Cheminformatic Profiling and Hit Prioritization of Natural Products with Activities against Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Descriptors and Physicochemical Properties of AMNPs and CDs
2.1.1. Molecular Weight
2.1.2. Calculated Octanol/Water Partition Coefficient (cLogP)
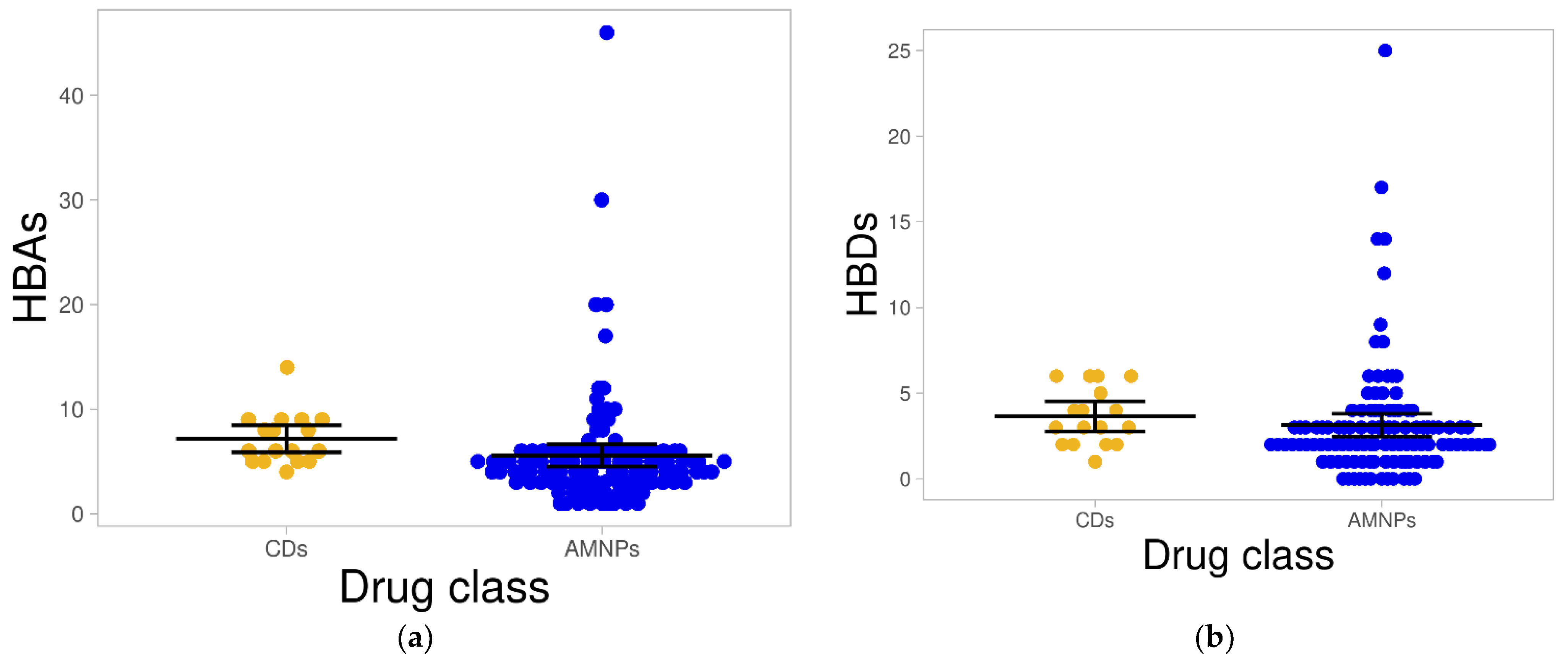
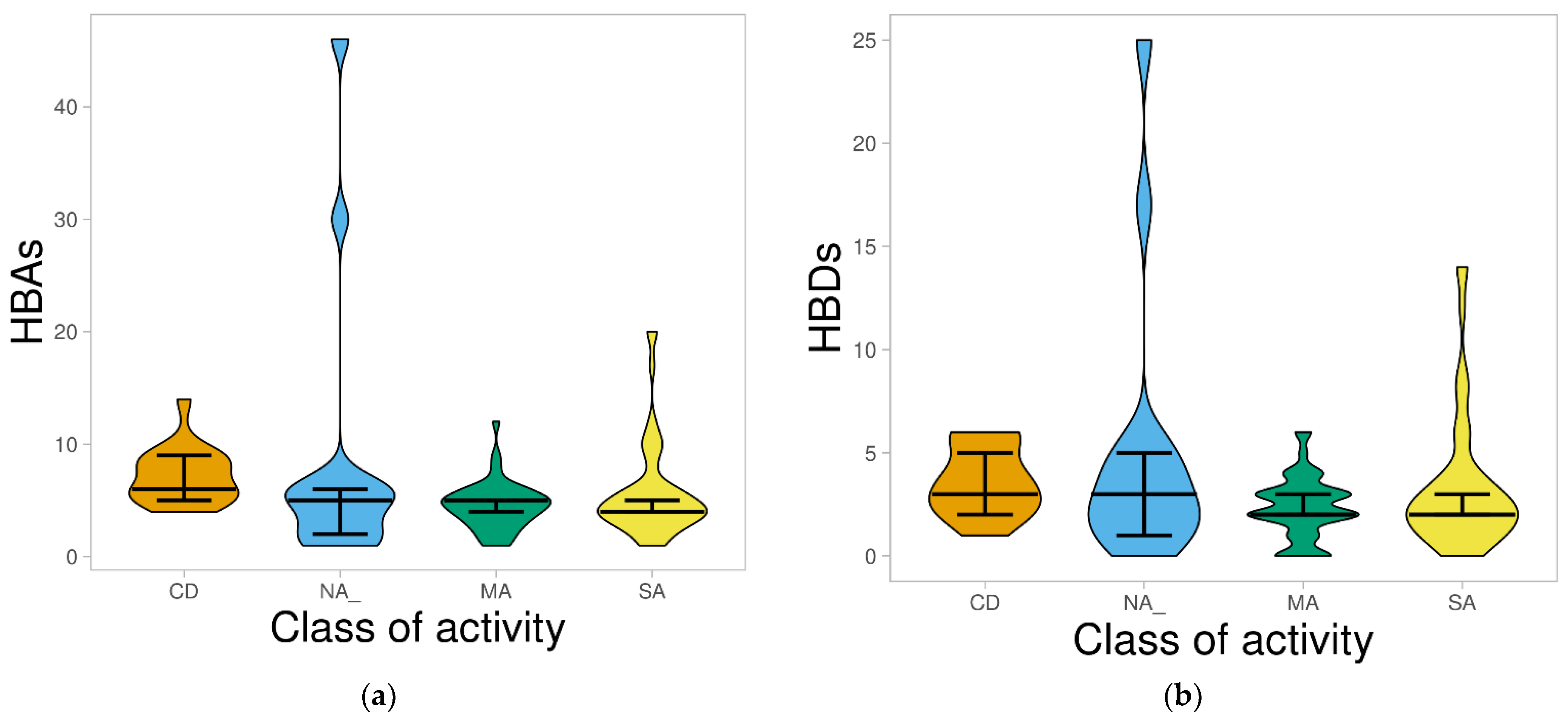
2.1.3. Hydrogen Bond Acceptors and Donors
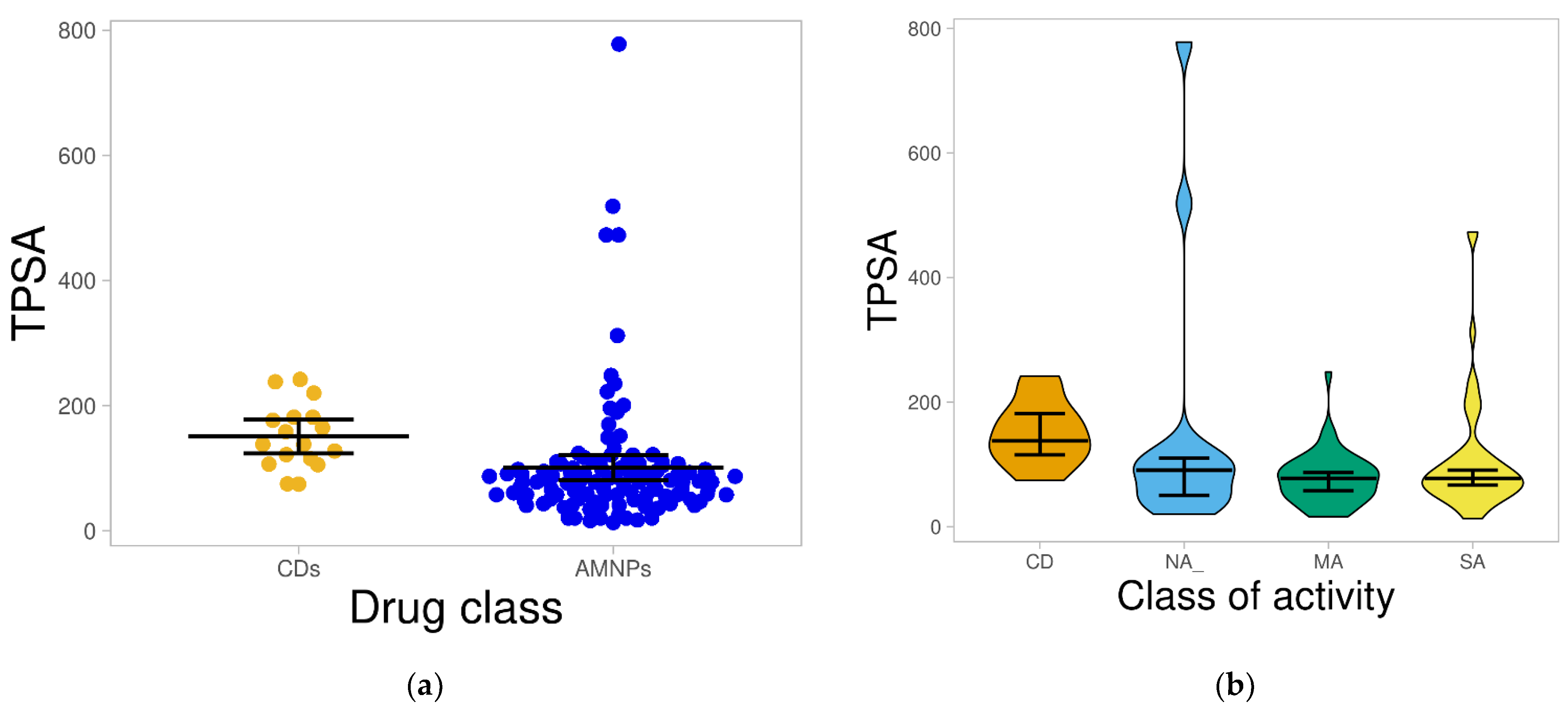
2.1.4. Total Polar Surface Area (TPSA)
2.1.5. Rotatable Bond (RTB) Count
2.2. Profiling Drug-Likeness of AMNPs
2.2.1. Prediction of Absorption and Distribution
2.2.2. Predicted Metabolism of AMNPs and Identification of Their Metabolites
2.2.3. Predicted CYP450 Inhibitory Potential of AMNPs
2.2.4. Toxicity Profiling of AMNPs
2.3. Synthetic Accessibility Score
2.4. Hit Prioritization of AMNPs
2.5. Desirability Scoring Function Allows In Silico Hit Optimization Strategies
2.6. Exploration of the Molecular Similarity/Diversity within the AMNPs
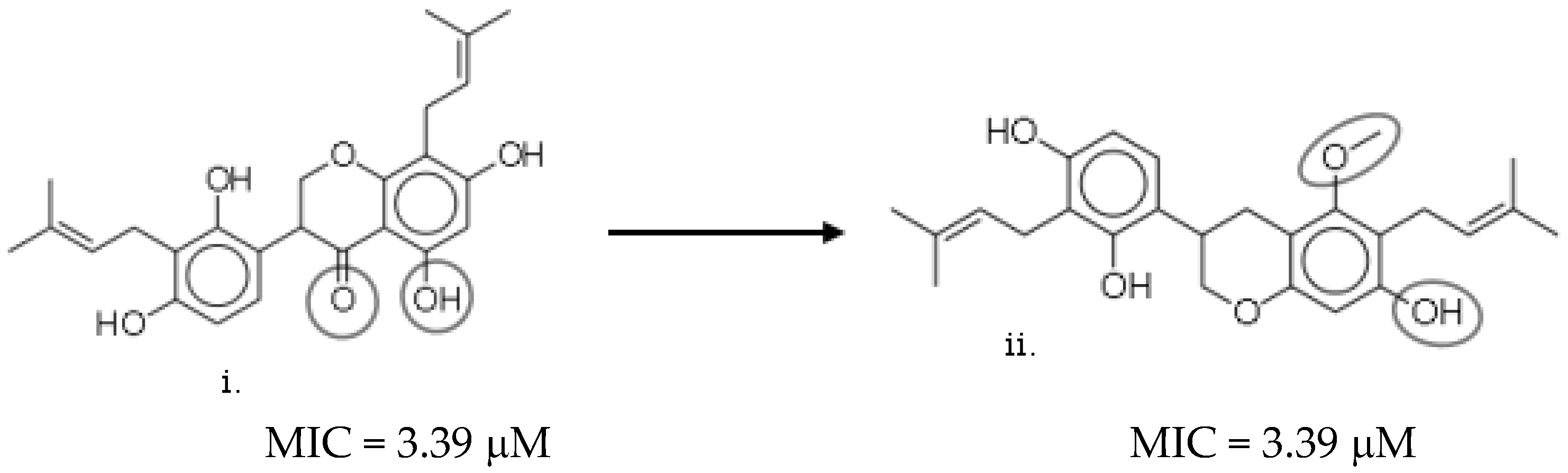
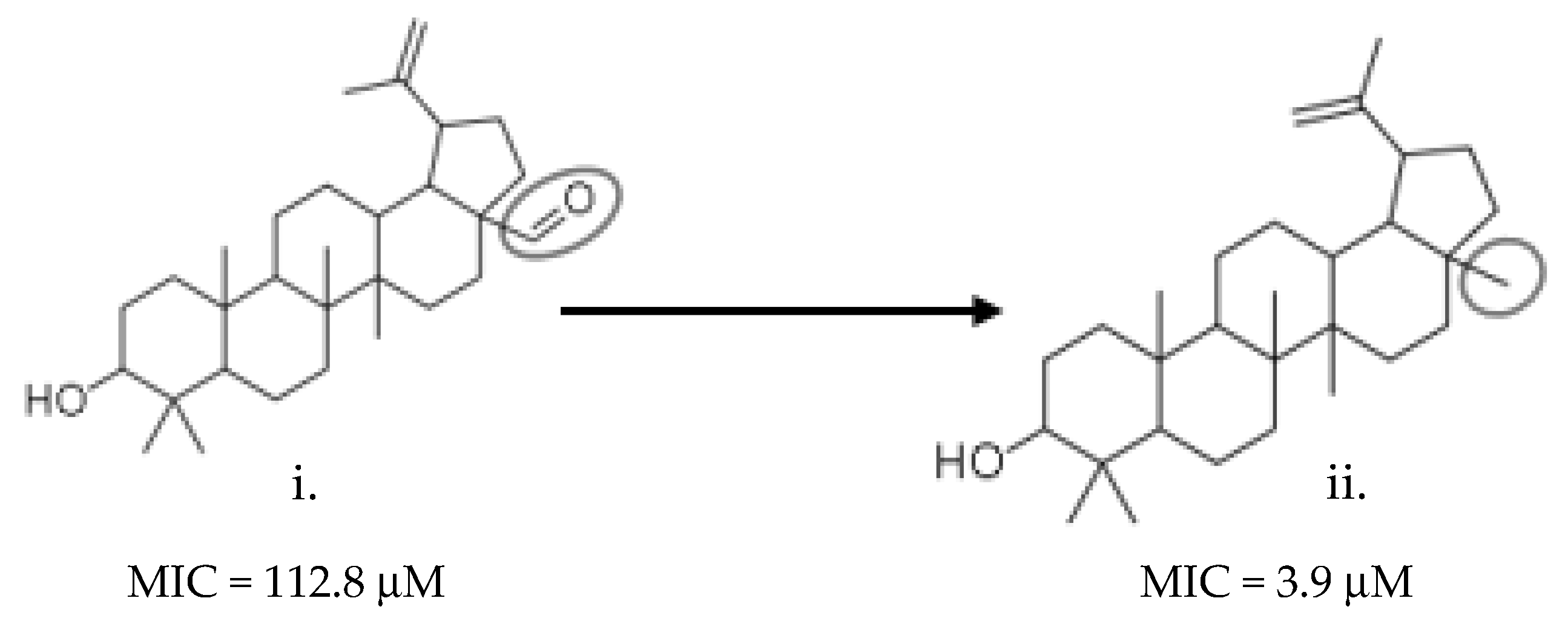
2.7. Structure–Activity Relationship (SAR) Landscape
3. Materials and Methods
3.1. Data Collection and Preparation
3.2. Estimation of Molecular Descriptors and Physicochemical and Pharmacokinetic Properties of the Datasets
3.3. Exploration of Chemical Space (Chemical Diversity)
3.4. Data Analysis and Visualization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Seebaluck-sandoram, R.; Lall, N.; Fibrich, B.; Blom, A.; Staden, V.; Mahomoodally, F. Antibiotic-potentiating activity, phytochemical profile, and cytotoxicity of Acalypha integrifolia Willd. (Euphorbiaceae). Perspect. Med. 2018, 11, 53–59. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645. [Google Scholar] [CrossRef] [PubMed]
- Monte, J.; Abreu, A.C.; Borges, A.; Simões, L.C.; Simões, M. Antimicrobial Activity of Selected Phytochemicals against Escherichia coli and Staphylococcus aureus and Their Biofilms. Pathogens 2014, 3, 473–498. [Google Scholar] [CrossRef] [PubMed]
- Wikaningtyas, P.; Sukandar, E.Y. The antibacterial activity of selected plants towards resistant bacteria isolated from clinical specimens. Asian Pac. J. Trop. Biomed. 2016, 6, 16–19. [Google Scholar] [CrossRef]
- Krishnamoorthy, R.; Athinarayanan, J.; Periasamy, V.S.; Adisa, A.R.; Al-Shuniaber, M.A.; Gassem, M.A.; Alshatwi, A.A. Microbial Pathogenesis Antimicrobial activity of nanoemulsion on drug-resistant bacterial pathogens. Microb. Pathog. 2018, 120, 85–96. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Medeiros Barreto, H.; Cerqueira Fontinele, F.; Pereira de Oliveira, A.; Arcanjo, D.D.; Cavalcanti Dos Santos, B.H.; de Abreu, A.P.; Douglas Melo Coutinho, H.; Alves Carvalho da Silva, R.; Oliveira de Sousa, T.; Freire de Medeiros, M.D.; et al. Phytochemical Prospection and Modulation of Antibiotic Activity In Vitro by Lippia origanoides H. B. K. in Methicillin Resistant Staphylococcus aureus. Biomed. Res. Int. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Tayel, A.A.; Shaban, S.M.; Moussa, S.H.; Elguindy, N.M.; Diab, A.M.; Mazrou, K.E.; Ghanem, R.A.; El-Sabbagh, S.M. Annals of Agricultural Sciences Bioactivity and application of plant seeds’ extracts to fight resistant strains of Staphylococcus aureus. Ann. Agric. Sci. 2018, 63, 47–53. [Google Scholar] [CrossRef]
- Seyed, M.A. A comprehensive review on Phyllanthus derived natural products as potential chemotherapeutic and immunomodulators for a wide range of human diseases. Biocatal. Agric. Biotechnol. 2019, 17, 529–537. [Google Scholar] [CrossRef]
- Medjeldi, S.; Bouslama, L.; Benabdallah, A.; Essid, R.; Haou, S.; Elkahouib, S. Biological activities, and phytocompounds of northwest Algeria Ajuga iva (L) extracts: Partial identification of the antibacterial fraction. Microb. Pathog. 2018, 121, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Liu, C.; Wang, Q.; Lin, P.; Cheng, F. In silico polypharmacology of natural products. Brief Bioinform. 2018, 19, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Du, J.; Yan, X.; Zhong, J.; Cui, L.; Lin, J.; Zeng, L.; Ding, P.; Chen, P.; Zhou, X.; et al. TCMAnalyzer: A Chemo- and Bioinformatics Web Service for Analyzing Traditional Chinese Medicine. J. Chem. Inf. Model. 2018, 58, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Nazir, M.; Ali, S.; Hussain, H.; Lee, S. Antimicrobial natural products: An update on future antibiotic drug candidates. Nat. Prod. Rep. 2010, 27, 238–254. [Google Scholar] [CrossRef]
- Catteau, L. Natural and hemi-synthetic pentacyclic triterpenes as antimicrobials and resistance modifying agents against Staphylococcus aureus: A review. Phytochem. Rev. 2018, 17, 1129–1163. [Google Scholar] [CrossRef]
- Egieyeh, S.A.; Syce, J.; Malan, S.F.; Christoffels, A. Prioritization of anti-malarial hits from nature: Chemo—Informatic profiling of natural products with in vitro antiplasmodial activities and currently registered anti-malarial drugs. Malar. J. 2016, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Yang, H. ADMET-score—A comprehensive scoring function for evaluation of chemical drug-likeness. Medchemcomm 2019, 10, 148–157. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Lifongo, L.L.; Mbah, J.A.; Owono Owono, L.C.; Megnassan, E.; Mbaze, L.M.; Judson, P.N.; Sippl, W.; Efange, S.M. In silico drug metabolism and pharmacokinetic profiles of natural products from medicinal plants in the Congo basin. Silico Pharmacol. 2013, 1–11. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Onguéné, P.A.; Lifongo, L.L.; Ndom, J.C.; Sippl, W.; Mbaze, L.M.A. The potential of anti-malarial compounds derived from African medicinal plants, part II: A pharmacological evaluation of non-alkaloids and non-terpenoids. Malar. J. 2014, 13, 1–9. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef]
- Doak, B.C.; Kihlberg, J. Drug discovery beyond the rule of 5—Opportunities and challenges. In Expert Opinion on Drug Discovery; Taylor and Francis Ltd: Oxford, UK, 2017; Volume 12, pp. 115–119. [Google Scholar]
- Nalini, C.N.; Deepthi, S.R.; Ramalakshmi, N.; Uma, G. Toxicity risk assesment of isatins. Rasayan J. Chem. 2011, 4, 829–833. [Google Scholar]
- Bhal, S.K. LogP—Making Sense of the Value; Advanced Chemistry Development, Inc.: Toronto, ON, Canada, 2007; pp. 1–4. [Google Scholar]
- Waring, M.J. Lipophilicity in drug discovery. Expert Opin. Drug Discov. 2010, 5, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef]
- Lipinski, C. Physicochemical properties and the discovery of orally active drugs: Technical and people issues. In Proceedings of the Molecular Informatics: Confronting Complexity, Proceedings of the Beilstein-Institut Workshop, Frankfurt, Germany, 13–16 May 2003; pp. 59–78. [Google Scholar]
- Whitty, A.; Zhong, M.; Viarengo, L.; Beglov, D.; Hall, D.R.; Vajda, S. Quantifying the chameleonic properties of macrocycles and other high-molecular-weight drugs. Drug Discov. Today 2016, 21, 712–717. [Google Scholar] [CrossRef]
- Sadgrove, N.J.; Jones, G.L. From petri dish to patient: Bioavailability estimation and mechanism of action for antimicrobial and immunomodulatory natural products. Front. Microbiol. 2019, 10, 2470. [Google Scholar] [CrossRef]
- Craciun, D.; Modra, D.; Isvoran, A. ADME-Tox profiles of some food additives and pesticides ADME-Tox Profiles of Some Food Additives and Pesticides. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2015; p. 040007. [Google Scholar]
- Di, L.; Kerns, E.H. Drug-Like Properties: Concepts, Structure Design and Methods from ADME to Toxicity Optimization, 1st ed.; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Dahlgren, D.; Lennernäs, H. Pharmaceutics Intestinal Permeability and Drug Absorption: Predictive Experimental, Computational and In Vivo Approaches. Pharmaceutics 2019, 11, 411. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Nazarbahjat, N.; Arif, A.; Abdullah, Z. Chwmistry, Synthesis, characterization, drug-likeness properties and determination of the in vitro antioxidant and cytotoxic activities of new 1, 3, 4-oxadiazole derivatives. Med. Chem. Res. 2016, 25, 2015–2029. [Google Scholar] [CrossRef]
- Prachayasittikul, V.; Prachayasittikul, V. P-glycoprotein transporter in drug development. EXCLI J. 2016, 15, 113–118. [Google Scholar] [PubMed]
- Wen, H.; Jung, H.; Li, X. Drug Delivery Approaches in Addressing Clinical Pharmacology-Related Issues: Opportunities and Challenges. AAPS J. 2015, 17, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Mannan, A.; Unnisa, F. Role of biochemicals and other factors affecting biotransformation and drug absorption. Pharma. Innov. J. 2019, 8, 198–206. [Google Scholar]
- Zhang, Z.; Tang, W. Drug metabolism in drug discovery and development. Acta Pharm. Sin. B. 2018, 8, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Sychev, D.A.; Ashraf, G.M.; Svistunov, A.A.; Maksimov, M.L.; Tarasov, V.V.; Chubarev, V.N.; Otdelenov, V.A.; Denisenko, N.P.; Barreto, G.E.; Aliev, G. The cytochrome P450 isoenzyme and some new opportunities for the prediction of negative drug interaction in vivo. Drug Des. Devel. Ther. 2018, 12, 1147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, R.; Roychowdhury, A. Modulation of cytochrome-P450 inhibition (CYP) in drug discovery: A medicinal chemistry perspective. Curr. Med. Chem. 2012, 19, 3605–3621. [Google Scholar] [CrossRef] [PubMed]
- Živković, J.V.; Kalauzović, S.G.; Milosavljević, M.R.; Kristina, G.K. In silico evaluation of selected benzimidazole derivatives in the discovery of new potent antimicrobial agents. J. Med. Chem. 2019, 58, 106–115. [Google Scholar]
- Kramer, J.A.; Sagartz, J.E.; Morris, D.L. The application of discovery toxicology and pathology towards the design of safer pharmaceutical lead candidates. Nat. Rev. Drug Discov. 2007, 6, 636–649. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug- likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Wunberg, T.; Hendrix, M.; Hillisch, A.; Lobell, M.; Meier, H.; Schmeck, C.; Wild, H.; Hinzen, B. Improving the hit-to-lead process: Data-driven assessment of drug-like and lead-like screening hits. Drug Discov. Today 2006, 11, 175–180. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Harrold, M.W.; Zavod, R.M.; Harrold, M.W.; Zavod, R.M. Functional group characteristics and roles. Basic Concepts Med. Chem. 2013, 15–50. [Google Scholar] [CrossRef]
- Joubert, R.; Steyn, J.D.; Heystek, H.J.; Steenekamp, J.H.; Du Preez, J.L.; Hamman, J.H. In vitro oral drug permeation models: The importance of taking physiological and physico- chemical factors into consideration. Expert Opin Drug Deliv. 2016, 14, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Morris-Natschke, S.L.; Lee, K.H. Strategies for the Optimization of Natural Leads to Anticancer Drugs or Drug Candidates. Med. Res. Rev. 2016, 36, 32–91. [Google Scholar] [CrossRef]
- Begam, B.F.; Kumar, J.S. Visualization of Chemical Space Using Principal Component Analysis. Appl. Sci. J. 2014, 29, 53–59. [Google Scholar]
- Rose, J.; Gottfries, J.; Muresan, S.; Backlund, A.; Oprea, T.I. Novel Chemical Space Exploration via Natural Products. J. Med. Chem. 2009, 52, 1953–1962. [Google Scholar] [CrossRef]
- Stratton, C.F.; Newman, D.J.; Tan, D.S. Bioorganic & Medicinal Chemistry Letters Cheminformatic comparison of approved drugs from natural product versus synthetic origins. Bioorg. Med. Chem. Lett. 2015, 25, 4802–4807. [Google Scholar]
- Stumpfe, D.; Hu, H.; Bajorath, J. Advances in exploring activity cliffs. J. Comput.-Aided Mol. Des. 2020, 34, 929–942. [Google Scholar] [CrossRef]
- Ekins, S.; Mestres, J.; Testa, B. In silico pharmacology for drug discovery: Methods for virtual ligand screening and profiling. Br. J. Pharmacol. 2007, 152, 9–20. [Google Scholar] [CrossRef]
- Kuete, V. Medicinal Spices and Vegetables from Africa: Therapeutic Potential against Metabolic, Inflammatory, Infectious and Systemic Diseases; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Tagousop, C.N.; de Tamokou, J.D.; Kengne, I.C.; Ngnokam, D.; Voutquenne-Nazabadioko, L. Antimicrobial activities of saponins from Melanthera elliptica and their synergistic effects with antibiotics against pathogenic phenotypes. Chem. Cent. J. 2018, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ndjateu, F.S.T.; Tsafack, R.B.N.; Nganou, B.K.; Awouafack, M.D.; Wabo, H.K.; Tene, M.; Eloff, J.N. Antimicrobial and antioxidant activities of extracts and ten compounds from three Cameroonian medicinal plants: Dissotis perkinsiae (Melastomaceae), Adenocarpus mannii (Fabaceae) and Barteria fistulosa (Passifloraceae). S. Afr. J. Bot. 2014, 91, 37–42. [Google Scholar] [CrossRef]
- Hähnke, V.D.; Kim, S.; Bolton, E.E. PubChem chemical structure standardization. J. Cheminform. 2018, 10, 1–40. [Google Scholar] [CrossRef]
- CCGI MOE. Molecular Operating Environment (MOE); Chem Comput Gr Inc.: Montreal, QC, Canada, 2016. [Google Scholar]
- Feunang, Y.D.; Fiamoncini, J.; Gil, A.; Fuente, D.; Greiner, R. BioTransformer: A comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminform. 2019, 11, 1–25. [Google Scholar]
- Sander, T.; Freyss, J.; Kor, M.; Von Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Postma, M.; Goedhart, J. PlotsOfData—A web app for visualizing data together with their summaries. PLoS Biol. 2019, 17, e3000202. [Google Scholar] [CrossRef]



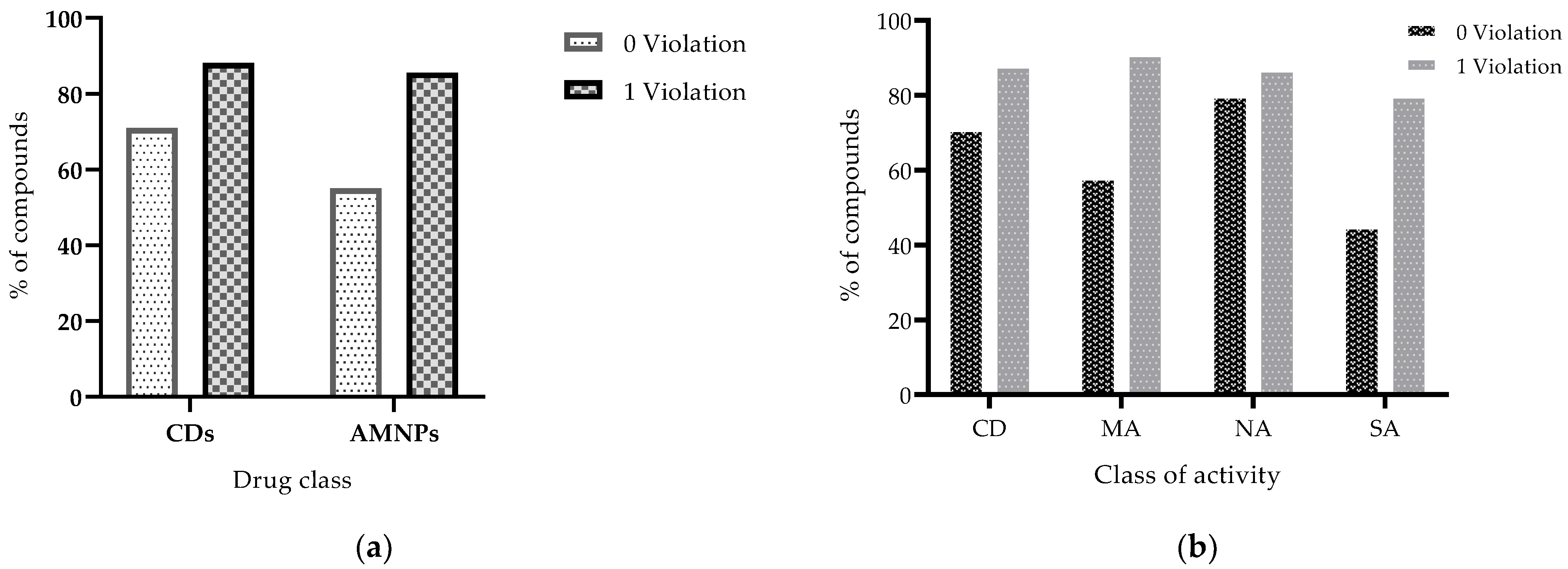
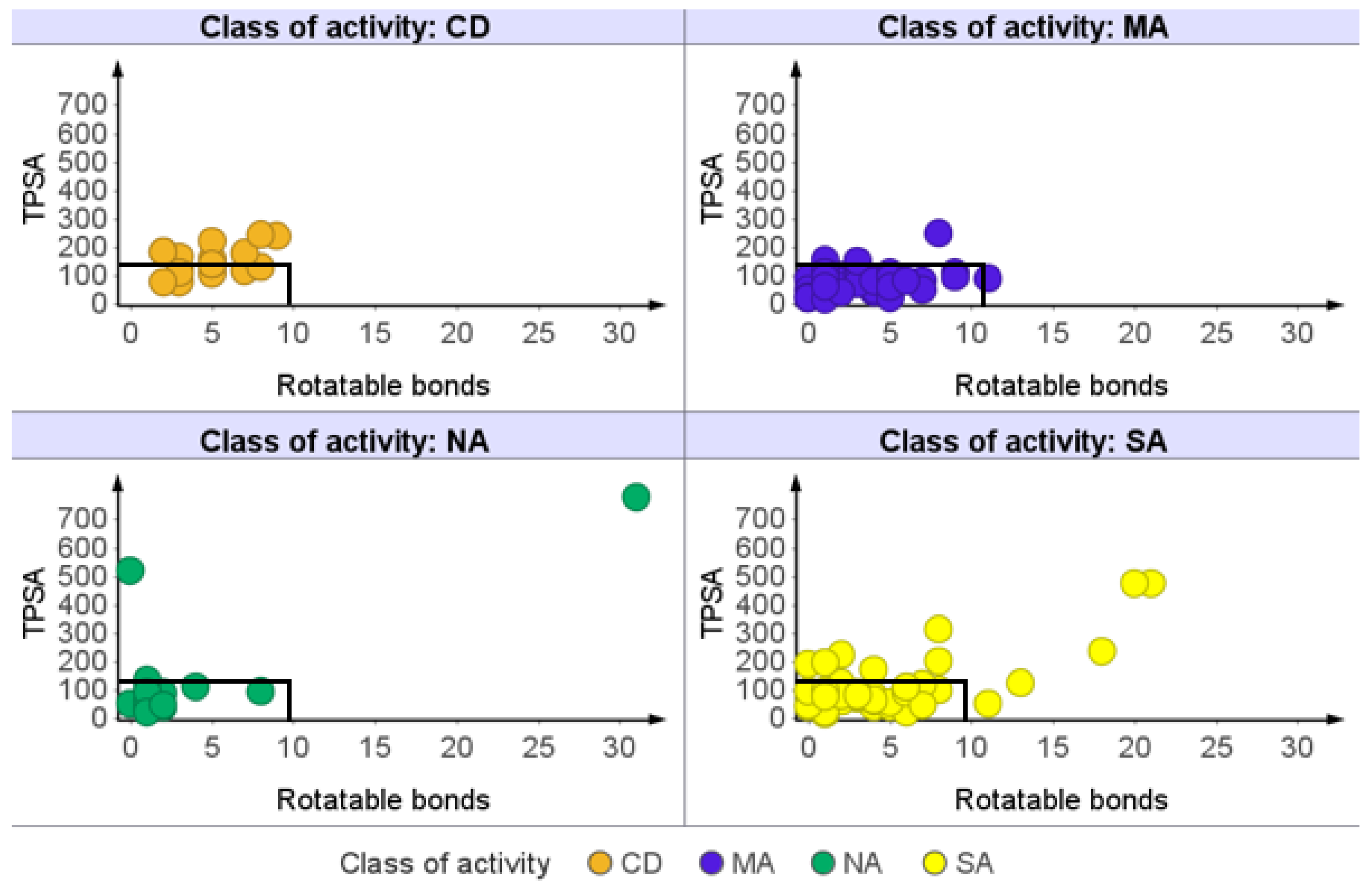
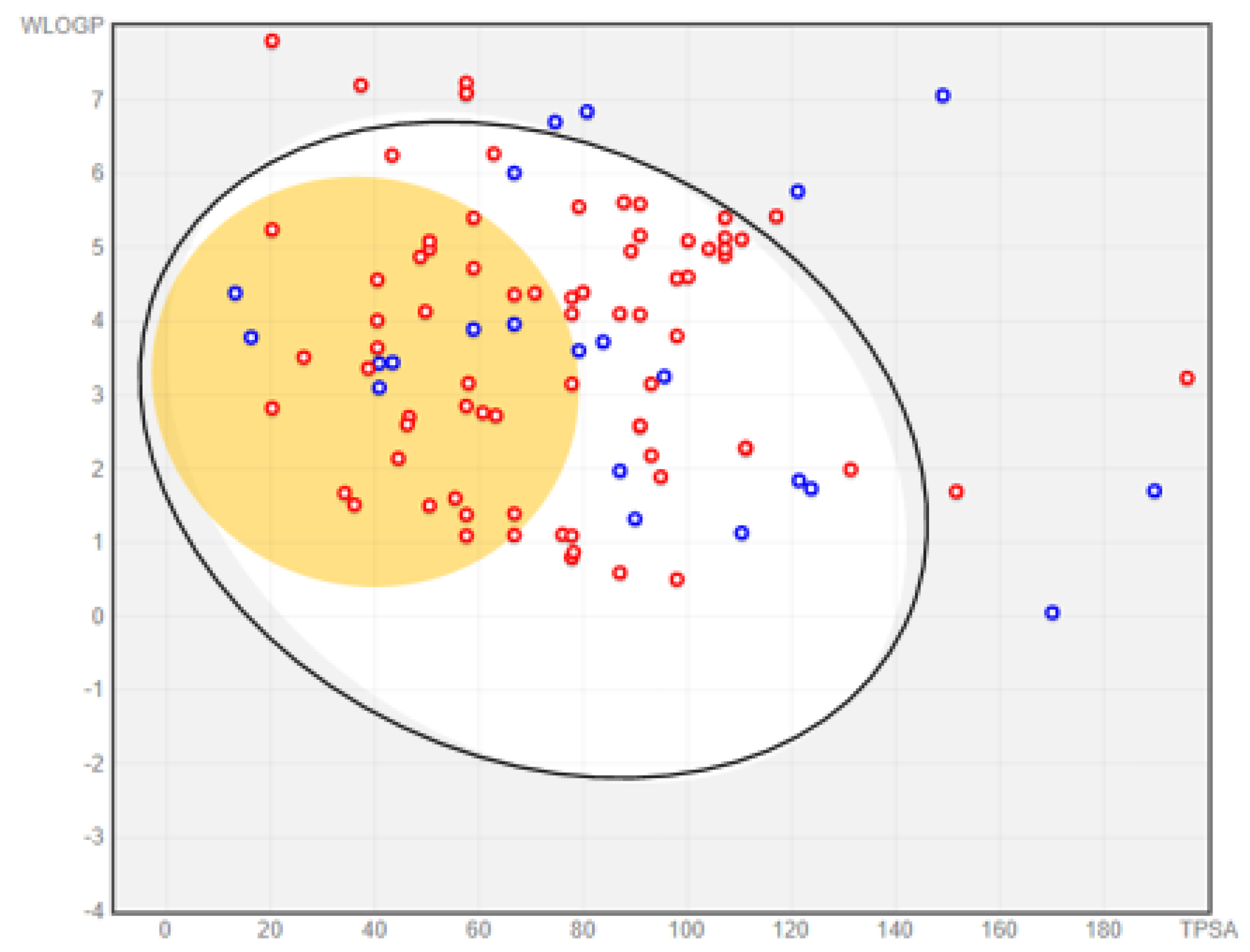
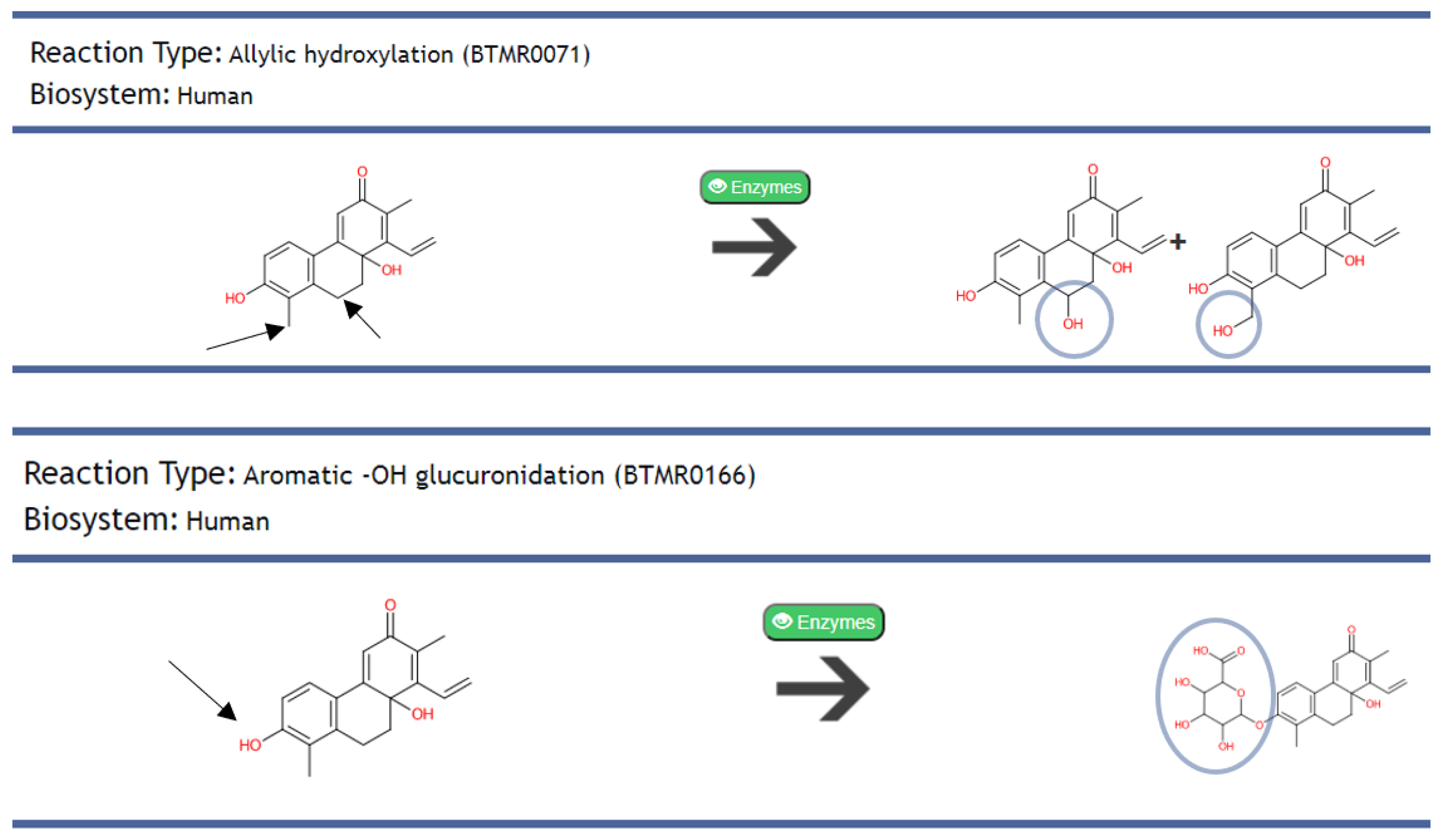
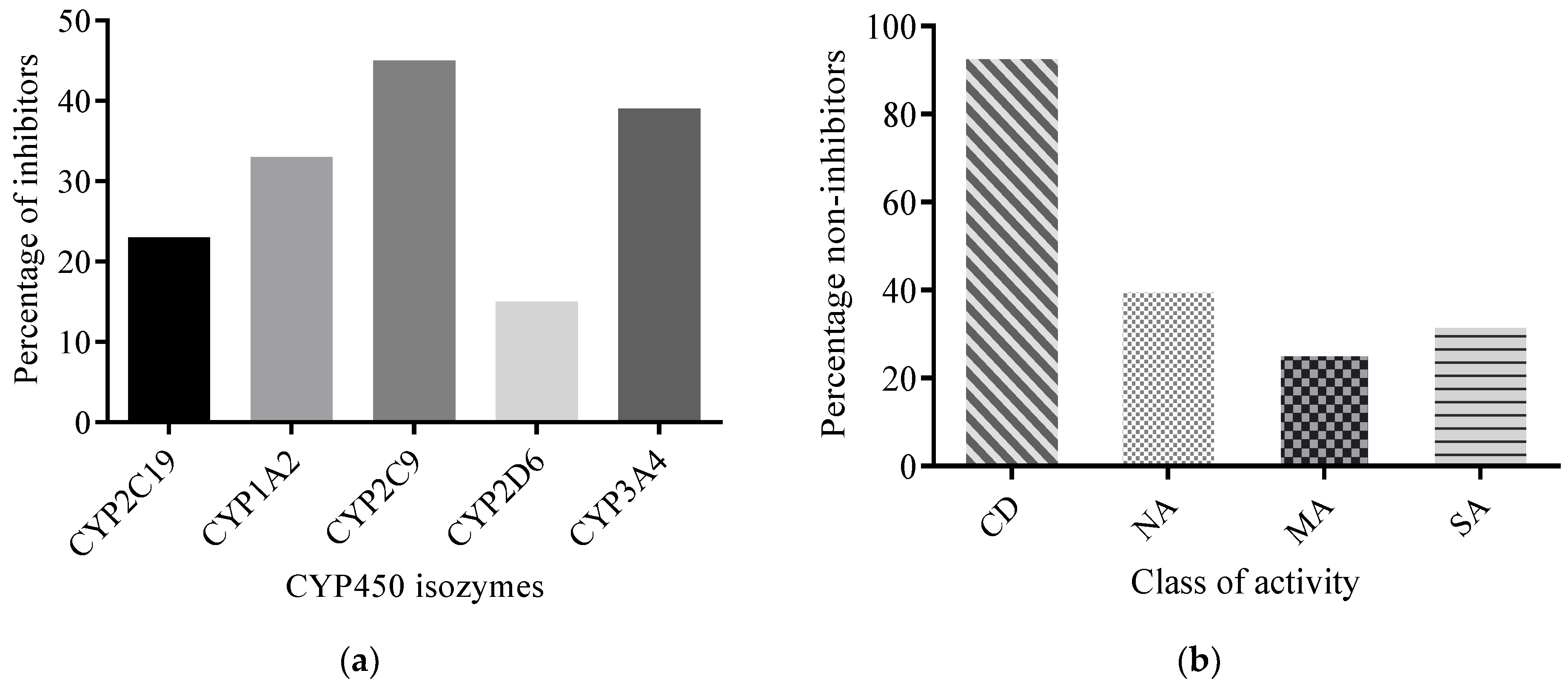
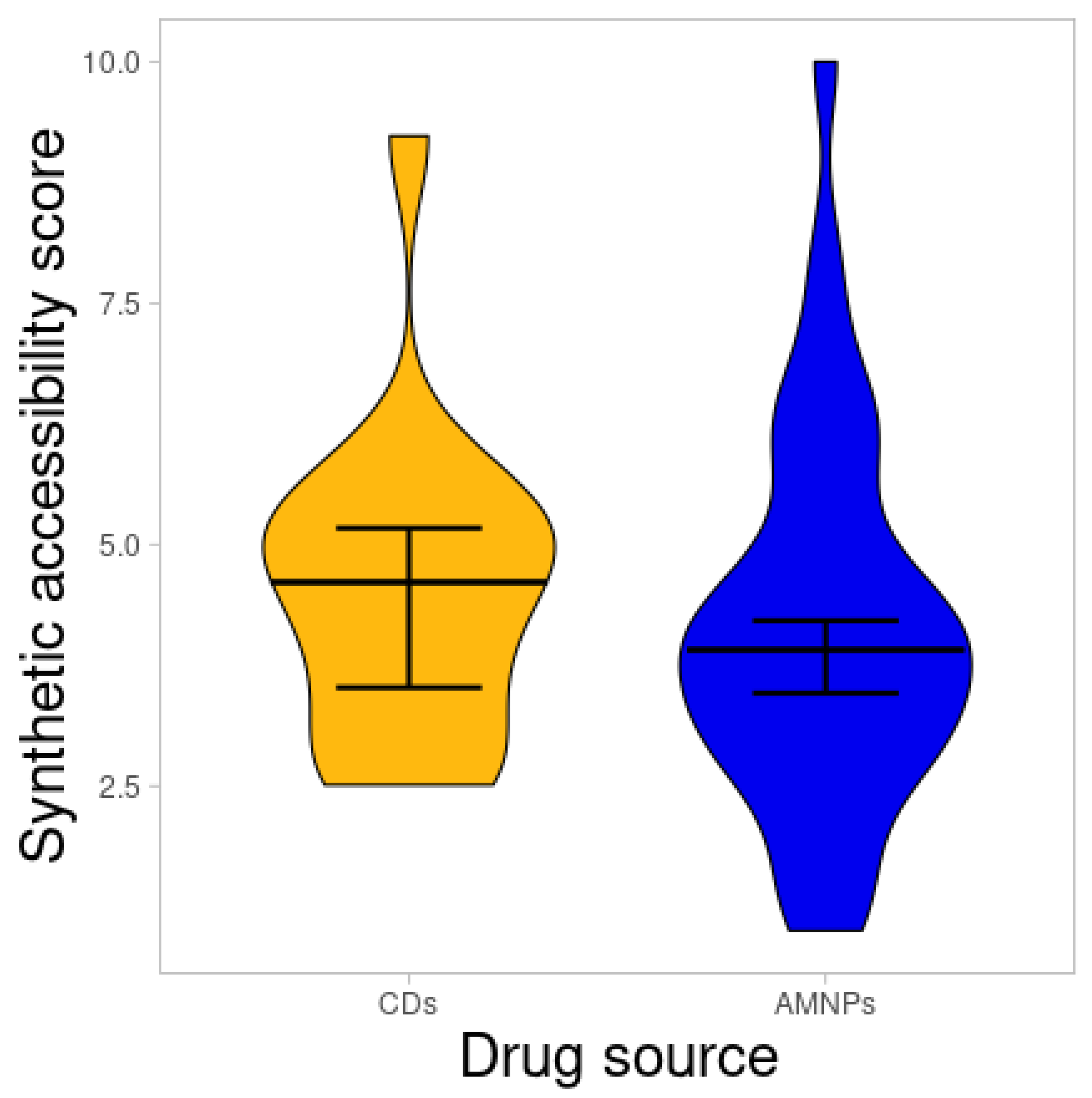
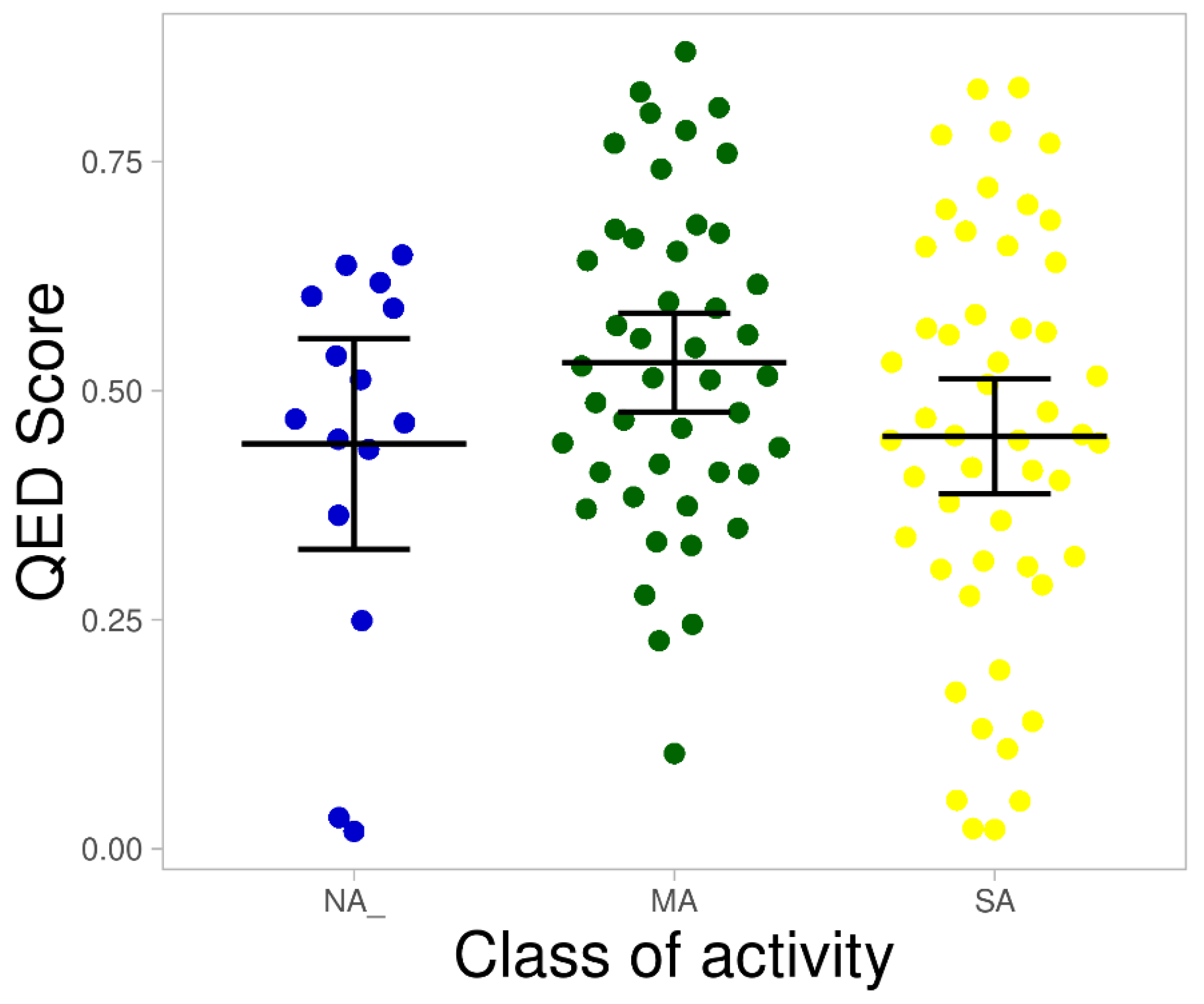

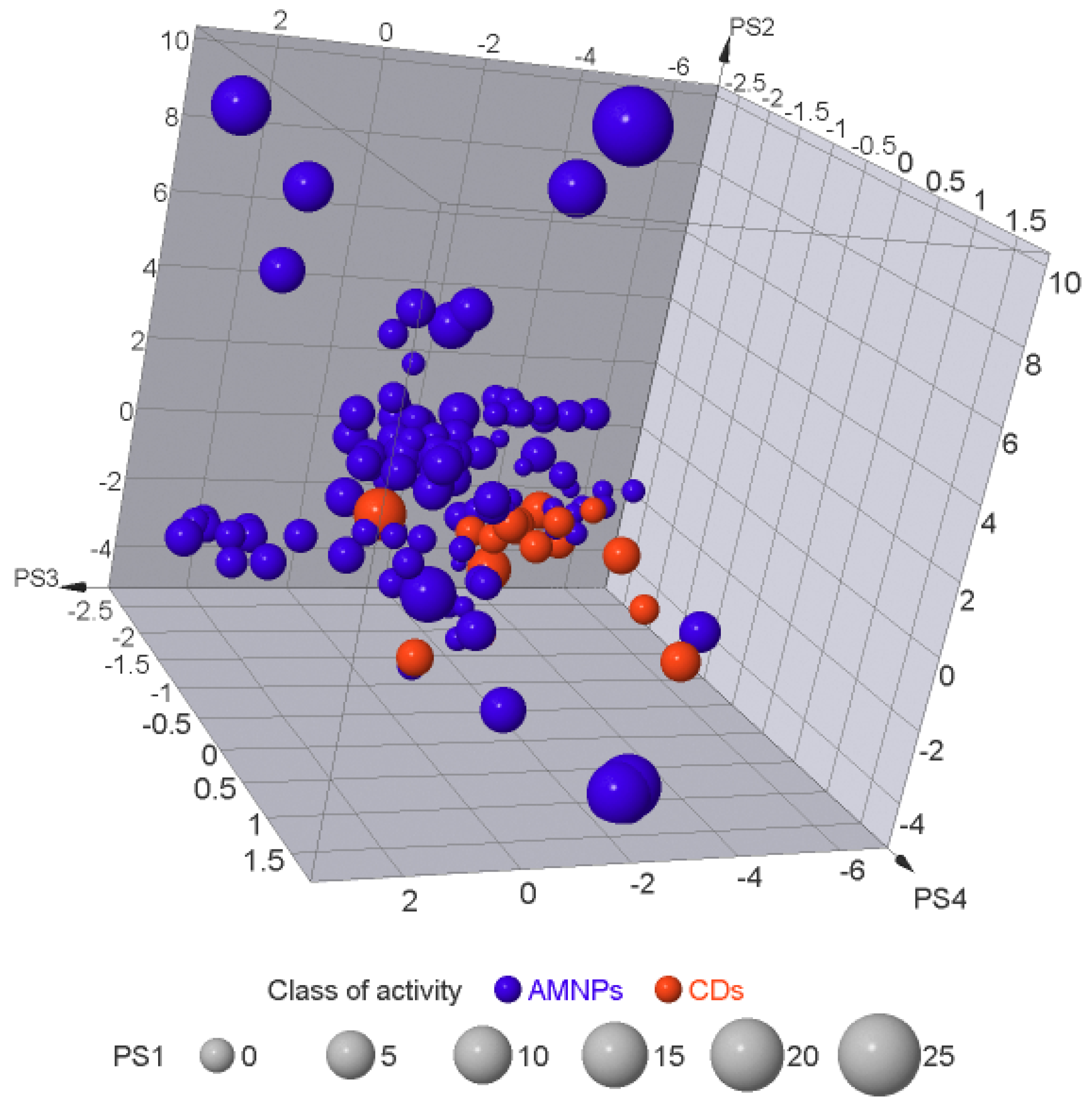
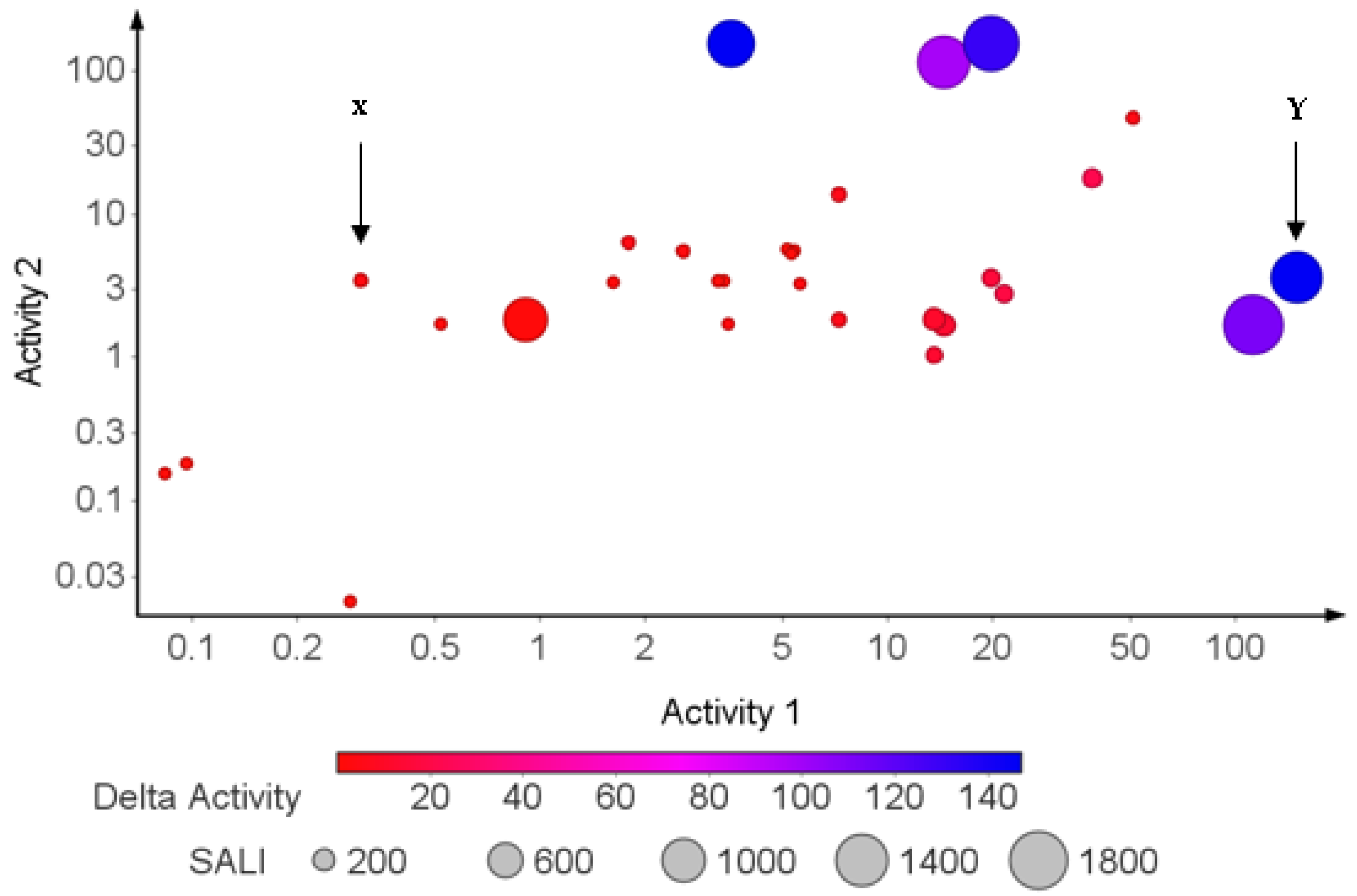
| Class of Activity | MW Violated | cLogP Violated | HBDs Violated | HBAs Violated |
|---|---|---|---|---|
| NA (n = 15) | 12 | 13 | 12 | 12 |
| MA (n = 45) | 27 | 41 | 28 | 27 |
| SA (n = 51) | 25 | 41 | 26 | 25 |
| AMNPs (n = 111) | 64 | 95 | 66 | 64 |
| CD (n = 17) | 13 | 13 | 15 | 13 |
| 1 | No. of AMNPs that Formed Metabolites (n = 111) |
|---|---|
| Phase 1 | 66 |
| Phase 2 | 79 |
| Both phase 1 and 2 | 56 |
| Without Metabolites | 22 |
| Class of Activity | Mutagenicity | Tumorigenicity | Reproductive Effects | Irritant Effects |
|---|---|---|---|---|
| CD (n = 17) | 13 | 14 | 12 | 16 |
| MA (n = 45) | 40 | 41 | 36 | 38 |
| NA (n = 15) | 9 | 13 | 11 | 13 |
| SA (n = 51) | 44 | 49 | 37 | 46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oselusi, S.O.; Egieyeh, S.A.; Christoffels, A. Cheminformatic Profiling and Hit Prioritization of Natural Products with Activities against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2021, 26, 3674. https://doi.org/10.3390/molecules26123674
Oselusi SO, Egieyeh SA, Christoffels A. Cheminformatic Profiling and Hit Prioritization of Natural Products with Activities against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules. 2021; 26(12):3674. https://doi.org/10.3390/molecules26123674
Chicago/Turabian StyleOselusi, Samson O., Samuel A. Egieyeh, and Alan Christoffels. 2021. "Cheminformatic Profiling and Hit Prioritization of Natural Products with Activities against Methicillin-Resistant Staphylococcus aureus (MRSA)" Molecules 26, no. 12: 3674. https://doi.org/10.3390/molecules26123674
APA StyleOselusi, S. O., Egieyeh, S. A., & Christoffels, A. (2021). Cheminformatic Profiling and Hit Prioritization of Natural Products with Activities against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules, 26(12), 3674. https://doi.org/10.3390/molecules26123674







