Optimization of Multiparameters for Increased Yields of Cytochrome B5 in Bioreactors
Abstract
1. Introduction
2. Results
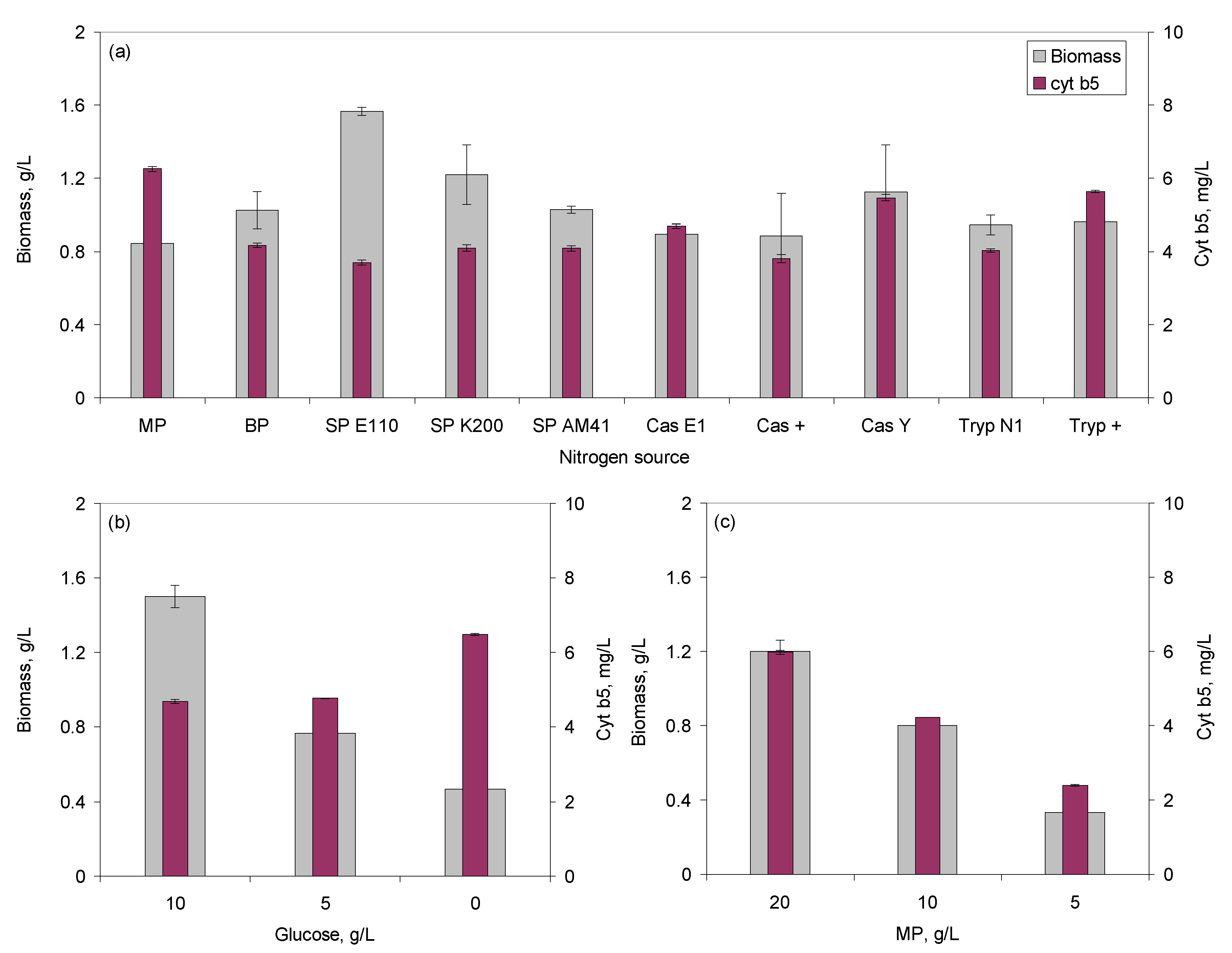
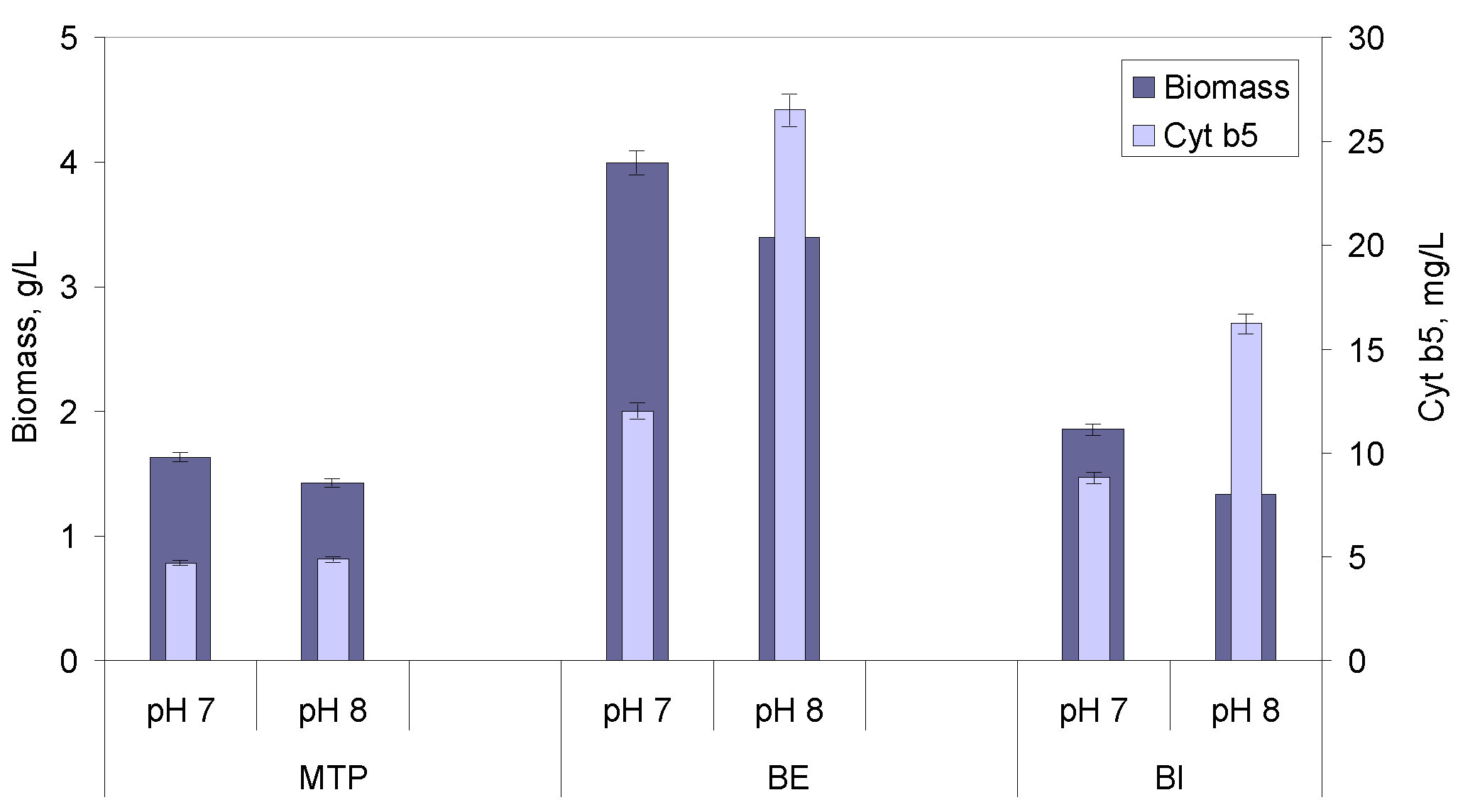
2.1. Media Optimization Using Microtitre Plates
2.2. Scale-Up to Bioreactor
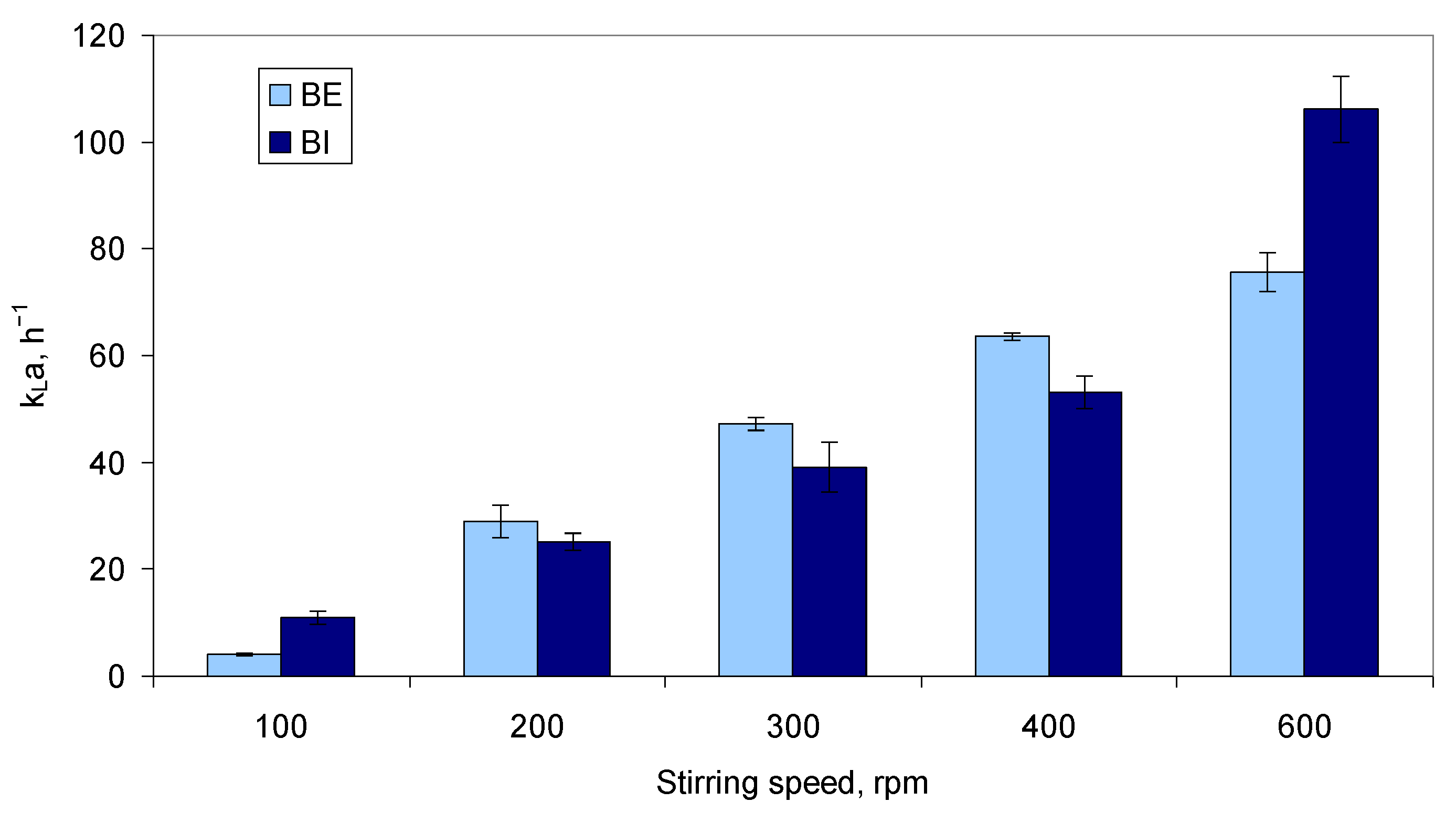
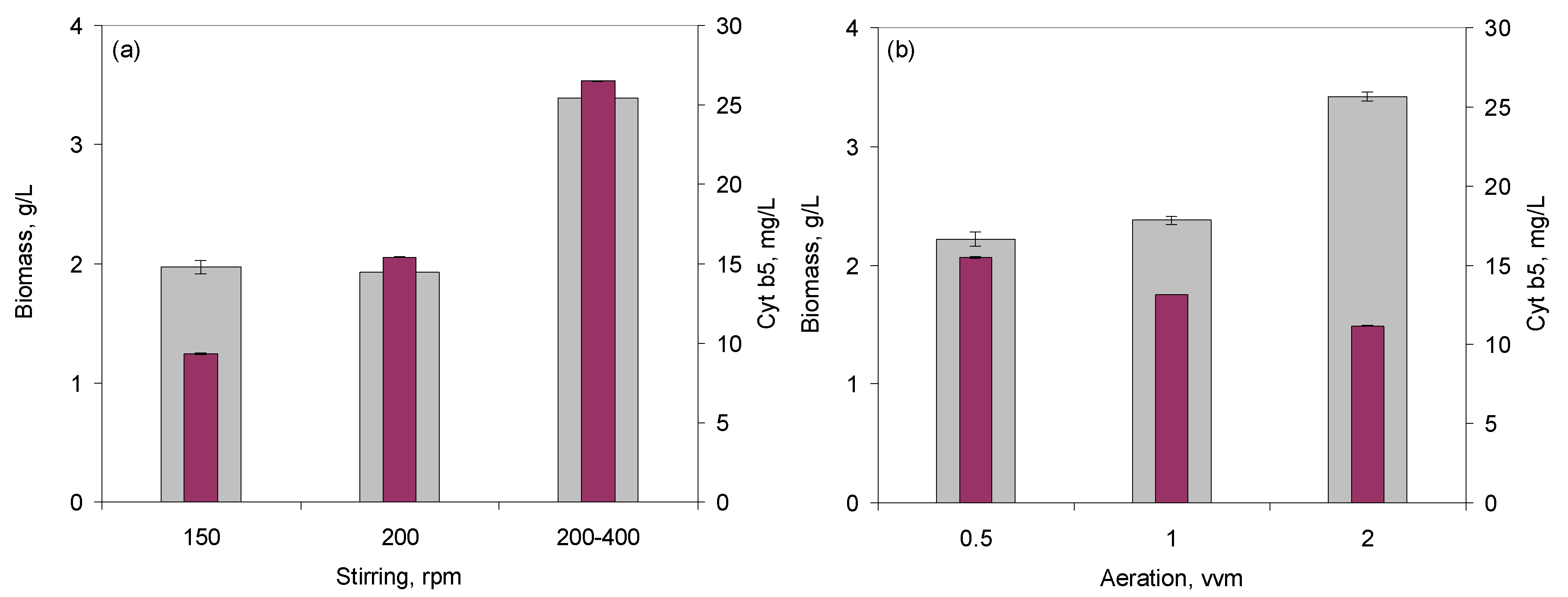
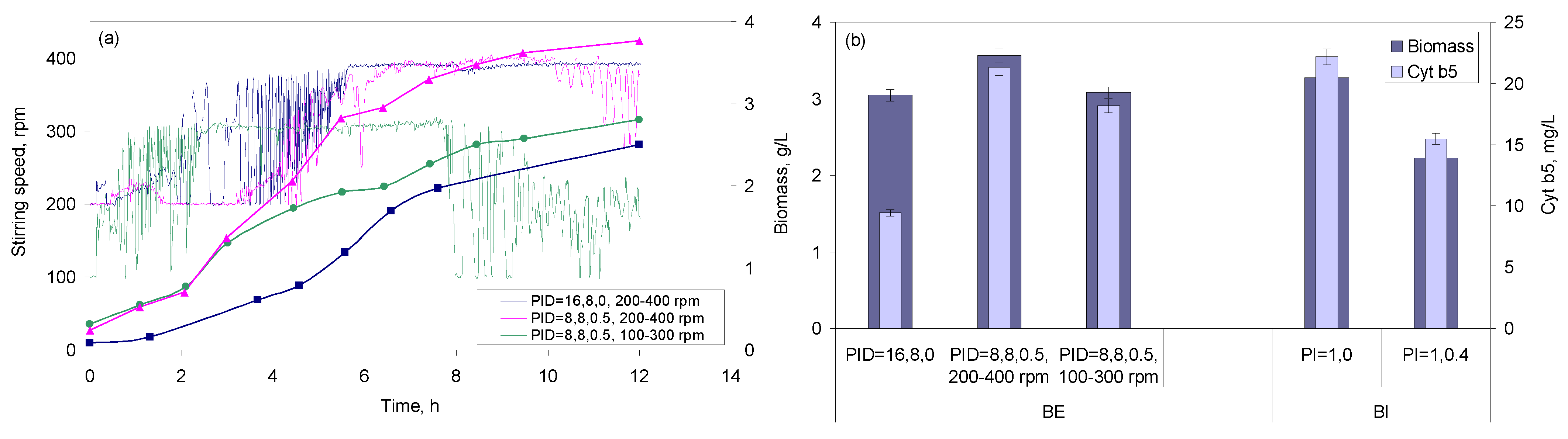
2.2.1. Effect of Stirring, Air Flow, and PID Control
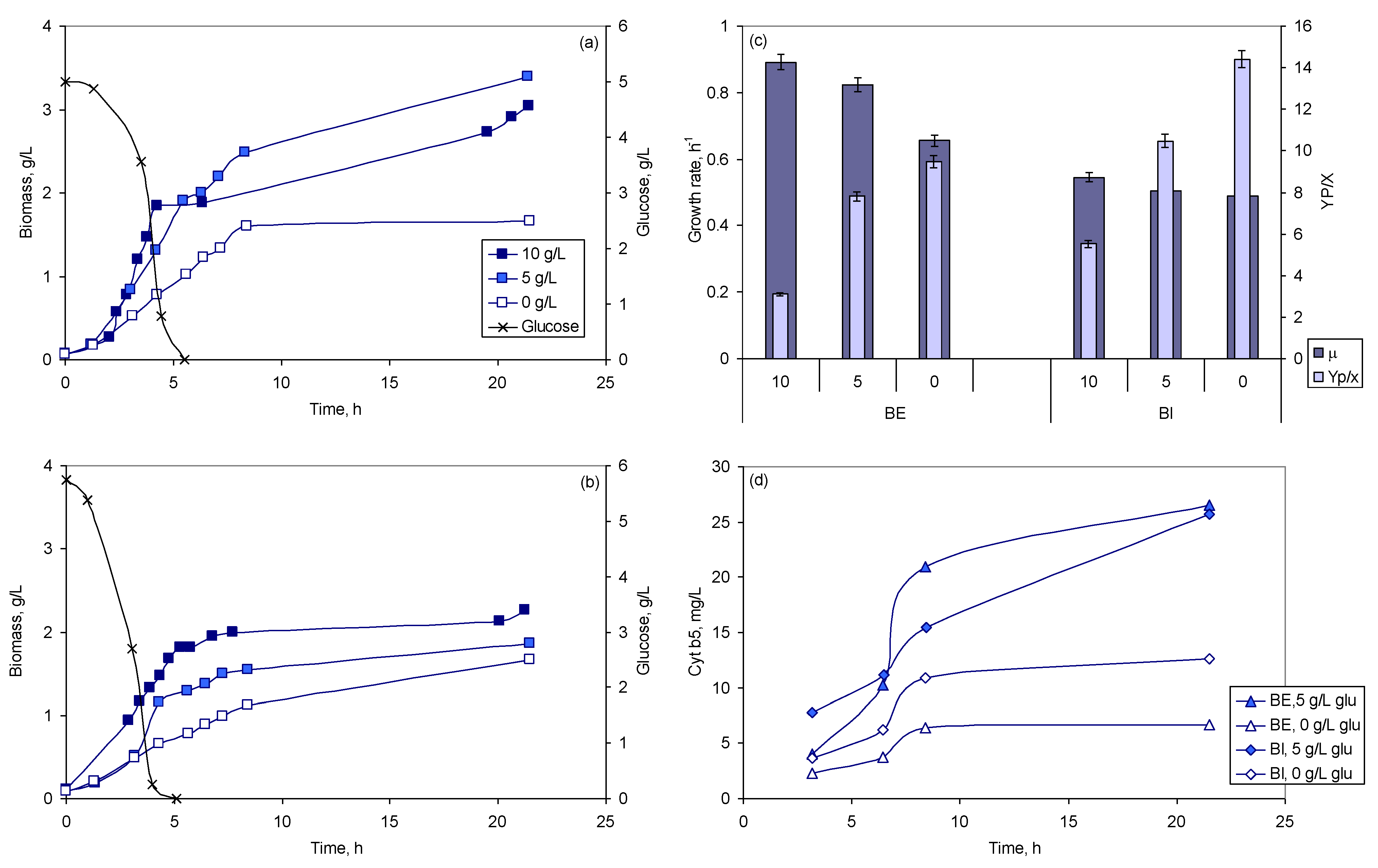
2.2.2. Effect of Glucose Concentration
2.2.3. Effect of pH
2.2.4. Multivariate Analysis of the Different Conditions
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Growth Conditions
4.2. Effect of Media Composition and pH
4.3. Cell Growth in 2 L Bioreactors
4.4. Analytical Methods
4.5. Analytical Methods
4.6. Multivariate Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sanborn, R.; William, C.M. The cytochrome system in the cecropia silkworm, with special reference to the properties of a new component. J. Gen. Physiol. 1950, 33, 579–588. [Google Scholar] [CrossRef]
- Vergères, G.; Waskell, L. Cytochrome b5, its functions, structure and membrane topology. Biochimie 1995, 77, 604–620. [Google Scholar] [CrossRef]
- Gostinčar, C.; Turk, M.; Gunde-Cimerman, N. The evolution of fatty acid desaturases and cytochrome b5 in eukaryotes. J. Membr. Biol. 2010, 233, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Runnegar, B. Derivation of the globins from type b cytochromes. J. Mol. Evol. 1984, 21, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Reid, E.L.; Weynberg, K.D.; Love, J.; Isupov, M.N.; Littlechild, J.A.; Wilson, W.H.; Kelly, S.L.; Lamb, D.C.; Allen, M.J. Functional and structural characterisation of a viral cytochrome b5. FEBS Lett. 2013, 587, 3633–3639. [Google Scholar] [CrossRef]
- Von Bodman, S.B.; Schuler, M.A.; Jollie, D.R.; Sligar, S.G. Synthesis, bacterial expression, and mutagenesis of the gene coding for mammalian cytochrome b5. Proc. Natl. Acad. Sci. USA 1986, 83, 9443–9447. [Google Scholar] [CrossRef]
- Porter, T.D. The roles of cytochrome b5 in cytochrome P450 reactions. J. Biochem. Mol. Toxicol. 2002, 16, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Reid, L.S.; Gray, H.B.; Dalvit, C.; Wright, P.E.; Saltman, P. Electron transfer from cytochrome b5 to iron and copper complexes. Biochemistry 1987, 26, 7102–7107. [Google Scholar] [CrossRef]
- Borgese, N.; Gazzoni, I.; Barberi, M.; Colombo, S.; Pedrazzini, E. Targeting of a tail-anchored protein to endoplasmic reticulum and outer membrane by independent but competing pathways. Mol. Biol. Cell 2001, 12, 2482–2496. [Google Scholar] [CrossRef]
- Lederer, F.; Ghrir, R.; Cortial, S.; Ito, A. Two homologous cytochromes b(5) in a single cell. Eur. J. Biochem. 1983, 132, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Schenkman, J.B.; Jansson, I. The many roles of cytochrome b5. Pharmacol. Ther. 2003, 97, 139–152. [Google Scholar] [CrossRef]
- Martin-Montalvo, A.; Sun, Y.; Diaz-Ruiz, A.; Ali, A.; Gutierrez, V.; Palacios, H.H.; Curtis, J.; Siendones, E.; Ariza, J.; Abulwerdi, G.A.; et al. Cytochrome b5 reductase and the control of lipid metabolism and healthspan. NPJ Aging Mech. Dis. 2016, 2, 16006. [Google Scholar] [CrossRef]
- Elahian, F.; Sepehrizadeh, Z.; Moghimi, B.; Mirzaei, S.A. Human cytochrome b5 reductase: Structure, function, and potential applications. Crit. Rev. Biotechnol. 2014, 34, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.J.; McLaughlin, L.A.; Wolf, C.R. Evidence that cytochrome b5 and cytochrome b5 reductase can act as sole electron donors to the hepatic cytochrome P450 system. Mol. Pharmacol. 2013, 83, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Stiborová, M.; Indra, R.; Moserová, M.; Frei, E.; Schmeiser, H.H.; Kopka, K.; Philips, D.H.; Arlt, V.M. NADH:Cytochrome b5 reductase and cytochrome b5 can act as sole electron donors to human cytochrome P450 1A1-mediated oxidation and DNA adduct formation by benzo[a]pyrene. Chem. Res. Toxicol. 2016, 29, 1325–1334. [Google Scholar] [CrossRef]
- Kurian, J.R.; Bajad, S.U.; Miller, J.L.; Chin, N.A.; Trepanier, L.A. NADH cytochrome b5 reductase and cytochrome b5 catalyze the microsomal reduction of xenobiotic hydroxylamines and amidoximes in humans. J. Pharmacol. Exp. Ther. 2004, 311, 1171–1178. [Google Scholar] [CrossRef]
- Henderson, C.J.; McLaughlin, L.A.; Finn, R.D.; Ronseaux, S.; Kapelyukh, Y.; Wolf, C.R. A role for cytochrome b5 in the in vivo disposition of anticancer and cytochrome P450 probe drugs in mice. Drug Metab. Dispos. 2014, 42, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Krishnarjuna, B.; Ravula, T.; Ramamoorthy, A. Detergent-free extraction, reconstitution and characterization of membrane-anchored cytochrome-b5 in native lipids. Chem. Commun. 2020, 56, 6511–6514. [Google Scholar] [CrossRef]
- Liu, Y.; Denisov, I.G.; Sligar, S.G.; Kincaid, J.R. Substrate-specific allosteric effects on the enhancement of CYP17A1 lyase efficiency by cytochrome b5. J. Am. Chem. Soc. 2021, 143, 3729–3733. [Google Scholar] [CrossRef]
- Zhang, M.; Le Clair, S.V.; Huang, R.; Ahuja, S.; Im, S.-C.; Waskell, L.; Ramamoorthy, A. Insights into the role of substrates on the interaction between Cytochrome b5 and Cytochrome P450 2B4 by NMR. Sci. Rep. 2015, 5, 8392. [Google Scholar] [CrossRef]
- Reed, L.; Indra, R.; Mrizova, I.; Moserova, M.; Schmeiser, H.H.; Wolf, C.R.; Henderson, C.J.; Stiborova, M.; Phillips, D.H.; Arlt, V.M. Application of hepatic cytochrome b 5 /P450 reductase null (HBRN) mice to study the role of cytochrome b 5 in the cytochrome P450-mediated bioactivation of the anticancer drug ellipticine. Toxicol. Appl. Pharmacol. 2019, 366, 64–74. [Google Scholar] [CrossRef]
- Kim, D.; Kim, V.; McCarty, K.D.; Guengerich, F.P. Tight binding of cytochrome b5 to cytochrome P450 17A1 is a critical feature of stimulation of C21 steroid lyase activity and androgen synthesis. J. Biol. Chem. 2021, 296. [Google Scholar] [CrossRef]
- Emoto, C.; Iwasaki, K. Approach to predict the contribution of cytochrome P450 enzymes to drug metabolism in the early drug-discovery stage: The effect of the expression of cytochrome b5 with recombinant P450 enzymes. Xenobiotica 2007, 37, 986–999. [Google Scholar] [CrossRef] [PubMed]
- Chudaev, M.V.; Usanov, S.A. Expression of functionally active cytochrome b5 in Escherichia coli: Isolation, purification, and use of the immobilized recombinant heme protein for affinity chromatography of electron-transfer proteins. Biochemistry 1997, 62, 401–411. [Google Scholar] [PubMed]
- Tambourgi, E.B.; Fischer, G.A.; Fileti, A.M.F. Neural modeling for cytochrome b5 extraction. Process. Biochem. 2006, 41, 1272–1275. [Google Scholar] [CrossRef]
- Belo, I.; Mota, M. Batch and fed-batch cultures of E. coli TB1 at different oxygen transfer rates. Bioprocess. Eng. 1998, 18, 451–455. [Google Scholar] [CrossRef]
- Santos, J.A.; Belo, I.; Mota, M.; Cabral, J.M. Freeze/thawing and sonication of Escherichia coli TB1 cells for cytochrome b5 recovery. Bioseparation 1996, 6, 81–89. [Google Scholar]
- Gomes, A.M.V.; Carmo, T.S.; Carvalho, L.S.; Bahia, F.M.; Parachin, N.S. Comparison of yeasts as hosts for recombinant protein production. Microorganisms 2018, 6, 38. [Google Scholar] [CrossRef]
- Gupta, S.K.; Shukla, P. Sophisticated cloning, fermentation, and purification technologies for an enhanced therapeutic protein production: A review. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef]
- Taipa, M.Â.; Fernandes, P.; de Carvalho, C.C.C.R. Production and purification of therapeutic enzymes. In Therapeutic Enzymes: Function and Clinical Implications; Labrou, N., Ed.; Springer: Singapore, 2019; pp. 1–24. [Google Scholar]
- Saez, N.J.; Vincentelli, R. High-throughput expression screening and purification of recombinant proteins in E. coli. In Structural Genomics: General Applications; Chen, Y.W., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 33–53. [Google Scholar]
- Gutiérrez-González, M.; Farías, C.; Tello, S.; Pérez-Etcheverry, D.; Romero, A.; Zúñiga, R.; Ribeiro, C.H.; Lorenzo-Ferreiro, C.; Molina, M.C. Optimization of culture conditions for the expression of three different insoluble proteins in Escherichia coli. Sci. Rep. 2019, 9, 16850. [Google Scholar] [CrossRef]
- Gronemeyer, P.; Ditz, R.; Strube, J. Trends in Upstream and Downstream process development for antibody manufacturing. Bioengineering 2014, 1, 188–212. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.K.; Shrivastava, A. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Baneyx, F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999, 10, 411–421. [Google Scholar] [CrossRef]
- Makrides, S.C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996, 60, 512–538. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Murado, M.A. Mathematical tools for objective comparison of microbial cultures: Application to evaluation of 15 peptones for lactic acid bacteria productions. Biochem. Eng. J. 2008, 39, 276–287. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Denham, M.C. Implementing partial least squares. Stat. Comput. 1995, 5, 191–202. [Google Scholar] [CrossRef]
- Tripathi, N.K. Production and purification of recombinant proteins from Escherichia coli. ChemBioEng Rev. 2016, 3, 116–133. [Google Scholar] [CrossRef]
- Nancib, N.; Branlant, C.; Boudrant, J. Metabolic roles of peptone and yeast extract for the culture of a recombinant strain of Escherichia coli. J. Ind. Microbiol. 1991, 8, 165–169. [Google Scholar] [CrossRef]
- Schär-Zammaretti, P.; Dillmann, M.-L.; D’Amico, N.; Affolter, M.; Ubbink, J. Influence of fermentation medium composition on physicochemical surface properties of Lactobacillus acidophilus. Appl. Environ. Microbiol. 2005, 71, 8165–8173. [Google Scholar] [CrossRef]
- Hettmann, T.; Schmidt, C.L.; Anemüller, S.; Zähringer, U.; Moll, H.; Petersen, A.; Schäfer, G. Cytochrome b 558/566 from the Archaeon Sulfolobus acidocaldarius: A novel highly glycosylated, membrane-bound b-type hemoprotein. J. Biol. Chem. 1998, 273, 12032–12040. [Google Scholar] [CrossRef]
- Konz, J.O.; King, J.; Cooney, C.L. Effects of oxygen on recombinant protein expression. Biotechnol. Prog. 1998, 14, 393–409. [Google Scholar] [CrossRef]
- Vail, R.B.; Homann, M.J.; Hanna, I.; Zaks, A. Preparative synthesis of drug metabolites using human cytochrome P450s 3A4, 2C9 and 1A2 with NADPH-P450 reductase expressed in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2005, 32, 67–74. [Google Scholar] [CrossRef]
- Zhang, J.D.; Li, A.T.; Xu, J.H. Improved expression of recombinant cytochrome P450 monooxygenase in Escherichia coli for asymmetric oxidation of sulfides. Bioprocess. Biosyst. Eng. 2010, 33, 1043–1049. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Caramujo, M.-J. Fatty acids as a tool to understand microbial diversity and their role in food webs of Mediterranean temporary ponds. Molecules 2014, 19, 5570–5598. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Pons, M.-n.; da Fonseca, M.M.R. Principal Components Analysis as a tool to summarise biotransformation data: Influence on cells of solvent type and phase ratio. Biocatal. Biotransformation 2003, 21, 305–314. [Google Scholar] [CrossRef]
- De Alwis, D.M.; Dutton, R.L.; Scharer, J.; Moo-Young, M. Statistical methods in media optimization for batch and fed-batch animal cell culture. Bioprocess. Biosyst. Eng. 2007, 30, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.A.; Menezes, J.C. Industrial fermentation end-product modelling with multilinear PLS. Chemom. Intell. Lab. Syst. 2003, 68, 75–81. [Google Scholar] [CrossRef]
- Patel, N.; Corbett, B.; Trygg, J.; McCready, C.; Mhaskar, P. Subspace based model identification for an industrial bioreactor: Handling infrequent sampling using missing data algorithms. Processes 2020, 8, 1686. [Google Scholar] [CrossRef]
- Burke, P.V.; Kwast, K.E.; Everts, F.; Poyton, R.O. A fermentor system for regulating oxygen at low concentrations in cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1998, 64, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Åkesson, M.; Hagander, P. Control of Dissolved Oxygen in Stirred Bioreactors; Technical Reports TFRT-7571; Lund Institute of Technology (LTH): Lund, Sweden, 1998. [Google Scholar]
- Babuška, R.; Damen, M.R.; Hellinga, C.; Maarleveld, H. Intelligent adaptive control of bioreactors. J. Intell. Manuf. 2003, 14, 255–265. [Google Scholar] [CrossRef]
- Kuprijanov, A.; Gnoth, S.; Simutis, R.; Lübbert, A. Advanced control of dissolved oxygen concentration in fed batch cultures during recombinant protein production. Appl. Microbiol. Biotechnol. 2009, 82, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.P.C.; Walshe, K.; Doyle, S.; Fernandes, P.; de Carvalho, C.C.C.R. Anchoring high-throughput screening methods to scale-up bioproduction of siderophores. Process. Biochem. 2012, 47, 416–421. [Google Scholar] [CrossRef][Green Version]
- Tobajas, M.; García-Calvo, E. Comparison of experimental methods for determination of the volumetric mass transfer coefficient in fermentation processes. Heat Mass Transf. 2000, 36, 201–207. [Google Scholar] [CrossRef]
- Garcia-Ochoa, F.; Gomez, E. Bioreactor scale-up and oxygen transfer rate in microbial processes: An overview. Biotechnol. Adv. 2009, 27, 153–176. [Google Scholar] [CrossRef]
- Cesário, M.T.; Raposo, R.S.; de Almeida, M.C.M.D.; van Keulen, F.; Ferreira, B.S.; da Fonseca, M.M.R. Enhanced bioproduction of poly-3-hydroxybutyrate from wheat straw lignocellulosic hydrolysates. New Biotechnol. 2014, 31, 104–113. [Google Scholar] [CrossRef]

| Bioreactor Code | BE | BI |
|---|---|---|
| Model | Fermac 360 (Electrolab) | Minifors (Infors HT) |
| Volume (L) | 2.6 | 2.5 |
| Diameter (cm) | 12.0 | 11.0 |
| Height (cm) | 23.0 | 26.3 |
| Height/Diameter ratio | 1.9 | 2.4 |
| Baffles | ||
| Number | 2 | 3 |
| Height (cm) | 15.4 | 21.0 |
| Width (cm) | 1.0 | 1.5 |
| Impellers | ||
| Type | Rushton turbine | Rushton turbine |
| Number | 2 | 2 |
| Diameter (cm) | 5.7 | 4.7 |
| Blade height (cm) | 1.6 | 1.1 |
| Blade width (cm) | 1.7 | 1.0 |
| Bioreactor/Condition | H/D | vvm | Stirring | pH | Glu, g/L | Amp, ug/mL | P | I | D | X, g/L | Cit, mg/L | YP/X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BE/1 | 1.9 | 0.5 | cascade | 7 | 10 | 100 | 8 | 8 | 0.5 | 3.57 | 21.28 | 5.96 |
| BE/2 | 1.9 | 1 | cascade | 7 | 10 | 100 | 16 | 8 | 0 | 2.77 | 30.70 | 11.08 |
| BI/1 | 2.4 | 0.5 | cascade | 7 | 10 | 100 | 1 | 0 | 0 | 4.69 | 36.02 | 7.68 |
| BI/2 | 2.4 | 1 | cascade | 7 | 10 | 100 | 1 | 0 | 0 | 2.91 | 72.72 | 24.97 |
| Name | Abbreviation | Concentration (g/L) | Supplier |
|---|---|---|---|
| BactoTM Yeast Extract | Yeast extract | 5 | BD Biosciences, San Jose, CA, USA |
| Sodium chloride | NaCl | 5 | Panreac, AppliChem GmbH, Darmstadt, Germany |
| D(+)-Glucose | Glu | 10 | Merck, Darmstadt, Germany |
| Ampicillin sodium salt | Amp | 0.1 | Sigma-Aldrich, St. Louis, MO, USA |
| Nitrogen source tested | |||
| Meat Peptone, enzymatic digest for microbiology | MP | 10 | Millipore Sigma, St. Louis, MO, USA |
| Casein Yeast Peptone | Cas Y | ||
| Bacteriological Peptone | BP | Thermo Fisher Scientific, Inc., Waltham, MA, USA | |
| Soy Peptone | SP E110 | OrganoTechnie, SAS, La Courneuve, France | |
| SP K200 | |||
| SP AM41 | |||
| Casein Peptone | Cas E1 | ||
| Cas + | |||
| Tryptone | Tryp N1 | ||
| Tryp + | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, R.F.S.; de Carvalho, C.C.C.R. Optimization of Multiparameters for Increased Yields of Cytochrome B5 in Bioreactors. Molecules 2021, 26, 4148. https://doi.org/10.3390/molecules26144148
Pereira RFS, de Carvalho CCCR. Optimization of Multiparameters for Increased Yields of Cytochrome B5 in Bioreactors. Molecules. 2021; 26(14):4148. https://doi.org/10.3390/molecules26144148
Chicago/Turabian StylePereira, Ricardo F. S., and Carla C. C. R. de Carvalho. 2021. "Optimization of Multiparameters for Increased Yields of Cytochrome B5 in Bioreactors" Molecules 26, no. 14: 4148. https://doi.org/10.3390/molecules26144148
APA StylePereira, R. F. S., & de Carvalho, C. C. C. R. (2021). Optimization of Multiparameters for Increased Yields of Cytochrome B5 in Bioreactors. Molecules, 26(14), 4148. https://doi.org/10.3390/molecules26144148







