Response of Osteosarcoma Cell Metabolism to Platinum and Palladium Chelates as Potential New Drugs
Abstract
1. Introduction
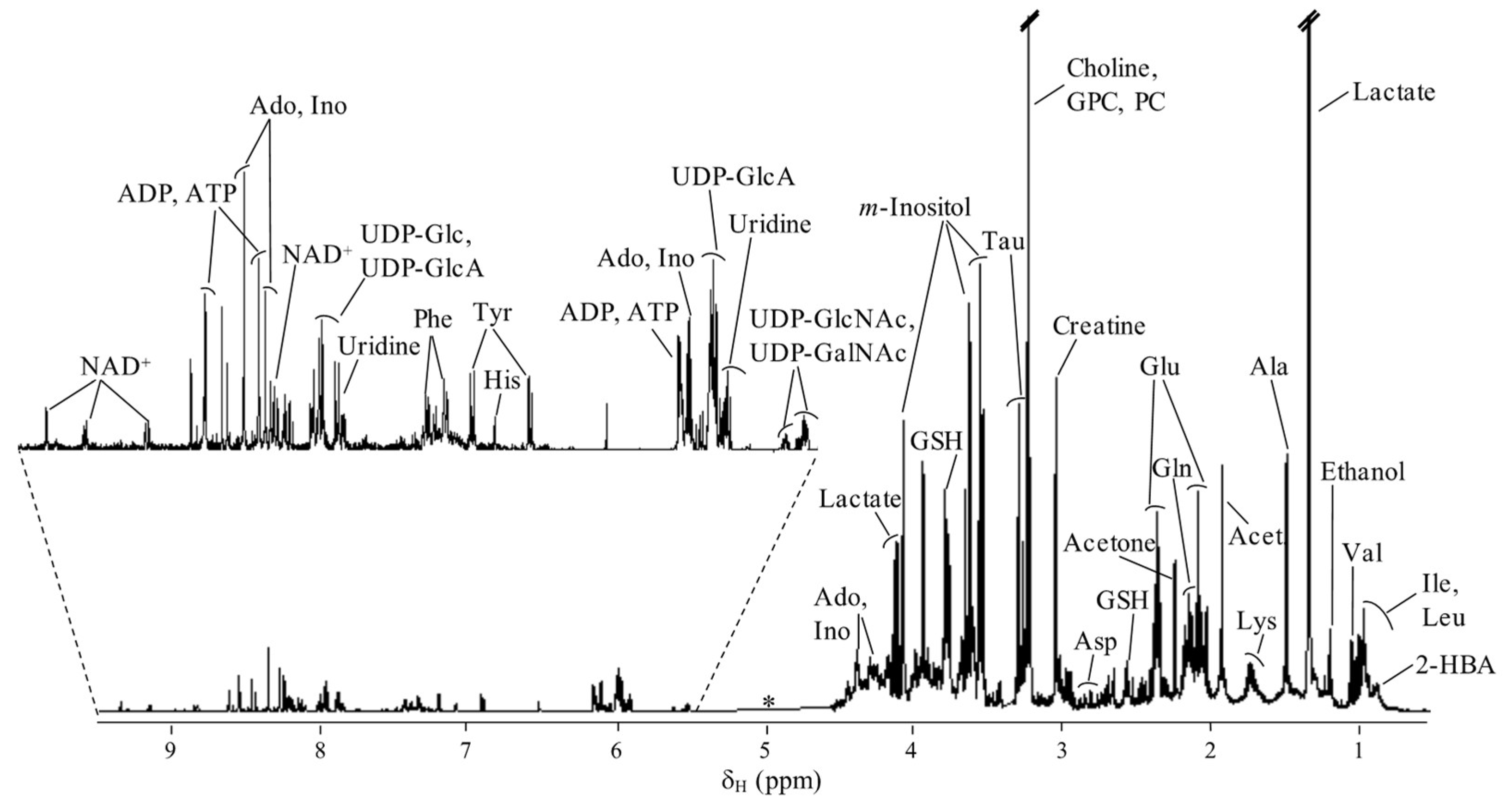
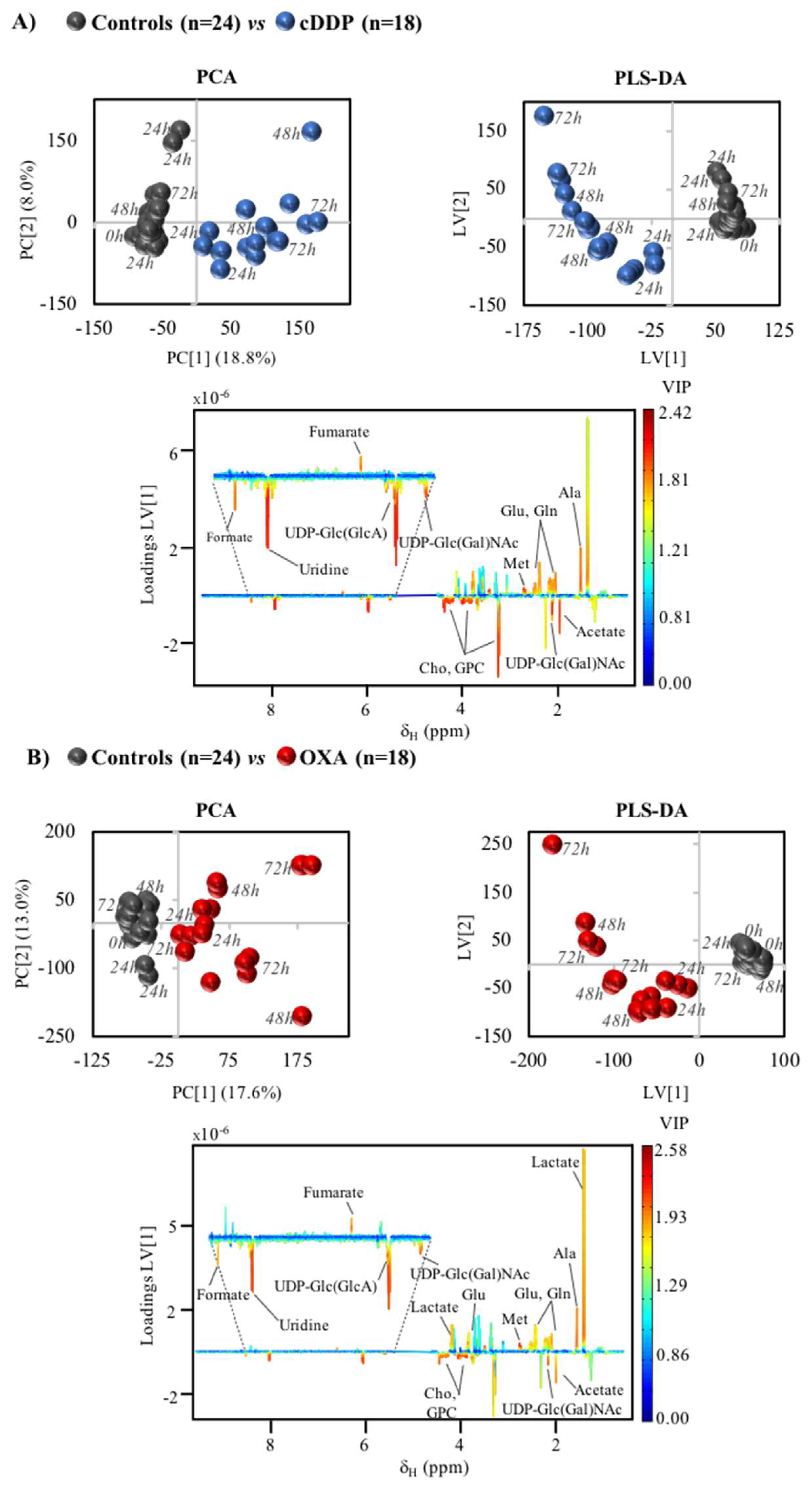
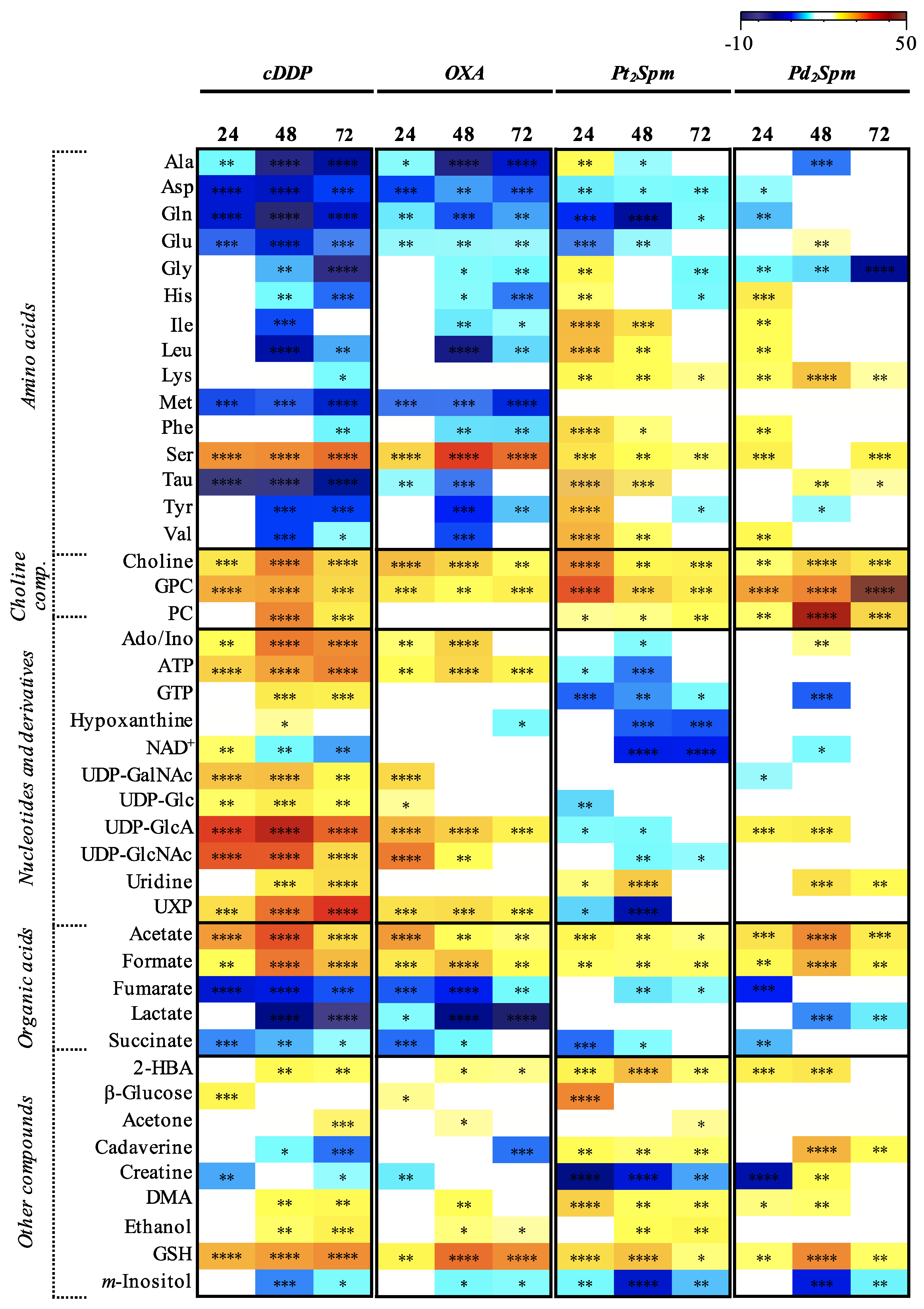
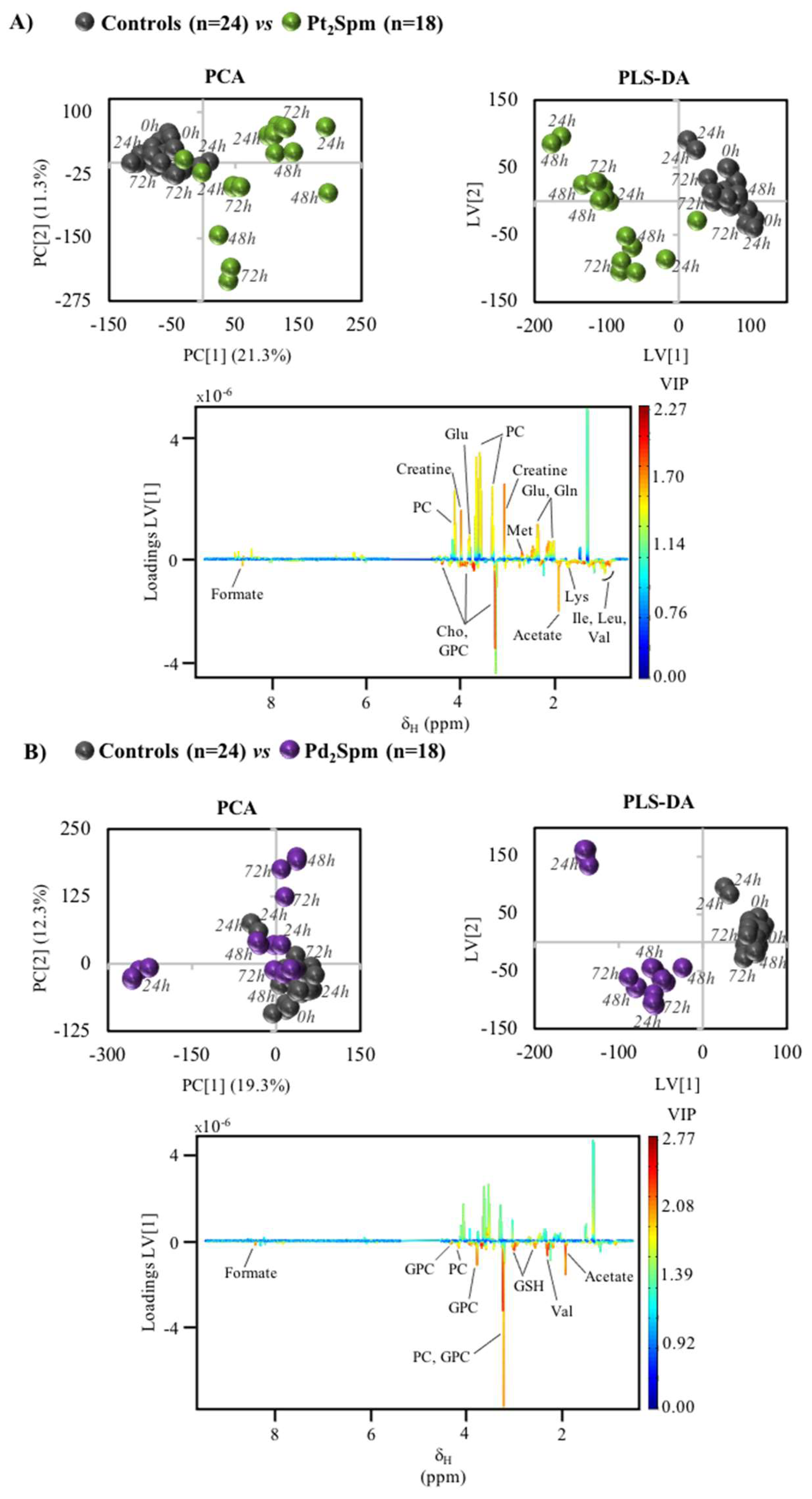
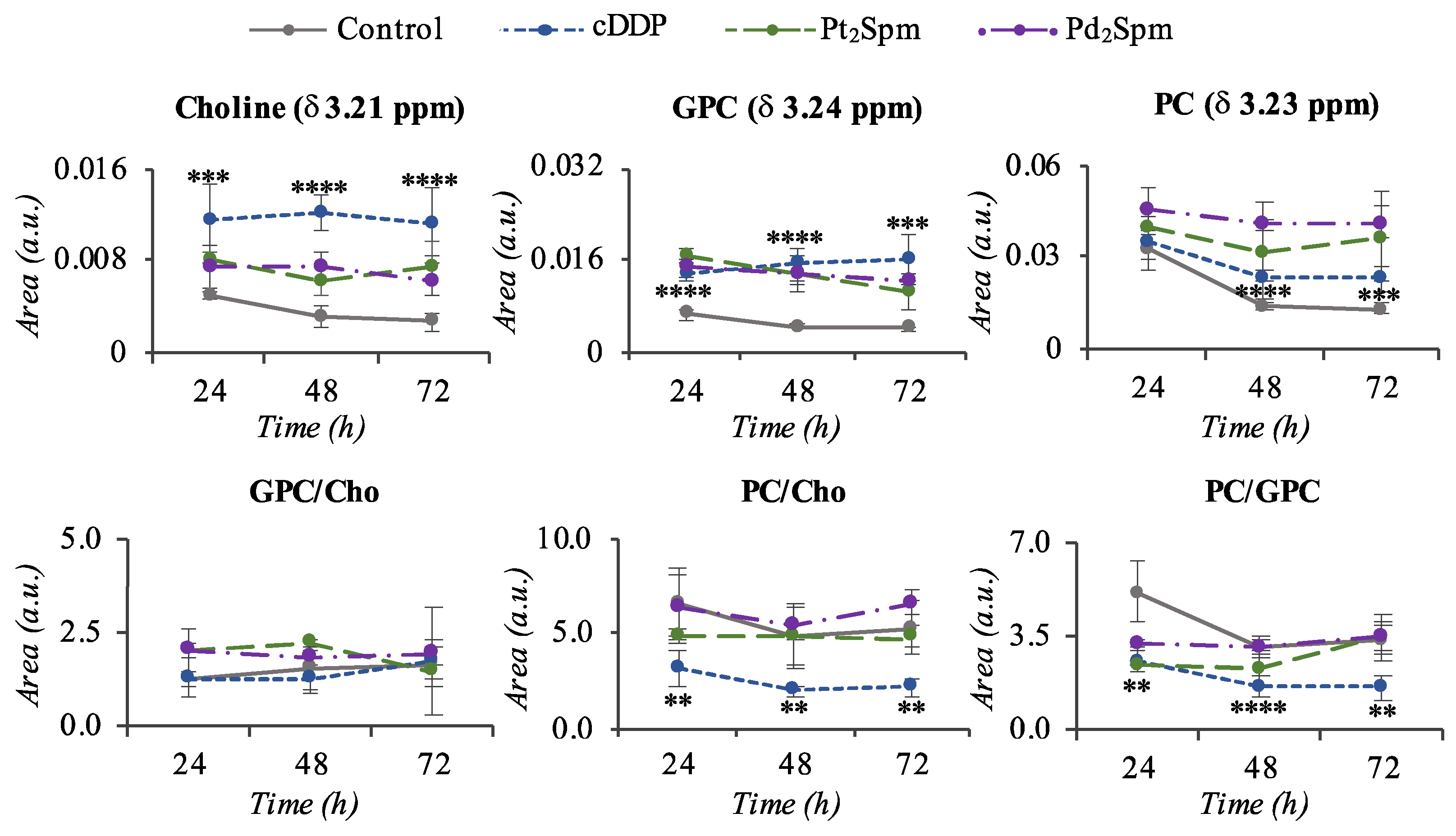
2. Results
3. Discussion
3.1. MG-63 Cell Metabolism Associated to cDDP and OXA Treatment
3.2. MG-63 Cell Metabolism Associated to Pt2Spm and Pd2Spm Treatment
4. Materials and Methods
4.1. Chemicals and Solutions
4.2. Cell Culture
4.3. Drug Administration and Sample Collection
4.4. Cell Extraction for Sample Preparation
4.5. NMR Spectroscopy
4.6. Data Processing and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics 2020. CA-Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Taran, S.J.; Taran, R.; Malipatil, N.B. Pediatric osteosarcoma: An updated review. Indian J. Med. Paediatr. Oncol. 2017, 38, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Wiltshaw, E. Cisplatin in the treatment of cancer. Platinum Met. Rev. 1979, 23, 90–98. [Google Scholar]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007, 107, 1387–1407. [Google Scholar] [CrossRef]
- Mauri, G.; Gori, V.; Bonazzina, E.; Amatu, A.; Tosi, F.; Bencardino, K.; Ruggieri, L.; Patelli, G.; Arena, S.; Bardelli, A.; et al. Oxaliplatin retreatment in metastatic colorectal cancer: Systematic review and future research opportunities. Cancer Treat. Rev. 2020, 91, 102112. [Google Scholar] [CrossRef]
- Marques, M.P.M. Platinum and palladium polyamine complexes as anticancer agents: The structural factor. ISRN Spectrosc. 2013, 2013, 287353. [Google Scholar] [CrossRef]
- Marques, M.P.M.; Girão, T.; Pedroso de Lima, M.C.; Gameiro, A.; Pereira, E.; Garcia, P. Cytotoxic effects of metal complexes of biogenic polyamines. I. Platinum(II) spermidine compounds: Prediction of their antitumour activity. Biochim. Biophys. Acta 2002, 1589, 63–70. [Google Scholar] [CrossRef][Green Version]
- Soares, A.S.; Fiuza, S.M.; Gonçalves, M.J.; Batista de Carvalho, L.A.E.; Marques, M.P.M.; Urbano, A.M. Effect of the metal cancer on the antitumor activity of the analogous dinuclear spermine chelates (PdCl2)2(spermine) and (PtCl2)2(spermine). Lett. Drug Des. Discov. 2007, 4, 460–463. [Google Scholar] [CrossRef]
- Tummala, R.; Diegelman, P.; Fiuza, S.M.; Batista de Carvalho, L.A.E.; Marques, M.P.M.; Kramer, D.L.; Clark, K.; Vujcic, S.; Porter, C.W.; Pendyala, L. Characterization of Pt-, Pd-spermine complexes for their effect on polyamine pathway and cisplatin resistance in A2780 ovarian carcinoma cells. Oncol. Rep. 2010, 24, 15–24. [Google Scholar]
- Batista de Carvalho, L.A.E.; Marques, M.P.M.; Martin, C.; Parker, S.F.; Tomkinson, J. Inelastic neutron scattering study of PtII complexes displaying anticancer properties. ChemPhysChem 2011, 12, 1334–1341. [Google Scholar] [CrossRef] [PubMed]
- Fiuza, S.M.; Holy, M.J.; Batista de Carvalho, L.A.E.; Marques, M.P.M. Biologic activity of a dinuclear Pd(II)–spermine complex toward human breast cancer. Chem. Biol. Drug Des. 2011, 77, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.M.; Oredsson, S.; Persson, L.; Woster, P.; Marques, M.P.M. Novel Pt(II) and Pd(II) complexes with polyamine analogues: Synthesis and vibrational analysis. J. Inorg. Biochem. 2012, 108, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.M.; Andersson, S.; Sukumaran, S.K.; Marques, M.P.M.; Persson, L.; Oredsson, S. Norspermidine and novel Pd(II) and Pt(II) polynuclear complexes of norspermidine as potential antineoplastic agents against breast cancer. PLoS ONE 2013, 8, e55651. [Google Scholar] [CrossRef]
- Marques, M.P.M.; Valero, R.; Parker, S.F.; Tomkinson, J.; Batista de Carvalho, L.A.E. Polymorphism in cisplatin anticancer drug. J. Phys. Chem. B. 2013, 117, 6421–6429. [Google Scholar] [CrossRef]
- Fiuza, S.M.; Amado, A.M.; Parker, S.F.; Marques, M.P.M.; Batista de Carvalho, L.A.E. Conformational insights and vibrational study of a promising anticancer agent: The role of the ligand in Pd(II)-amine complexes. New J. Chem. 2015, 39, 6274–6283. [Google Scholar] [CrossRef]
- Marques, M.P.M.; Gianolio, D.; Giannantonio, C.; Tomkinson, J.; Parker, S.F.; Valero, R.; Lopes, R.P.; Batista de Carvalho, L.A.E. A molecular view of cisplatin’s mode of action: Interplay with DNA bases and acquired resistance. Phys. Chem. Chem. Phys. 2015, 17, 5155–5171. [Google Scholar] [CrossRef]
- Batista de Carvalho, A.L.M.; Medeiros, P.S.C.; Costa, F.M.; Ribeiro, V.P.; Diniz, C.; Marques, M.P.M. Anti-invasive and anti-proliferative synergism between docetaxel and a polynuclear Pd-spermine agent. PLoS ONE 2016, 11, e0167218. [Google Scholar] [CrossRef]
- Batista de Carvalho, A.L.M.; Pilling, M.; Gardner, P.; Doherty, J.; Cinque, G.; Wehbe, K.; Kelley, C.; Batista de Carvalho, L.A.E.; Marques, M.P.M. Chemotherapeutic response to cisplatin-like drugs in human breast cancer cells probed by vibrational microspectroscopy. Faraday Discuss. 2016, 187, 273–298. [Google Scholar] [CrossRef]
- Marques, M.P.M.; Batista de Carvalho, A.L.M.; Sakai, V.G.; Hatter, L.; Batista de Carvalho, L.A.E. Intracellular water—An overlooked drug target? Cisplatin impact in cancer cells probed by neutrons. Phys. Chem. Chem. Phys. 2017, 19, 2702–2713. [Google Scholar] [CrossRef]
- Lamego, I.; Marques, M.P.M.; Duarte, I.F.; Martins, A.S.; Oliveira, H.; Gil, A.M. Impact of the Pd2Spermine chelate on osteosarcoma metabolism: An NMR metabolomics study. J. Proteome Res. 2017, 16, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Vojtek, M.; Marques, M.P.M.; Ferreira, I.M.P.L.V.O.; Mota-Filipe, H.; Diniz, C. Anticancer activity of palladium-based complexes against triple negative breast cancer. Drug Discov. Today 2019, 24, 1044–1058. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.S.; Batista de Carvalho, A.L.M.; Lamego, I.; Marques, M.P.M.; Gil, A.M. Cytotoxicity of platinum and palladium chelates against osteosarcoma. ChemistrySelect 2020, 5, 5993–6000. [Google Scholar] [CrossRef]
- Silva, T.M.; Fiuza, S.M.; Marques, M.P.M.; Persson, L.; Oredsson, S. Increased breast cancer cell toxicity by palladination of the polyamine analogue N1,N11 bis(ethyl)norspermine. Amino Acids 2014, 46, 339–352. [Google Scholar] [CrossRef]
- Corduneanu, O.; Chiorcea-Paquim, A.M.; Diculescu, V.; Fiuza, S.M.; Marques, M.P.M.; Oliveira-Brett, A.M. DNA Interaction with palladium chelates of biogenic polyamines using atomic force microscopy and voltammetric characterization. Anal. Chem. 2010, 82, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Corduneanu, O.; Chiorcea-Paquim, A.M.; Fiuza, S.M.; Marques, M.P.M.; Oliveira-Brett, A.M. Polynuclear palladium complexes with biogenic polyamines: AFM and voltammetric characterization. Bioelectrochemistry 2010, 78, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Tong, Y.; Wang, J.; Huang, Y. NMR studies of the relationship between the changes of membrane lipids and the cisplatin-resistance of A549/DDP cells. Cancer Cell Int. 2003, 3, 1–8. [Google Scholar] [CrossRef]
- Duarte, I.F.; Ladeirinha, A.F.; Lamego, I.; Gil, A.M.; Carvalho, L.; Carreira, I.M.; Melo, J.B. Potential markers of cisplatin treatment response unveiled by NMR metabolomics of human lung cells. Mol. Pharm. 2013, 10, 4242–4251. [Google Scholar] [CrossRef]
- Lindskog, M.; Spenger, C.; Jarvet, J.; Graslund, A.; Kogner, P. Predicting resistance or response to chemotherapy by proton magnetic resonance spectroscopy in neuroblastoma. J. Natl. Cancer Inst. 2004, 96, 1457–1466. [Google Scholar] [CrossRef][Green Version]
- Pan, X.; Wilson, M.; Mirbahai, L.; McConville, C.; Arvatinis, T.N.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. In vitro metabonomic study detects increases in UDP-GlcNAc and UDP-GalNAc, as early phase markers of cisplatin treatment response in brain tumor cells. J. Proteome Res. 2011, 10, 3493–3500. [Google Scholar] [CrossRef]
- Pan, X.; Wilson, M.; McConville, C.; Arvatinis, T.N.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. Increased unsaturation of lipids in cytoplasmic lipid droplets in DAOY cancer cells in response to cisplatin treatment. Metabolomics 2013, 9, 722–729. [Google Scholar] [CrossRef]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colagno, L.A. Impact of chemotherapy on metabolic reprogramming: Characterization of the metabolic profile of breast cancer MDA-MB-231 cells using 1H HR-MAS NMR spectroscopy. J. Pharm. Biomed. Anal. 2017, 146, 324–328. [Google Scholar] [CrossRef]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colagno, L.A. Effects of doxorubicin, cisplatin, and tamoxifen on the metabolic profile of human breast cancer MCF-7 cells as determined by 1H high-resolution magic angle spinning nuclear magnetic resonance. Biochemistry 2017, 56, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Del Coco, L.; Majellaro, M.; Boccarelli, A.; Cellamare, S.; Altomare, C.D.; Fanizzi, F.P. Novel antiproliferative biphenyl nicotinamide: NMR metabolomic study of its effect on the MCF-7 cell in comparison with cisplatin and vinblastine. Molecules 2020, 25, 3502. [Google Scholar] [CrossRef] [PubMed]
- DeCastro, F.; Benedetti, M.; Antonaci, G.; Del Coco, L.; De Pascalo, S.A.; Muscella, A.; Marsigliante, S.; Fanizzi, F.P. Response of cisplatin resistant Skov-3 cells to [Pt(O,O′-Acac)(γ-Acac)(DMS)] treatment revealed by a metabolomic 1H-NMR study. Molecules 2018, 23, 2301. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.F.; Lamego, I.; Marques, J.; Marques, M.P.M.; Blaise, B.J.; Gil, A.M. Nuclear magnetic resonance (NMR) study of the effect of cisplatin on the metabolic profile of MG-63 osteosarcoma cells. J. Proteome Res. 2010, 9, 5877–5886. [Google Scholar] [CrossRef] [PubMed]
- Lamego, I.; Duarte, I.F.; Marques, M.P.M.; Gil, A.M. Metabolic markers of MG-63 osteosarcoma cell line response to doxorubicin and methotrexate treatment: Comparison to cisplatin. J. Proteome Res. 2014, 19, 781–793. [Google Scholar] [CrossRef]
- Lin, C.; Dong, J.; Wei, Z.; Cheng, K.; Lie, J.; You, S.; Liu, Y.; Wang, X.; Chen, Z. 1H NMR-based metabolic profiles delineate the anticancer effect of vitamin C and oxaliplatin on hepatocellular carcinoma cells. J. Proteome Res. 2020, 13, 6033–6045. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Feng, Y.; Zhou, W.; Zhang, J.; Yu, Y.; Wang, X.; Zhang, P. 1H nuclear magnetic resonance-based extracellular metabolomic analysis of multidrug resistant Tca8113 oral squamous carcinoma cells. Oncol. Lett. 2015, 9, 2551–2559. [Google Scholar] [CrossRef]
- Santini, M.T.; Rainaldi, G.; Romano, R.; Ferrante, A.; Clemente, S.; Motta, A.; Indovina, P.L. MG-63 human osteosarcoma cells grown in monolayer and as three-dimensional tumor spheroids present a different metabolic profile: A 1H NMR study. FEBS Lett. 2004, 557, 148–154. [Google Scholar] [CrossRef]
- Santini, M.T.; Romano, R.; Rainaldi, G.; Ferrante, A.; Indovina, P.; Motta, A.; Indovina, P.L. 1H-NMR Evidence for a different response to the same dose (2 Gy) of ionizing radiation of MG-63 human osteosarcoma cells and three-dimensional spheroids. Anticancer Res. 2006, 26, 267–282. [Google Scholar]
- Naeni, M.B.; Bianconi, V.; Pirro, M.; Sahebkar, A. The role of phosphatidylserine recognition receptors in multiple biological functions. Cell. Mol. Biol. Lett. 2020, 25, 23. [Google Scholar] [CrossRef] [PubMed]
- Arfin, S.; Jha, N.K.; Jha, S.K.; Kesari, K.K.; Ruokolainen, J.; Roychoudhury, S.; Rathi, B.; Kumar, D. Oxidative stress in cancer cell metabolism. Antioxidants 2021, 10, 642. [Google Scholar] [CrossRef]
- Bizzari, M.; Dinicola, S.; Bevilacqua, A.; Cucina, A. Broad spectrum anticancer activity of myo-inositol and inositol hexakisphosphate. Int. J. Endocrinol. 2016, 2016, 5616807. [Google Scholar] [CrossRef]
- Oza, S.L.; Tetaud, E.; Ariyanayagam, M.R.; Warnon, S.S.; Fairlamb, A.H. A single enzyme catalyses formation of trypanothione from glutathione and spermidine in trypanosoma cruzi. J. Biol. Chem. 2002, 277, 35853–35861. [Google Scholar] [CrossRef] [PubMed]
- Bruno, P.M.; Liu, Y.; Park, G.Y.; Murai, J.; Koch, C.E.; Eisen, T.J.; Pritchard, J.R.; Pommier, Y.; Lippard, S.J.; Hemann, M.T. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 2017, 23, 461–471. [Google Scholar] [CrossRef]
- Bruton, V.G.; Grant, M.H.; Wallace, H.M. Spermine toxicity and glutathione depletion in BHK-21/C13 cells. Biochem. Pharmacol. 1990, 40, 1893–1990. [Google Scholar] [CrossRef]
- Codina, G.; Caubet, A.; Lopez, C.; Moreno, V.; Molins, E. Palladium (II) and Platinum (II) Polyamine Complexes: X-Ray Crystal Structures of (SP-4-2)-Chloro {N-[(3-amino-κN) propyl] propane-1, 3-diamine-κN, κN′} palladium (1+) Tetrachloropalladate (2–)(2: 1) and (R, S)-Tetrachloro [μ-(spermine)] dipalladium (II)(={μ-{N, N′-Bis [(3-amino-κN) propyl] butane-1, 4-diamine-κN: κN′}} tetrachlorodipalladium). Helv. Chim. Acta 1999, 82, 1025–1037. [Google Scholar]
- Demmter, K. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: Optimization of harvesting and extraction protocols. Anal. Bioanal. Chem. 2011, 399, 1127–1139. [Google Scholar]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Yang, K.; Yang, L.; Miao, Z.; Wang, Y.; Zhu, H. A 1H NMR-based metabonomic investigation of time-related metabolic trajectories of the plasma, urine and liver extracts of hyperlipidemic hamsters. PLoS ONE 2013, 8, e66786. [Google Scholar] [CrossRef]
- Gowda, G.A.N. Profiling redox and energy coenzymes in whole blood, tissue and cells using NMR spectroscopy. Metabolites 2018, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0—The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Veselkov, K.A. Recursive segment-wise peak alignment of biological 1H NMR spectra for improved metabolic biomarker recovery. Anal. Chem. 2009, 81, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.M.; Engberg, S. Effect size estimation: Methods and examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Ranstam, J. Multiple p-values and Bonferroni correction. Osteoarthr. Cartil. 2016, 24, 763–764. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, A.S.; Batista de Carvalho, A.L.M.; Marques, M.P.M.; Gil, A.M. Response of Osteosarcoma Cell Metabolism to Platinum and Palladium Chelates as Potential New Drugs. Molecules 2021, 26, 4805. https://doi.org/10.3390/molecules26164805
Martins AS, Batista de Carvalho ALM, Marques MPM, Gil AM. Response of Osteosarcoma Cell Metabolism to Platinum and Palladium Chelates as Potential New Drugs. Molecules. 2021; 26(16):4805. https://doi.org/10.3390/molecules26164805
Chicago/Turabian StyleMartins, Ana S., Ana L. M. Batista de Carvalho, Maria P. M. Marques, and Ana M. Gil. 2021. "Response of Osteosarcoma Cell Metabolism to Platinum and Palladium Chelates as Potential New Drugs" Molecules 26, no. 16: 4805. https://doi.org/10.3390/molecules26164805
APA StyleMartins, A. S., Batista de Carvalho, A. L. M., Marques, M. P. M., & Gil, A. M. (2021). Response of Osteosarcoma Cell Metabolism to Platinum and Palladium Chelates as Potential New Drugs. Molecules, 26(16), 4805. https://doi.org/10.3390/molecules26164805







