Proteomic Advances in Cereal and Vegetable Crops
Abstract
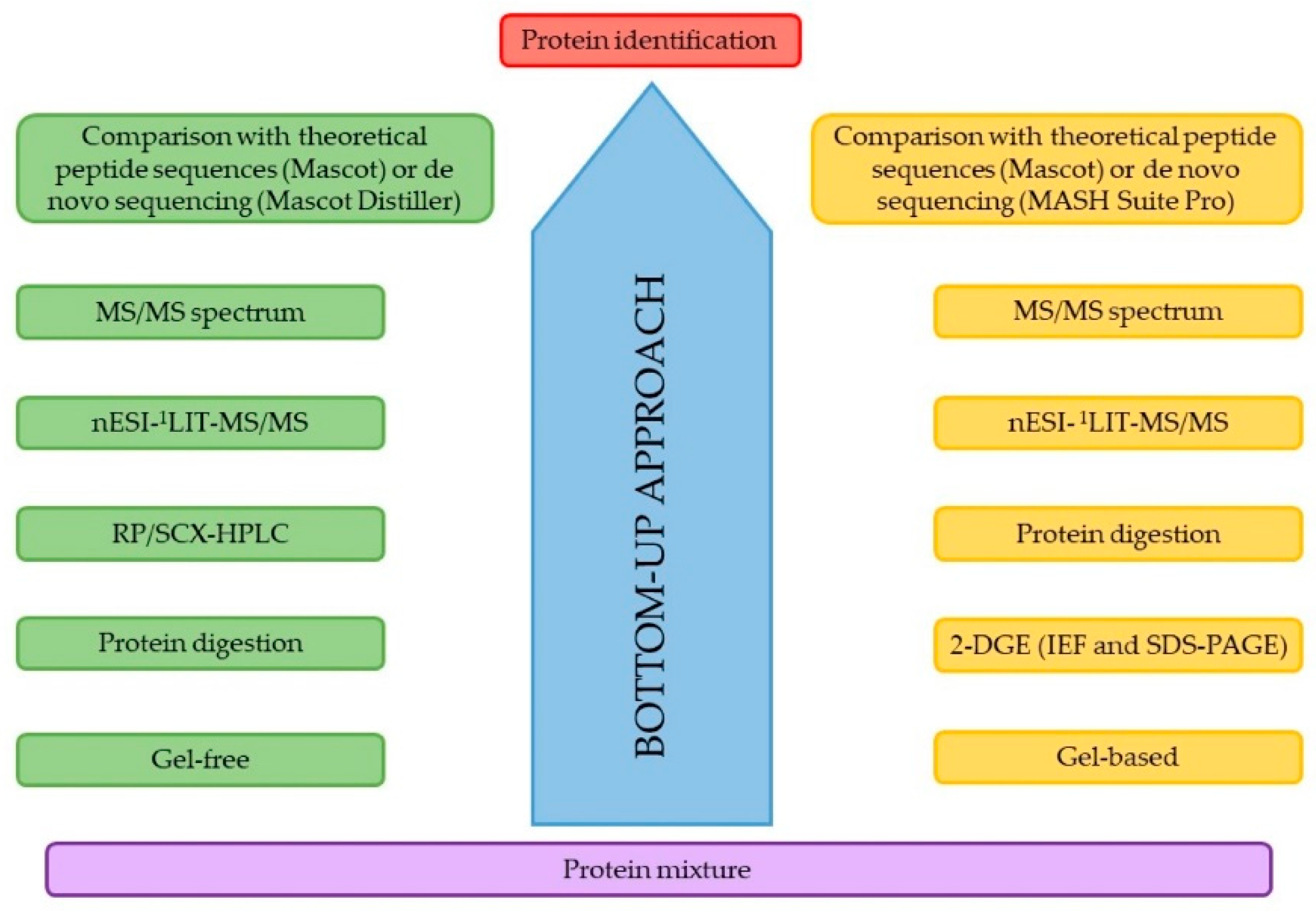
:1. Introduction
2. Role of Vegetable Proteins in Agriculture and Food
| AA (mg/g of Grain) | a Wheat | b Rice | c Oat | d Sorghum | e Barley | f Adult AA Requirements | |
|---|---|---|---|---|---|---|---|
| EAA | AA | Quantity (mg/kg per day) | |||||
| Histidine | 2.38 | 1.33 | 3.98 | 2.30 | 2.12 | Histidine | 10 |
| Isoleucine | 8.22 | 1.90 | 4.76 | 4.00 | 3.47 | Isoleucine | 20 |
| Leucine | 10.76 | 4.71 | 9.68 | 13.90 | 5.79 | Leucine | 39 |
| Lysine | 2.54 | 2.59 | 4.36 | 2.30 | 3.88 | Lysine | 30 |
| Methionine | 8.70 | 1.66 | 1.97 | 1.70 | 1.68 | Methionine | 10 |
| Methionine + cysteine | 15 | ||||||
| Phenylalanine | 6.13 | 3.08 | 6.03 | 5.30 | 4.06 | Phenylalanine + tyrosine | 25 |
| Threonine | 3.01 | 2.25 | 4.54 | 3.60 | 3.29 | Threonine | 15 |
| Tryptophan | - | - | - | - | Tryptophan | 4 | |
| Valine | 8.15 | 2.77 | 6.50 | 4.80 | 4.68 | Valine | 26 |
| ∑EAA | 49.87 | 20.29 | 41.79 | 37.90 | 28.97 | ||
| NEAA | |||||||
| Alanine | 4.87 | 3.24 | 5.87 | 9.90 | 4.03 | ||
| Arginine | 4.38 | 5.87 | 10.33 | 4.20 | 5.56 | ||
| Asparagine | 5.54 | 5.13 | 9.30 | 7.40 | 5.65 | ||
| Cysteine | 8.38 | - | - | 2.10 | 1.94 | Cysteine | 4 |
| Glutamine | 29.33 | 9.30 | 26.94 | 22.60 | 19.65 | ||
| Glycine | 4.44 | 2.87 | 6.23 | 3.30 | 4.15 | ||
| Proline | 10.41 | 2.63 | 7.63 | 8.50 | 8.79 | ||
| Serine | 4.56 | 3.04 | 5.84 | 5.10 | 4.03 | ||
| Tyrosine | 4.76 | 1.70 | 3.98 | 4.30 | 2.94 | ||
| ∑NEAA | 76.65 | 33.77 | 76.10 | 67.40 | 56.74 | ||
3. The Study of the Proteome. Technologies and Techniques
4. Benefits of Proteomics in the Production of Cereals and Vegetable Crops
4.1. Concept of Translational Plant Proteomics
4.2. Safety Assessment
4.2.1. Detection of Allergens
4.2.2. Detection of Pathogenic Microorganisms
4.3. Authenticity and Quality Assessment
| Food | Purpose of Analysis | Target | Proteomic Techniques | Reference |
|---|---|---|---|---|
| Beans | Comparison between transgenic (Embrapa 5.1) and natural bean | Grain proteome | 2-DGE: IEF and SDS-PAGE | Balsamo et al. [93] |
| MS: MALDI-TOF MS and MALDI-TOF MS/MS | ||||
| Maize | Comparison between transgenic (MON810) and natural maize | Seed proteome | 2-DGE: IEF and SDS-PAGE | Zolla et al. [94] |
| MS: a nHPLC-MS/MS | ||||
| Maize | Comparison between transgenic (MON810) and natural maize | Flour proteome | 2-DGE: IEF, SDS-PAGE, and 2D-DIGE | Vidal et al. [95] |
| MS: b nUPLC-c nESI-d QTOF-MS/MS | ||||
| Maize | Comparison between phytase transgenic and natural maize | Seed proteome | 2-DGE: IEF and SDS-PAGE | Tan et al. [96] |
| MS: MALDI-TOF MS/MS, iTRAQ, and e nLC-MS/MS | ||||
| Potato | Comparison between transgenic (S/CDI-expressing lines) and natural potato | Tuber proteome | 2-DGE: IEF and SDS-PAGE | Khalf et al. [97] |
| MS: LC-ESI-MS/MS | ||||
| Rice | Comparison between transgenic (Bar68-1 and 2036-1a) and natural rice | Seed proteome | 2-DGE: 2D-DIGE | Gong et al. [98] |
| MS: MALDI-TOF MS/MS | ||||
| Rice | Comparison between transgenic (Bt and PEPC) and natural rice | Seed proteome | 2-DGE: IEF and SDS-PAGE | Xue et al. [99] |
| MS: MALDI-TOF MS/MS | ||||
| Soybean | Comparison between transgenic (MSOY 7575 RR) and natural soybean | Seed proteome | 2-DGE: IEF and SDS-PAGE | Brandão et al. [89] |
| MS: MALDI-d QTOF MS | ||||
| Soybean | Comparison between transgenic (MSOY 7575 RR) and natural soybean | Seed proteome | 2-DGE: IEF, SDS-PAGE, and 2D-DIGE | Barbosa et al. [100] |
| MS: MALDI-d QTOF MS/MS and b nUPLC-c nESI-d QTOF MS/MS | ||||
| Tomato | Comparison between transgenic (TFM7) and natural tomato | Fruit proteome | 2-DGE: IEF and SDS-PAGE | Mora et al. [101] |
| MS: e nLC-ESI MS/MS |
4.4. Early Detection of Diseases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Radovich, T.J.K. Biology and Classification of vegetables. In Handbook of Vegetables and Vegetable Processing; Sinha, N.K., Ed.; Blackwell Publishing: Hoboken, NJ, USA, 2011; pp. 3–22. ISBN 9780813815411. [Google Scholar]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Rouf Shah, T.; Prasad, K.; Kumar, P. Maize-A potential source of human nutrition and health: A review. Cogent Food Agric. 2016, 2, 1166995. [Google Scholar] [CrossRef]
- Umadevi, M.; Pushpa, R.; Sampathkumar, K.P.; Bhowmik, D. Rice-Traditional Medicinal Plant in India. J. Pharmacogn. Phytochem. 2012, 1, 6–12. [Google Scholar]
- Kaur, K.D.; Jha, A.; Sabikhi, L.; Singh, A.K. Significance of coarse cereals in health and nutrition: A review. J. Food Sci. Technol. 2014, 51, 1429–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butnariu, M.; Butu, A. Chemical Composition of Vegetables and Their Products. In Handbook of Food Chemistry; Cheung, P.K., Metha, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–49. ISBN 9783642366055. [Google Scholar]
- Fox, P.F.; Brodkorb, A. The casein micelle: Historical aspects, current concepts and significance. Int. Dairy J. 2008, 18, 677–684. [Google Scholar] [CrossRef]
- Bendixen, E. Understanding the proteome. In Proteomics in Food: Principles and Applications; Toldrá, F., Nollet, L.M.L., Eds.; Springer: New York, NY, USA, 2013; pp. 3–19. [Google Scholar]
- Jorrín-Novo, J.; Valledor, L. Translational proteomics special issue. J. Proteom. 2013, 93, 1–4. [Google Scholar] [CrossRef]
- Panghal, A.; Khatkar, B.S.; Singh, U. Cereal Proteins and Their Role in Food Industry. Indian Food Ind. 2006, 25, 58–62. [Google Scholar]
- Shewry, P.R. Improving the protein content and composition of cereal grain. J. Cereal Sci. 2007, 46, 239–250. [Google Scholar] [CrossRef]
- Tacer-Caba, Z.; Nilufer-Erdil, D.; Ai, Y. Chemical Composition of Cereals and Their Products. In Handbook of Food Chemistry; Cheung, P.C.K., Mehta, B.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 301–330. ISBN 9783642366048. [Google Scholar]
- Hounsome, N.; Hounsome, B.; Tomos, D.; Edwards-Jones, G. Plant metabolites and nutritional quality of vegetables. J. Food Sci. 2008, 73, R48–R65. [Google Scholar] [CrossRef]
- Wrigley, C. Cereal-Grain Morphology and Composition. In Cereal Grains: Assessing and Managing Quality, 2nd ed.; Wrigley, C., Batey, I., Miskelly, D., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 55–87. ISBN 9780081007198. [Google Scholar]
- Rizzello, C.G.; Tagliazucchi, D.; Babini, E.; Sefora Rutella, G.; Taneyo Saa, D.L.; Gianotti, A. Bioactive peptides from vegetable food matrices: Research trends and novel biotechnologies for synthesis and recovery. J. Funct. Foods 2016, 27, 549–569. [Google Scholar] [CrossRef]
- Carbonaro, M.; Maselli, P.; Nucara, A. Structural aspects of legume proteins and nutraceutical properties. Food Res. Int. 2015, 76, 19–30. [Google Scholar] [CrossRef]
- Lal, R. Feeding 11 billion on 0.5 billion hectare of area under cereal crops. Food Energy Secur. 2016, 5, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Nam, D.S.; Kong, C. Variability in nutrient composition of cereal grains from different origins. Springerplus 2016, 5, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavazos, A.; Gonzalez de Mejia, E. Identification of Bioactive Peptides from Cereal Storage Proteins and Their Potential Role in Prevention of Chronic Diseases. Compr. Rev. Food Sci. Food Saf. 2013, 12, 364–380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wu, P.; Tian, J. Genetic analysis of amino acid content in wheat grain. J. Genet. 2014, 93, 451–458. [Google Scholar] [CrossRef]
- Sekhar, B.P.S.; Reddy, G.M. Amino Acid Profiles in Some Scented Rice Varieties. Theor. Appl. Genet. 1982, 62, 35–37. [Google Scholar] [CrossRef]
- Sterna, V.; Zute, S.; Brunava, L. Oat Grain Composition and its Nutrition Benefice. Agric. Agric. Sci. Procedia 2016, 8, 252–256. [Google Scholar] [CrossRef] [Green Version]
- Waggle, D.H.; Deyoe, C.W.; Sanford, P.E. Relationship of Protein Level of Sorghum Grain to its Nutritive Value as Measured by Chick Performance and Amino Acid Composition 1. Poult. Sci. 1967, 46, 655–659. [Google Scholar] [CrossRef]
- Smith, D.B. The amino acid composition of barley grain protein during development and germination. J. Agric. Sci. 1972, 78, 265–273. [Google Scholar] [CrossRef]
- Millward, D.J. Identifying recommended dietary allowances for protein and amino acids: A critique of the 2007 WHO/FAO/UNU report. Br. J. Nutr. 2012, 108, S3–S21. [Google Scholar] [CrossRef] [Green Version]
- Marko-Varga, G. Proteomics principles and challenges. Pure Appl. Chem. 2004, 76, 829–837. [Google Scholar] [CrossRef] [Green Version]
- Bradshaw, R.A. Proteomics Today, Proteomics Tomorrow. In Proteomics: Biomedical and Pharmaceutical Applications; Hondermarck, H., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 1–17. ISBN 978-1-4020-2323-1. [Google Scholar]
- Wilkins, M.R.; Sanchez, J.C.; Gooley, A.A.; Appel, R.D.; Humphery-Smith, I.; Hochstrasser, D.F.; Williams, K.L. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 1996, 13, 19–50. [Google Scholar] [CrossRef] [Green Version]
- Tyers, M.; Mann, M. From genomics to proteomics. Nature 2003, 422, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Lawrance, I.C.; Klopcic, B.; Wasinger, V.C. Proteomics: An Overview Proteomics: What does it Mean? Inflamm. Bowel Dis. 2005, 11, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Goez, M.M.; Torres-Madroñero, M.C.; Röthlisberger, S.; Delgado-Trejos, E. Preprocessing of 2-Dimensional Gel Electrophoresis Images Applied to Proteomic Analysis: A Review. Genom. Proteom. Bioinforma. 2018, 16, 63–72. [Google Scholar] [CrossRef]
- Schulze, W.X.; Usadel, B. Quantitation in mass-spectrometry-based proteomics. Annu. Rev. Plant Biol. 2010, 61, 491–516. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Huang, J.; Lu, J.; Zhang, X. Reversible Lysine Derivatization Enabling Improved Arg-C Digestion, a Highly Specific Arg-C Digestion Using Trypsin. Anal. Chem. 2018, 90, 1554–1559. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Heck, A.J.R. Proteomics beyond trypsin. FEBS J. 2015, 282, 2612–2626. [Google Scholar] [CrossRef]
- Dupree, E.J.; Jayathirtha, M.; Yorkey, H.; Mihasan, M.; Petre, B.A.; Darie, C.C. A critical review of bottom-up proteomics: The good, the bad, and the future of this field. Proteomes 2020, 8, 14. [Google Scholar] [CrossRef]
- Cunsolo, V.; Muccilli, V.; Saletti, R.; Foti, S. Mass spectrometry in food proteomics: A tutorial. J. Mass Spectrom. 2014, 49, 768–784. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, R.; Holland, J.W.; Deeth, H.C.; Alewood, P. Milk proteomics. Int. Dairy J. 2004, 14, 1013–1023. [Google Scholar] [CrossRef]
- Xie, F.; Smith, R.D.; Shen, Y. Advanced proteomic liquid chromatography. J. Chromatogr. A 2012, 1261, 78–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisser, H.; Choudhary, J.S. Targeted Feature Detection for Data-Dependent Shotgun Proteomics. J. Proteome Res. 2017, 16, 2964–2974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcus, K.; Lelong, C.; Rabilloud, T. What room for two-dimensional gel-based proteomics in a shotgun proteomics world? Proteomes 2020, 8, 17. [Google Scholar] [CrossRef]
- Gao, Y.; Yates, J.R. Protein Analysis by Shotgun Proteomics. In Mass Spectrometry-Based Chemical Proteomics; Tao, W.A., Zhang, Y., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 1–38. ISBN 9781118969557. [Google Scholar]
- Bubis, J.A.; Levitsky, L.I.; Ivanov, M.V.; Tarasova, I.A.; Gorshkov, M.V. Comparative evaluation of label-free quantification methods for shotgun proteomics. Rapid Commun. Mass Spectrom. 2017, 31, 606–612. [Google Scholar] [CrossRef]
- Borràs, E.; Sabidó, E. What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics 2017, 17, 17–18. [Google Scholar] [CrossRef]
- Arora, A.; Somasundaram, K. Targeted Proteomics Comes to the Benchside and the Bedside: Is it Ready for Us? Bioessays 2019, 41, 1800042. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Song, E.; Nie, S.; Rodland, K.D.; Liu, T.; Qian, W.J.; Smith, R.D. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16, 2160–2182. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.K.; Pedreschi, R.; Barkla, B.J.; Bindschedler, L.V.; Cramer, R.; Sarkar, A.; Renaut, J.; Job, D.; Rakwal, R. Translational plant proteomics: A perspective. J. Proteom. 2012, 75, 4588–4601. [Google Scholar] [CrossRef]
- Breiteneder, H.; Mills, E.N.C. Plant food allergens—Structural and functional aspects of allergenicity. Biotechnol. Adv. 2005, 23, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Radauer, C. A classification of plant food allergens. J. Allergy Clin. Immunol. 2004, 113, 821–830. [Google Scholar] [CrossRef]
- Ahsan, N.; Rao, R.S.P.; Gruppuso, P.A.; Ramratnam, B.; Salomon, A.R. Targeted proteomics: Current status and future perspectives for quantification of food allergens. J. Proteomics 2016, 143, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Inomata, N. Wheat allergy. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Grishina, G.; Bardina, L.; Grishin, A. 2D-electrophoresis and immunoblotting in food allergy. In Food Allergens: Methods and Protocols, Methods in Molecular Biology; Lin, J., Alcocer, M., Eds.; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1592, pp. 59–69. ISBN 978-1-4939-6923-4. [Google Scholar]
- Akagawa, M.; Handoyo, T.; Ishii, T.; Kumazawa, S.; Morita, N.; Suyama, K. Proteomic analysis of wheat flour allergens. J. Agric. Food Chem. 2007, 55, 6863–6870. [Google Scholar] [CrossRef]
- López-Pedrouso, M.; Lorenzo, J.M.; Gagaoua, M.; Franco, D. Current trends in proteomic advances for food allergen analysis. Biology 2020, 9, 247. [Google Scholar] [CrossRef]
- Ma, X.; Li, H.; Zhang, J.; Huang, W.; Han, J.; Ge, Y.; Sun, J.; Chen, Y. Comprehensive quantification of sesame allergens in processed food using liquid chromatography-tandem mass spectrometry. Food Control 2020, 107, 106744. [Google Scholar] [CrossRef]
- Boo, C.C.; Parker, C.H.; Jackson, L.S. A targeted LC-MS/MS method for the simultaneous detection and quantitation of egg, milk, and peanut allergens in sugar cookies. J. AOAC Int. 2018, 101, 108–117. [Google Scholar] [CrossRef]
- Biesiekierski, J.R. What is gluten? J. Gastroenterol. Hepatol. 2017, 32, 78–81. [Google Scholar] [CrossRef] [Green Version]
- Fiedler, K.L.; McGrath, S.C.; Callahan, J.H.; Ross, M.M. Characterization of grain-specific peptide markers for the detection of gluten by mass spectrometry. J. Agric. Food Chem. 2014, 62, 5835–5844. [Google Scholar] [CrossRef]
- Martínez-Esteso, M.J.; Nørgaard, J.; Brohée, M.; Haraszi, R.; Maquet, A.; O’Connor, G. Defining the wheat gluten peptide fingerprint via a discovery and targeted proteomics approach. J. Proteom. 2016, 147, 156–168. [Google Scholar] [CrossRef]
- García-Molina, M.D.; Muccilli, V.; Saletti, R.; Foti, S.; Masci, S.; Barro, F. Comparative proteomic analysis of two transgenic low-gliadin wheat lines and non-transgenic wheat control. J. Proteom. 2017, 165, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, C.; Planchon, S.; Renaut, J.; Oliveira, M.M.; Batista, R. Characterization of maize allergens—MON810 vs. its non-transgenic counterpart. J. Proteom. 2012, 75, 2027–2037. [Google Scholar] [CrossRef] [PubMed]
- Batista, R.; Martins, I.; Jenö, P.; Ricardo, C.P.; Oliveira, M.M. A proteomic study to identify soya allergens—The human response to transgenic versus non-transgenic soya samples. Int. Arch. Allergy Immunol. 2007, 144, 29–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, C.Y.; Wang, T. Proteomic evaluation of genetically modified crops: Current status and challenges. Front. Plant Sci. 2013, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Berger, C.N.; Sodha, S.V.; Shaw, R.K.; Griffin, P.M.; Pink, D.; Hand, P.; Frankel, G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010, 12, 2385–2397. [Google Scholar] [CrossRef]
- Saranraj, P.; Stella, D.; Reetha, D. Microbial spoilage of vegetables and its control measures: A review. Int. J. Nat. Prod. Sci. 2012, 2, 1–12. [Google Scholar]
- D’Alessandro, A.; Zolla, L. We are what we eat: Food safety and proteomics. J. Proteome Res. 2012, 11, 26–36. [Google Scholar] [CrossRef]
- Warriner, K. Pathogens in vegetables. In Improving the Safety of Fresh Fruit and Vegetables; Jongen, W., Ed.; Woodhead Publishing: Cambridge, UK, 2005; pp. 3–43. ISBN 9781855739567. [Google Scholar]
- Gharechahi, J.; Zeinolabedini, M.; Salekdeh, G.H. Proteomics in detection of contaminations and adulterations in agricultural foodstuffs. In Agricultural Proteomics Volume 1: Crops, Horticulture, Farm Animals, Food, Insect and Microorganisms; Salekdeh, H.G., Ed.; Springer International Publishing: New York, NY, USA, 2016; pp. 67–85. ISBN 9783319432755. [Google Scholar]
- Piras, C.; Roncada, P.; Rodrigues, P.M.; Bonizzi, L.; Soggiu, A. Proteomics in food: Quality, safety, microbes, and allergens. Proteomics 2016, 16, 799–815. [Google Scholar] [CrossRef]
- Fratamico, P.; Gunther, N.W. Advances in genomics and proteomics-based methods for the study of foodborne bacterial pathogens. In Advances in Microbial Food Safety; Sofos, J., Ed.; Woodhead Publishing: Cambridge, UK, 2013; Volume 1, pp. 462–497. ISBN 9780857094384. [Google Scholar]
- Righetti, P.G.; Fasoli, E.; D’Amato, A.; Boschetti, E. Making progress in plant proteomics for improved food safety. In Comprehensive Analytical Chemistry. Applications of Advanced Omics Technologies: From Genes to Metabolites; García-Cañas, V., Cifuentes, A., Simó, C., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2014; Volume 64, pp. 131–155. ISBN 9780444626509. [Google Scholar]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell, A.G.M. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3106–3129. [Google Scholar] [CrossRef]
- Breuer, T.; Benkel, D.H.; Shapiro, R.L.; Hall, W.N.; Winnett, M.M.; Linn, M.J.; Neimann, J.; Barrett, T.J.; Dietrich, S.; Downes, F.P.; et al. A Multistate Outbreak of Escherichia coli O157:H7 Infections Linked to Alfalfa Sprouts Grown from Contaminated Seeds. Emerg. Infect. Dis. 2001, 7, 977–982. [Google Scholar] [CrossRef] [Green Version]
- Kulasekara, B.R.; Jacobs, M.; Zhou, Y.; Wu, Z.; Sims, E.; Saenphimmachak, C.; Rohmer, L.; Ritchie, J.M.; Radey, M.; McKevitt, M.; et al. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 2009, 77, 3713–3721. [Google Scholar] [CrossRef] [Green Version]
- Marder, E.P.; Garman, K.N.; Ingram, L.A.; Dunn, J.R. Multistate outbreak of Escherichia coli O157:H7 associated with bagged salad. Foodborne Pathog. Dis. 2014, 11, 593–595. [Google Scholar] [CrossRef]
- Fagerquist, C.K.; Garbus, B.R.; Miller, W.G.; Williams, K.E.; Yee, E.; Bates, A.H.; Boyle, S.; Harden, L.A.; Cooley, M.B.; Mandrell, R.E. Rapid identification of protein biomarkers of Escherichia coli O157:H7 by matrix-assisted laser desorption Lonization-time-of-flight-time-of-flight mass spectrometry and top-down proteomics. Anal. Chem. 2010, 82, 2717–2725. [Google Scholar] [CrossRef] [PubMed]
- Fagerquist, C.K.; Zaragoza, W.J.; Sultan, O.; Woo, N.; Quiñones, B.; Cooley, M.B.; Mandrell, R.E. Top-down proteomic identification of shiga toxin 2 subtypes from Shiga toxin-producing Escherichia coli by matrix-assisted laser desorption ionization-tandem time of flight mass spectrometry. Appl. Environ. Microbiol. 2014, 80, 2928–2940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Liu, G.; Huang, X.; Li, L.; Lin, H.; Xu, D. MALDI-TOF MS-Based identification of bacteria and a survey of fresh vegetables with pathogenic bacteria in Beijing, China. Food Biosci. 2021, 41, 100746. [Google Scholar] [CrossRef]
- Böhme, K.; Fernández-No, I.C.; Calo-Mata, P.; Barros-Velázquez, J. Proteomics of Food Spoilage Pathogens. In Proteomics in Food Science: From Farm to Fork; Colgrave, M.L., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 417–431. ISBN 9780128040577. [Google Scholar]
- Calo-Mata, P.; Carrera, M.; Böhme, K.; Caamaño-Antelo, S.; Gallardo, J.M.; Barros-Velázquez, J.; Cañas, B. Novel Peptide Biomarker Discovery for Detection and Identification of Bacterial Pathogens by LC-ESI-MS/MS. J. Anal. Bioanal. Tech. 2016, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wigmann, É.F.; Behr, J.; Vogel, R.F.; Niessen, L. MALDI-TOF MS fingerprinting for identification and differentiation of species within the Fusarium fujikuroi species complex. Appl. Microbiol. Biotechnol. 2019, 103, 5323–5337. [Google Scholar] [CrossRef]
- Quéro, L.; Girard, V.; Pawtowski, A.; Tréguer, S.; Weill, A.; Arend, S.; Cellière, B.; Polsinelli, S.; Monnin, V.; van Belkum, A.; et al. Development and application of MALDI-TOF MS for identification of food spoilage fungi. Food Microbiol. 2019, 81, 76–88. [Google Scholar] [CrossRef]
- Ortea, I.; O’Connor, G.; Maquet, A. Review on proteomics for food authentication. J. Proteom. 2016, 147, 212–225. [Google Scholar] [CrossRef]
- Bansal, S.; Singh, A.; Mangal, M.; Mangal, A.K.; Kumar, S. Food adulteration: Sources, health risks, and detection methods. Crit. Rev. Food Sci. Nutr. 2017, 57, 1174–1189. [Google Scholar] [CrossRef]
- Russo, R.; Cusano, E.; Perissi, A.; Ferron, F.; Severino, V.; Parente, A.; Chambery, A. Ultra-high performance liquid chromatography tandem mass spectrometry for the detection of durum wheat contamination or adulteration. J. Mass Spectrom. 2014, 49, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Agregán, R.; Echegaray, N.; López-Pedrouso, M.; Kharabsheh, R.; Franco, D.; Lorenzo, J.M. Proteomic Advances in Milk and Dairy Products. Molecules 2021, 26, 3832. [Google Scholar] [CrossRef]
- Bönick, J.; Huschek, G.; Rawel, H.M. Determination of wheat, rye and spelt authenticity in bread by targeted peptide biomarkers. J. Food Compos. Anal. 2017, 58, 82–91. [Google Scholar] [CrossRef]
- European Parliament. E.C. Regulation (EC) No 1830/2003 of September 2003 concerning the traceability and labelling of genetically modified organisms and the traceability of food and feed products produced from genetically modified organisms and amending Directive 2001/18/EC. J. Eur. Union 2003, L268, 24–28. [Google Scholar]
- Development Assistance Committee. OCDE/GD(91)208, D.A.C.O. DAC Principles for Evaluation of Development Assistance, OCDE/GD(91)208; Development Assistance Committee, OECD: Paris, France, 1991. [Google Scholar]
- Brandão, A.R.; Barbosa, H.S.; Arruda, M.A.Z. Image analysis of two-dimensional gel electrophoresis for comparative proteomics of transgenic and non-transgenic soybean seeds. J. Proteom. 2010, 73, 1433–1440. [Google Scholar] [CrossRef]
- Min, C.W.; Gupta, R.; Agrawal, G.K.; Rakwal, R.; Kim, S.T. Concepts and strategies of soybean seed proteomics using the shotgun proteomics approach. Expert Rev. Proteom. 2019, 16, 795–804. [Google Scholar] [CrossRef]
- Liu, W.; Xu, W.; Li, L.; Dong, M.; Wan, Y.; He, X.; Huang, K.; Jin, W. iTRAQ-based quantitative tissue proteomic analysis of differentially expressed proteins (DEPs) in non-transgenic and transgenic soybean seeds. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Liu, W.; Li, L.; Zhang, Z.; Dong, M.; Jin, W. iTRAQ-based quantitative proteomic analysis of transgenic and non-transgenic maize seeds. J. Food Compos. Anal. 2020, 92, 103564. [Google Scholar] [CrossRef]
- Balsamo, G.M.; Valentim-Neto, P.A.; Mello, C.S.; Arisi, A.C.M. Comparative Proteomic Analysis of Two Varieties of Genetically Modified (GM) Embrapa 5.1 Common Bean (Phaseolus vulgaris L.) and Their Non-GM Counterparts. J. Agric. Food Chem. 2015, 63, 10569–10577. [Google Scholar] [CrossRef]
- Zolla, L.; Rinalducci, S.; Antonioli, P.; Righetti, P.G. Proteomics as a complementary tool for identifying unintended side effects occurring in transgenic maize seeds as a result of genetic modifications. J. Proteome Res. 2008, 7, 1850–1861. [Google Scholar] [CrossRef] [Green Version]
- Vidal, N.; Barbosa, H.; Jacob, S.; Arruda, M. Comparative study of transgenic and non-transgenic maize (Zea mays) flours commercialized in Brazil, focussing on proteomic analyses. Food Chem. 2015, 180, 288–294. [Google Scholar] [CrossRef]
- Tan, Y.; Tong, Z.; Yang, Q.; Sun, Y.; Jin, X.; Peng, C.; Guo, A.; Wang, X. Proteomic analysis of phytase transgenic and non-transgenic maize seeds. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Khalf, M.; Goulet, C.; Vorster, J.; Brunelle, F.; Anguenot, R.; Fliss, I.; Michaud, D. Tubers from potato lines expressing a tomato Kunitz protease inhibitor are substantially equivalent to parental and transgenic controls. Plant Biotechnol. J. 2010, 8, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.Y.; Li, Q.; Yu, H.T.; Wang, Z.; Wang, T. Proteomics insight into the biological safety of transgenic modification of rice as compared with conventional genetic breeding and spontaneous genotypic variation. J. Proteome Res. 2012, 11, 3019–3029. [Google Scholar] [CrossRef]
- Xue, K.; Yang, J.; Liu, B.; Xue, D. The integrated risk assessment of transgenic rice Oryza sativa: A comparative proteomics approach. Food Chem. 2012, 135, 314–318. [Google Scholar] [CrossRef]
- Barbosa, H.S.; Arruda, S.C.C.; Azevedo, R.A.; Arruda, M.A.Z. New insights on proteomics of transgenic soybean seeds: Evaluation of differential expressions of enzymes and proteins. Anal. Bioanal. Chem. 2012, 402, 299–314. [Google Scholar] [CrossRef]
- Mora, L.; Bramley, P.M.; Fraser, P.D. Development and optimisation of a label-free quantitative proteomic procedure and its application in the assessment of genetically modified tomato fruit. Proteomics 2013, 13, 2016–2030. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Chen, J.; Dai, L.; Ma, H.; Zhang, H.; Yang, J.; Wang, F.; Yan, C. Proteomic dissection of plant responses to various pathogens. Proteomics 2015, 15, 1525–1543. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Fernandez, R.; Jorrin-Novo, J.V. Contribution of proteomics to the study of plant pathogenic fungi. J. Proteome Res. 2012, 11, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Han, Z.; Wang, S.; Wang, X.; Sun, A.; Zu, X.; Chen, Y. Comparative proteomic analysis of the plant-virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J. Proteomics 2013, 89, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Barnabas, L.; Ashwin, N.M.R.; Kaverinathan, K.; Trentin, A.R.; Pivato, M.; Sundar, A.R.; Malathi, P.; Viswanathan, R.; Rosana, O.B.; Neethukrishna, K.; et al. Proteomic analysis of a compatible interaction between sugarcane and Sporisorium scitamineum. Proteomics 2016, 16, 1111–1122. [Google Scholar] [CrossRef]
- Lin, S.Q.; Yang, Z.J.; Huang, B.F.; Bi, C.Y.; Huang, X.F.; Chen, G.T.; Nijiati, N.; Chen, X.Y. Comparative proteomic analysis of the sweetpotato provides insights into response mechanisms to Fusarium oxysporum f. sp. batatas. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.; Koh, J.; Yoo, M.J.; Zhu, N.; Feole, M.; Yi, S.; Chen, S. Quantitative proteomics of tomato defense against Pseudomonas syringae infection. Proteomics 2013, 13, 1934–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampitsch, C.; Bykova, N.V. Proteomics and plant disease: Advances in combating a major threat to the global food supply. Proteomics 2012, 12, 673–690. [Google Scholar] [CrossRef]
- Das, P.P.; Lin, Q.; Wong, S.-M. Comparative proteomics of Tobacco mosaic virus-infected Nicotiana tabacum plants identified major host proteins involved in photosystems and plant defence. J. Proteom. 2019, 194, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.R.; Sun, H.D.; Ali, A.; Rott, P.C.; Javed, T.; Fu, H.Y.; Gao, S.J. Quantitative proteomic analysis of the sugarcane defense responses incited by Acidovorax avenae subsp. avenae causing red stripe. Ind. Crops Prod. 2021, 162, 113275. [Google Scholar] [CrossRef]
| 2-DGE-Based Proteomics | Shotgun Proteomics | |
|---|---|---|
| Sample consuming | ++(+) * | + |
| Time consuming | +++ | ++ |
| Analysis depth | ++ | +++ |
| Separation/identification | ||
| Separation/detection of proteoforms | ||
| Identification on protein level | Multiple identifications | Only by inference from peptides |
| Detection of proteoforms | +++ | - |
| Details at peptide level (e.g., sequence coverage) | +++ | + |
| Number of modulated proteins identified | + | +++ |
| Coupling with biochemical methods | ||
| Antibodies | +++ | + |
| Enzymes (zymography) | + | - |
| Robustness of quantification | ||
| Sensitivity | ++ | +++ |
| Linearity | +++ | + |
| Need of validation | +++ | +++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agregán, R.; Echegaray, N.; López-Pedrouso, M.; Aadil, R.M.; Hano, C.; Franco, D.; Lorenzo, J.M. Proteomic Advances in Cereal and Vegetable Crops. Molecules 2021, 26, 4924. https://doi.org/10.3390/molecules26164924
Agregán R, Echegaray N, López-Pedrouso M, Aadil RM, Hano C, Franco D, Lorenzo JM. Proteomic Advances in Cereal and Vegetable Crops. Molecules. 2021; 26(16):4924. https://doi.org/10.3390/molecules26164924
Chicago/Turabian StyleAgregán, Rubén, Noemí Echegaray, María López-Pedrouso, Rana Muhammad Aadil, Christophe Hano, Daniel Franco, and José M. Lorenzo. 2021. "Proteomic Advances in Cereal and Vegetable Crops" Molecules 26, no. 16: 4924. https://doi.org/10.3390/molecules26164924
APA StyleAgregán, R., Echegaray, N., López-Pedrouso, M., Aadil, R. M., Hano, C., Franco, D., & Lorenzo, J. M. (2021). Proteomic Advances in Cereal and Vegetable Crops. Molecules, 26(16), 4924. https://doi.org/10.3390/molecules26164924











