hAPE1 Activity? Molecules, 26(17), 5177. https://doi.org/10.3390/molecules26175177








