Engineering of Yeast Old Yellow Enzyme OYE3 Enables Its Capability Discriminating of (E)-Citral and (Z)-Citral
Abstract
:1. Introduction
2. Results and Discussion
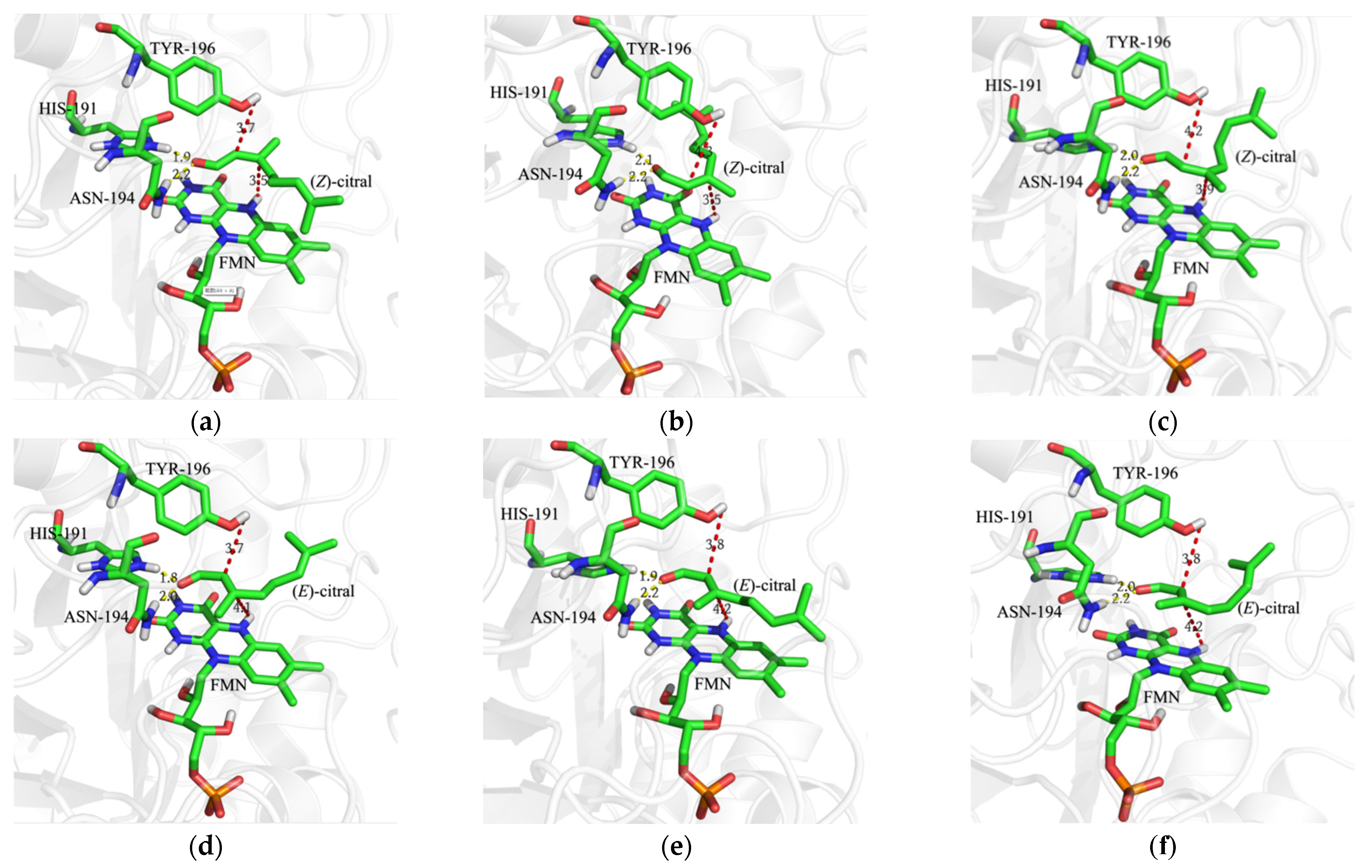
2.1. Identification of Key Residues for the Enantioselectivity of OYE3
2.2. Exploration of Double Substitution at the Sites W116 and S296 of OYE3
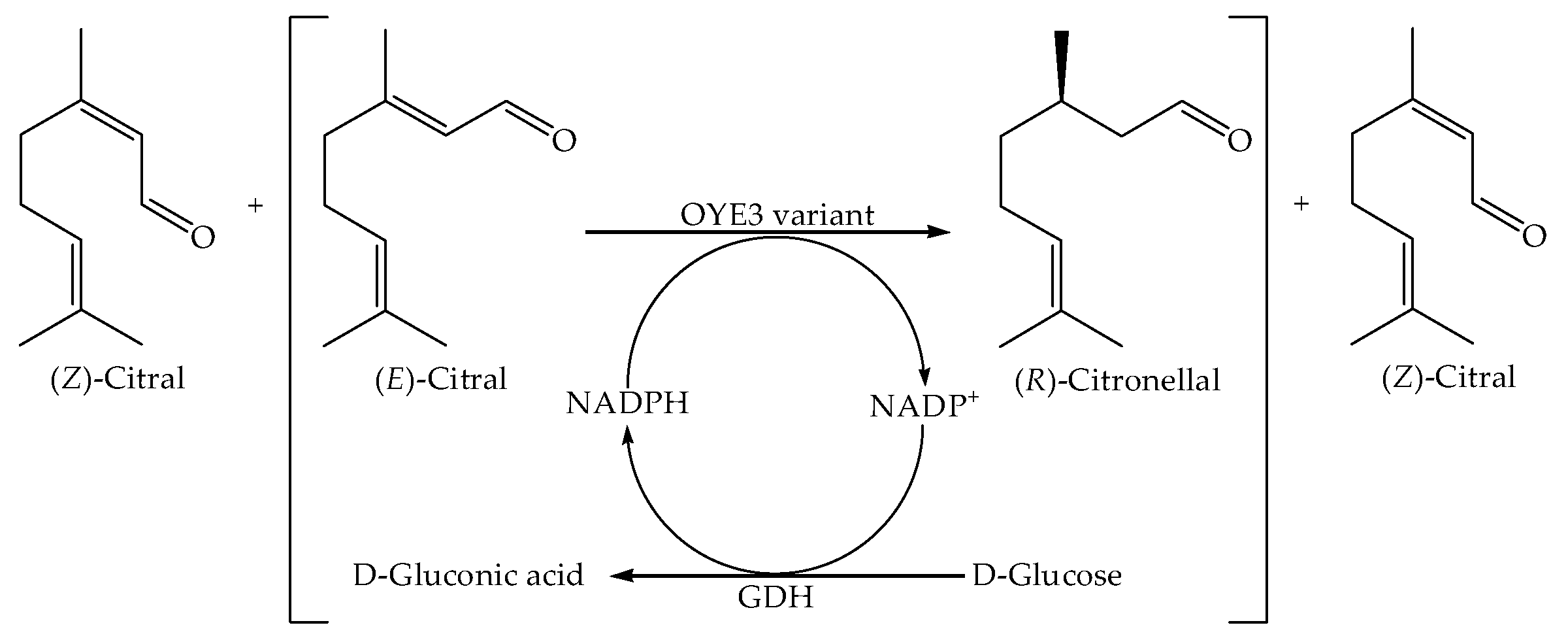
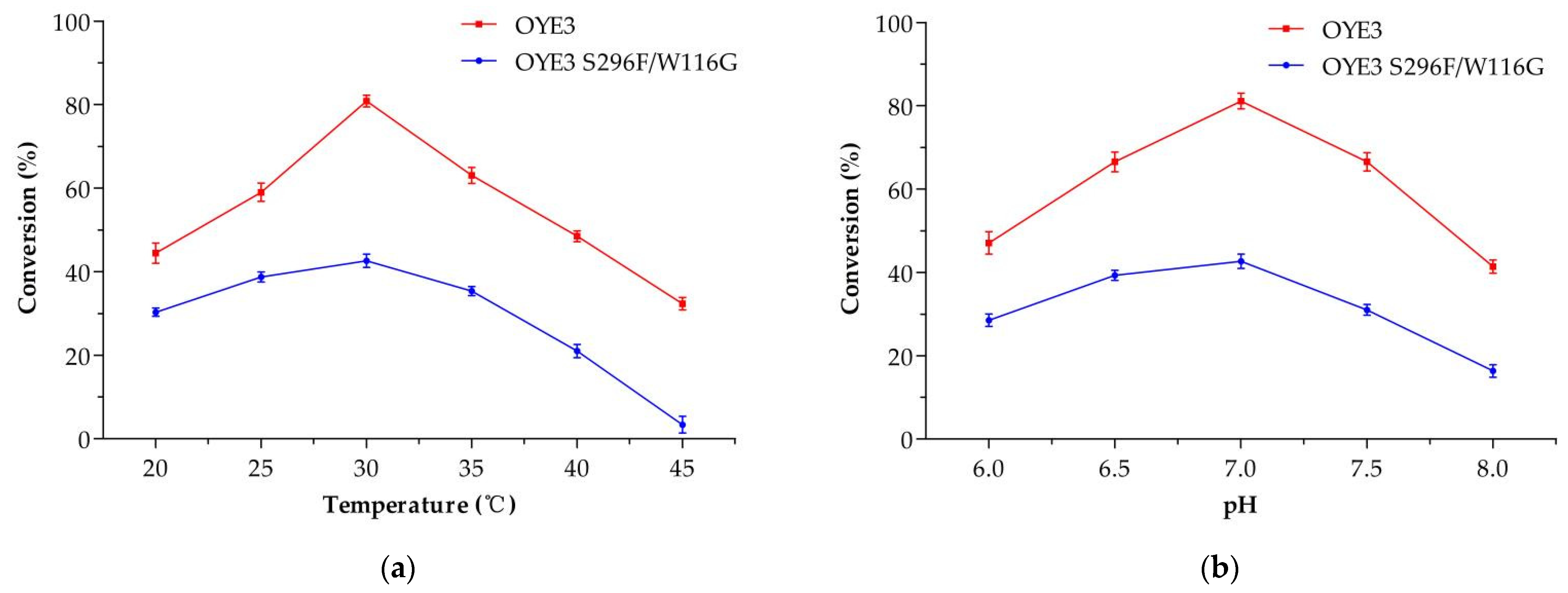
2.3. Asymmetric Synthesis of (R)-Citronellal and Simultaneous Resolution of (E)-Citral and (Z)-Citral Catalyzed by the OYE3 Variant S296F/W116G and Glucose Dehydrogenase
3. Materials and Methods
3.1. Genes, Organisms, and Chemicals
3.2. Over-Expression and Purification of OYE3 and GDH
3.3. Construction of OYE3 Variants by Site-Directed Mutagenesis
3.4. Assays of Glucose Dehydrogenase and Old Yellow Enzymes
3.5. Homology Modeling and Molecular Docking
3.6. Investigation of Catalytic Performance of OYE3 and Its Variants in the Reduction of (E)-, (Z)-, or (E/Z)-Citral
3.7. Identification of Key Factors for S296F/W116G Mediated-Reduction of (E)-Citral
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Lenardão, E.J.; Botteselle, G.V.; de Azambuja, F.; Perin, G.; Jacob, R.G. Citronellal as key compound in organic synthesis. Tetrahedron 2007, 63, 6671–6712. [Google Scholar] [CrossRef]
- Siedenburg, G.; Jendrossek, D.; Breuer, M.; Juhls, B.; Pleiss, J.; Seitz, M.; Klebensberger, J.; Hauer, B. Activation-independent cyclization of monoterpenoids. Appl. Environ. Microbiol. 2012, 78, 1055–1062. [Google Scholar] [CrossRef] [Green Version]
- Siedenburg, G.; Breuer, M.; Jendrossek, D. Prokaryotic squalene-hopene cyclases can be converted to citronellal cyclases by single amino acid exchange. Appl. Microbiol. Biotechnol. 2013, 97, 1571–1580. [Google Scholar] [CrossRef]
- Itoh, H.; Maeda, H.; Yamada, S.; Hori, Y.; Mino, T.; Sakamoto, M. Kinetic resolution of citronellal by chiral aluminum catalysts: L-menthol synthesis from citral. Org. Chem. Front. 2014, 1, 1107–1115. [Google Scholar] [CrossRef]
- Tani, K.; Yamagata, T.; Akutagawa, S.; Kumobayashi, H.; Taketomi, T.; Takaya, H.; Miyashita, A.; Noyori, R.; Otsuka, S. Highly enantioselective isomerization of prochiral allylamines catalyzed by chiral diphosphine Rhodium(I) complexes: Preparation of optically active enamines. J. Am. Chem. Soc. 1984, 106, 5208–5217. [Google Scholar] [CrossRef]
- Sayo, N.; Matsumoto, T. Method for Producing L-Menthol. U.S. Patent US6342644, 29 January 2002. [Google Scholar]
- Nowak, R.; Michler, M.; Reiss, I.; Winkel, R. Menthol-Containing Solids Composition. U.S. Patent US8524257, 3 September 2013. [Google Scholar]
- Zheng, L.; Lin, J.; Zhang, B.; Kuang, Y.; Wei, D. Identification of a yeast old yellow enzyme for highly enantioselective reduction of citral isomers to (R)-citronellal. Bioresour. Bioprocess. 2018, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- de María, P.D.; de Gonzalo, G.; Alcántara, A.R. Biocatalysis as useful tool in asymmetric synthesis: An assessment of recently granted patents (2014–2019). Catalysts 2019, 9, 802. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Yu, S.; Huang, M.; Wei, R.; Meng, S.; Cheng, F.; Yu, M.; Ying, M.; Zhao, M.; Wang, Z. Engineering the enantioselectivity of yeast old yellow enzyme OYE2y in asymmetric reduction of (E/Z)-citral to (R)-citronellal. Molecules 2019, 24, 1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholtissek, A.; Tischler, D.; Westphal, A.H.; Van Berkel, W.J.H.; Paul, C.E. Old yellow enzyme-catalysed asymmetric hydrogenation: Linking family roots with improved catalysis. Catalysts 2017, 7, 130. [Google Scholar] [CrossRef]
- Tentori, F.; Bavaro, T.; Brenna, E.; Colombo, D.; Monti, D.; Semproli, R.; Ubiali, D. Immobilization of Old Yellow Enzymes via covalent or coordination bonds. Catalysts 2020, 10, 260. [Google Scholar] [CrossRef] [Green Version]
- Wolken, W.A.M.; ten Have, R.; van der Werf, M.J. Amino acid-catalyzed conversion of citral: Cis-trans isomerization and its conversion into 6-methyl-5-hepten-2-one and acetaldehyde. J. Agric. Food Chem. 2000, 48, 5401–5405. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Hauer, B.; Rosche, B. Asymmetric alkene reduction by yeast old yellow enzymes and by a novel Zymomonas mobilis reductase. Biotechnol. Bioeng. 2007, 98, 22–29. [Google Scholar] [CrossRef]
- Amato, E.D.; Stewart, J.D. Applications of protein engineering to members of the old yellow enzyme family. Biotechnol. Adv. 2015, 33, 624–631. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, M.; Miyakawa, T.; Shimizu, S.; Tanokura, M. Enzymes useful for chiral compound synthesis: Structural biology, directed evolution, and protein engineering for industrial use. Appl. Microbiol. Biotechnol. 2016, 100, 5747–5757. [Google Scholar] [CrossRef]
- Toogood, H.S.; Scrutton, N.S. Discovery, characterization, engineering, and applications of ene-reductases for industrial biocatalysis. ACS Catal. 2018, 8, 3532–3549. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Wang, H.; Liu, J.; Li, S.; Guo, J.; Li, H.; Jia, X.; Huo, H.; Zheng, Z.; You, S.; et al. Old yellow enzymes: Structures and structure-guided engineering for stereocomplementary bioreduction. Appl. Microbiol. Biotechnol. 2020, 104, 8155–8170. [Google Scholar] [CrossRef] [PubMed]
- Padhi, S.K.; Bougioukou, D.J.; Stewart, J.D. Site-saturation mutagenesis of tryptophan 116 of Saccharomyces pastorianus old yellow enzyme uncovers stereocomplementary variants. J. Am. Chem. Soc. 2009, 131, 3271–3280. [Google Scholar] [CrossRef] [PubMed]
- Brenna, E.; Crotti, M.; Gatti, F.G.; Monti, D.; Parmeggiani, F.; Powell, R.W., III; Santangelo, S.; Stewart, J.D. Opposite enantioselectivity in the bioreduction of (Z)-β-aryl-bcyanoacrylates mediated by the tryptophan 116 mutants of old yellow enzyme 1: Synthetic approach to (R)- and (S)-β-aryl-glactams. Adv. Synth. Catal. 2015, 357, 1849–1860. [Google Scholar] [CrossRef]
- Crotti, M.; Parmeggiani, F.; Ferrandi, E.E.; Gatti, F.G.; Sacchetti, A.; Riva, S.; Brenna, E.; Monti, D. Stereoselectivity switch in the reduction of α-alkyl-β-arylenones by structure-guided designed variants of the ene reductase OYE1. Front. Bioeng. Biotechnol. 2019, 7, 89. [Google Scholar] [CrossRef]
- Walton, A.Z.; Sullivan, B.; Patterson-Orazem, A.C.; Stewart, J.D. Residues controlling facial selectivity in an alkene reductase and semirational alterations to create stereocomplementary variants. ACS Catal. 2015, 4, 2307–2318. [Google Scholar] [CrossRef] [Green Version]
- Rüthlein, E.; Classen, T.; Dobnikar, L.; Schölzel, M.; Pietruszka, J. Finding the selectivity switch—A rational approach towards stereocomplementary variants of the ene reductase YqjM. Adv. Synth. Catal. 2015, 357, 1775–1786. [Google Scholar] [CrossRef]
- Yin, B.; Deng, J.; Lim, L.; Yuan, Y.A.; Wei, D. Structural insights into stereospecific reduction of α, β-unsaturated carbonyl substrates by old yellow enzyme from Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 2015, 79, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Kress, N.; Rapp, J.; Hauer, B. Enantioselective reduction of citral isomers in NCR ene reductase: Analysis of an active site mutant library. ChemBioChem 2017, 18, 717–720. [Google Scholar] [CrossRef]
- Bougioukou, D.J.; Walton, A.Z.; Stewart, J.D. Towards preparative-scale, biocatalytic alkene reductions. Chem. Commun. 2010, 46, 8558–8560. [Google Scholar] [CrossRef]
- Müller, A.; Hauer, B.; Rosche, B. Enzymatic reduction of the α,β-unsaturated carbon bond in citral. J. Mol. Catal. B Enzym. 2006, 38, 126–130. [Google Scholar] [CrossRef]
- Hall, M.; Stueckler, C.; Hauer, B.; Stuermer, R.; Friedrich, T.; Breuer, M.; Kroutil, W.; Faber, K. Asymmetric bioreduction of activated C=C bonds using Zymomonas mobilis NCR enoate reductase and old yellow enzymes OYE 1-3 from yeasts. Eur. J. Org. Chem. 2008, 2008, 1511–1516. [Google Scholar] [CrossRef]
- Peters, C.; Frasson, D.; Sievers, M.; Buller, R. Novel old yellow enzyme subclasses. ChemBioChem 2019, 20, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Pompeu, Y.A.; Sullivan, B.; Stewart, J.D. X-ray crystallography reveals how subtle changes control the orientation of substrate binding in an alkene reductase. ACS Catal. 2013, 3, 2376–2390. [Google Scholar] [CrossRef]
- Haridas, M.; Abdelraheem, E.M.M.; Hanefeld, U. 2-Deoxy-D-ribose-5-phosphate aldolase (DERA): Applications and modifications. Appl. Microbial. Biotechnol. 2018, 102, 9959–9971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Gao, L.; Zhang, L.; Bai, Y.; Chen, L.; Yu, M.; Cheng, F.; Sun, J.; Wang, Z.; Ying, X. Asymmetric reduction of ketopantolactone using a strictly (R)-stereoselective carbonyl reductase through efficient NADPH regeneration and the substrate constant-feeding strategy. Biotechnol. Lett. 2017, 39, 1741–1746. [Google Scholar] [CrossRef]
- Ying, X.; Wang, C.; Shao, S.; Wang, Q.; Zhou, X.; Bai, Y.; Chen, L.; Lu, C.; Zhao, M.; Wang, Z. Efficient Oxidation of Methyl Glycolate to Methyl Glyoxylate Using a Fusion Enzyme of Glycolate Oxidase, Catalase and Hemoglobin. Catalysts 2020, 10, 943. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, C.; Zeng, Y.; Wang, T.; Qiao, J.; Lu, C.; Wang, Z.; Ying, X. Efficient whole-cell oxidation of α,β-unsaturated alcohols to α,β-unsaturated aldehydes through the cascade biocatalysis of alcohol dehydrogenase, NADPH oxidase and hemoglobin. Microb. Cell Fact. 2021, 20, 17. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, Q.; Qiao, J.; Feng, B.; Zhou, X.; Jin, L.; Feng, Y.; Yang, D.; Lu, C.; Ying, X. Cascading Old Yellow Enzyme, Alcohol Dehydrogenase and Glucose Dehydrogenase for Selective Reduction of (E/Z)-Citral to (S)-Citronellol. Catalysts 2021, 11, 931. [Google Scholar] [CrossRef]
- Tsuboi, S.; Ishii, N.; Sakai, T.; Tari, I.; Utaka, M. Oxidation of alcohols with electrolytic manganese dioxide. Its application for the synthesis of insect pheromones. Bull. Chem. Soc. Jpn. 1990, 63, 1888–1893. [Google Scholar] [CrossRef]
- Shapiro, A.L.; Viñuela, E.; Maizel, J.V. Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 1967, 28, 815–820. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.M.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An antomated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Conversion (%) | e.e. (%) | Conversion (%) | e.e. (%) | Conversion (%) | |
| OYE3 | 63 ± 1.1 (R) | 91.9 ± 3.0 | 47 ± 1.2 (S) | 88.8 ± 2.1 | 23 ± 1.0 (R) | 86.6 ± 1.7 |
| W116A | >99 (R) | 31.9 ± 1.2 | 43 ± 1.0 (R) | 7.91 ± 0.1 | >99 (R) | 9.8 ± 0.2 |
| S296F | >99 (R) | 71.1 ± 2.0 | 52 ± 0.9 (R) | 80.1 ± 1.1 | 72 ± 1.5 (R) | 80.3 ± 1.8 |
| H329R | 56 ± 0.9 (R) | 87.9 ± 2.1 | 32 ± 0.7 (S) | 86.5 ± 2.3 | 48 ± 0.9 (R) | 60.4 ± 1.3 |
| R38H | 55 ± 0.8 (R) | 89.3 ± 2.4 | 15 ± 0.6 (S) | 79.4 ± 1.8 | 38 ± 1.2 (R) | 69.2 ± 1.7 |
| M67L | 63 ± 1.3 (R) | 91.5 ± 2.8 | 37 ± 1.1 (S) | 89.0 ± 2.3 | 24 ± 0.8 (R) | 88.8 ± 2.4 |
| A79S | 43 ± 1.0 (R) | 88.5 ± 1.9 | 49 ± 1.2 (S) | 85.7 ± 2.5 | 22 ± 1.3 (R) | 86.6 ± 2.3 |
| V288A | 70 ± 1.9 (R) | 86.2 ± 1.8 | 38 ± 1.3 (S) | 84.9 ± 2.5 | 21 ± 1.2 (R) | 85.6 ± 1.6 |
| R12L | 63 ± 1.2 (R) | 94.1 ± 2.1 | 45 ± 1.4 (S) | 89.4 ± 1.8 | 21 ± 0.8 (R) | 92.8 ± 1.9 |
| I344K | 61 ± 1.5 (R) | 91.7 ± 1.7 | 44 ± 0.8 (S) | 94.0 ± 1.3 | 20 ± 0.7 (R) | 88.6 ± 1.4 |
| I75C | 62 ± 2.1 (R) | 91.8 ± 1.6 | 57 ± 1.6 (S) | 85.9 ± 2.0 | 14 ± 1.0 (R) | 92.9 ± 2.4 |
| P77C | 62 ± 1.1 (R) | 90.3 ± 2.4 | 77 ± 1.7 (S) | 96.7 ± 1.3 | 10 ± 0.6 (R) | 91.8 ± 2.6 |
| V113A | 60 ± 1.6 (R) | 97.2 ± 1.1 | 28 ± 0.7 (S) | 78.8 ± 1.5 | 10 ± 0.8 (R) | 89.3 ± 1.0 |
| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Conversion (%) | e.e. (%) | Conversion (%) | e.e. (%) | Conversion (%) | |
| OYE3 | 63 ± 1.1 (R) | 91.9 ± 3.0 | 47 ± 1.2 (S) | 88.8 ± 2.1 | 23 ± 1.0 (R) | 86.8 ± 1.7 |
| S296F | >99 (R) | 71.1 ± 1.2 | 52 ± 0.8 (R) | 80.1 ± 2.7 | 72 ± 1.8 (R) | 80.3 ± 1.9 |
| S296W | 56 ± 1.0 (R) | 81.6 ± 1.7 | 1.2 ± 0.5 (R) | 82.6 ± 1.9 | 32 ± 2.3 (R) | 75.6 ± 0.8 |
| S296Y | 58 ± 1.3 (R) | 75.1 ± 1.3 | 41 ± 1.4 (S) | 82.8 ± 1.4 | 22 ± 0.9 (R) | 88.5 ± 1.4 |
| S296A | 54 ± 0.8 (R) | 94.3 ± 2.0 | 49 ± 1.7 (S) | 88.5 ± 1.2 | 15 ± 1.4 (R) | 89.5 ± 2.3 |
| S296G | 51 ± 0.7 (R) | 95.6 ± 2.3 | 44 ± 1.1 (S) | 71.6 ± 0.9 | 12 ± 1.5 (R) | 91.1 ± 2.0 |
| W116A | >99 (R) | 4.1 ± 0.1 | >99 (R) | 2.9 ± 0.1 | >99 (R) | 8.4 ± 0.1 |
| W116V | 76 ± 1.3 (R) | 22.5 ± 0.4 | 57 ± 2.0 (R) | 7.1 ± 0.2 | >99 (R) | 1.4 ± 0.1 |
| W116I | 47 ± 1.2 (R) | 29.0 ± 0.7 | 21 ± 1.2 (S) | 15.6 ± 0.7 | 44 ± 1.8 (R) | 24.3 ± 0.5 |
| W116F | 47 ± 1.2 (R) | 88.0 ± 2.2 | 67 ± 0.9 (S) | 14.7 ± 0.4 | 1.7 ± 0.8 (R) | 67.5 ± 1.4 |
| W116Y | 31 ± 1.0 (R) | 80.8 ± 1.7 | 71 ± 1.5 (S) | 65.1 ± 1.9 | 5.2 ± 1.0 (S) | 57.1 ± 1.6 |
| W116S | >99 (R) | 14.7 ± 0.3 | / | / | >99 (R) | 9.0 ± 0.2 |
| W116G | >99 (R) | 11.1 ± 0.2 | / | / | / | / |
| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Conversion (%) | e.e. (%) | Conversion (%) | e.e. (%) | Conversion (%) | |
| S296F/W116V | >99 (R) | 11.8 ± 0.2 | / | / | / | / |
| S296F/W116S | >99 (R) | 29.3 ± 1.0 | / | / | / | / |
| S296F/W116A | >99 (R) | 8.0 ± 0.1 | / | / | / | / |
| S296F/W116G | >99 (R) | 31.8 ± 0.9 | / | / | >99 (R) | 25.3 ± 0.5 |
| Parameter | OYE3 | OYE3 S296F/W116G | ||
|---|---|---|---|---|
| (E)-Citral | (Z)-Citral | (E)-Citral | (Z)-Citral | |
| Km (mM) | 0.05 ± 0.01 | 0.07 ± 0.01 | 0.09 ± 0.01 | / |
| Ki (mM) | 4.53 ± 0.28 | 5.75 ± 0.37 | 5.85 ± 0.33 | / |
| Vmax (U/mg) | 0.16 ± 0.03 | 0.12 ± 0.02 | 0.14 ± 0.02 | / |
| Kcat (s−1) | 0.13 ± 0.02 | 0.10 ± 0.01 | 0.11 ± 0.01 | / |
| Kcat/Km (mM−1s−1) | 2.76 ± 0.11 | 1.49 ± 0.08 | 1.32 ± 0.09 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Wei, R.; Feng, Y.; Jin, L.; Jia, Y.; Yang, D.; Liang, Z.; Han, M.; Li, X.; Lu, C.; et al. Engineering of Yeast Old Yellow Enzyme OYE3 Enables Its Capability Discriminating of (E)-Citral and (Z)-Citral. Molecules 2021, 26, 5040. https://doi.org/10.3390/molecules26165040
Wang T, Wei R, Feng Y, Jin L, Jia Y, Yang D, Liang Z, Han M, Li X, Lu C, et al. Engineering of Yeast Old Yellow Enzyme OYE3 Enables Its Capability Discriminating of (E)-Citral and (Z)-Citral. Molecules. 2021; 26(16):5040. https://doi.org/10.3390/molecules26165040
Chicago/Turabian StyleWang, Tairan, Ran Wei, Yingting Feng, Lijun Jin, Yunpeng Jia, Duxia Yang, Zuonan Liang, Mengge Han, Xia Li, Chenze Lu, and et al. 2021. "Engineering of Yeast Old Yellow Enzyme OYE3 Enables Its Capability Discriminating of (E)-Citral and (Z)-Citral" Molecules 26, no. 16: 5040. https://doi.org/10.3390/molecules26165040
APA StyleWang, T., Wei, R., Feng, Y., Jin, L., Jia, Y., Yang, D., Liang, Z., Han, M., Li, X., Lu, C., & Ying, X. (2021). Engineering of Yeast Old Yellow Enzyme OYE3 Enables Its Capability Discriminating of (E)-Citral and (Z)-Citral. Molecules, 26(16), 5040. https://doi.org/10.3390/molecules26165040






