K-Nearest Neighbor and Random Forest-Based Prediction of Putative Tyrosinase Inhibitory Peptides of Abalone Haliotis diversicolor
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sun, C.L.; Chen, L.; Xu, J.; Qu, W.; Guan, L.; Liu, W.Y.; Akihisa, T.; Feng, F.; Zhang, J. Melanogenesis-inhibitory and antioxidant activities of Phenolics from Periploca forrestii. Chem. Biodivers. 2017, 14. [Google Scholar] [CrossRef]
- Simon, J.D.; Peles, D.; Wakamatsu, K.; Ito, S. Current challenges in understanding melanogenesis: Bridging chemistry, biological control, morphology, and function. Pigment Cell Melanoma Res. 2009, 22, 563–579. [Google Scholar] [CrossRef]
- Halaban, R.; Patton, R.S.; Cheng, E.; Svedine, S.; Trombetta, E.S.; Wahl, M.L.; Ariyan, S.; Hebert, D.N. Abnormal acidification of melanoma cells induces tyrosinase retention in the early secretory pathway. J. Biol. Chem. 2002, 277, 14821–14828. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.T.; Kwon, T.R.; Jang, Y.J.; Yoo, K.H.; Kim, B.J.; Kim, H. Inhibitory effects of Stichopus japonicus extract on melanogenesis of mouse cells via ERK phosphorylation. Mol. Med. Rep. 2017, 16, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Park, S.H.; Lee, H.B.; Goo, Y.A.; Kim, H.S.; Cho, S.H.; Lee, J.H.; Ahn, G.W.; Kim, J.P.; Kang, S.M.; et al. Kojic acid peptide: A new compound with anti-tyrosinase potential. Ann. Dermatol. 2016, 28, 555–561. [Google Scholar] [CrossRef]
- Karkouch, I.; Tabbene, O.; Gharbi, D.; Mlouka, M.A.B.; Elkahoui, S.; Rihouey, C.; Coquet, L.; Cosette, P.; Jouenne, T.; Limam, F. Antioxidant, antityrosinase and antibiofilm activities of synthesized peptides derived from Vicia faba protein hydrolysate: A powerful agents in cosmetic application. Ind. Crops Prod. 2017, 109, 310–319. [Google Scholar] [CrossRef]
- Schurink, M.; van Berkel, W.J.; Wichers, H.J.; Boeriu, C.G. Novel peptides with tyrosinase inhibitory activity. Peptides 2007, 28, 485–495. [Google Scholar] [CrossRef]
- Ubeid, A.A.; Zhao, L.; Wang, Y.; Hantash, B.M. Short-sequence oligopeptides with inhibitory activity against mushroom and human tyrosinase. J. Investig. Dermatol. 2009, 129, 2242–2249. [Google Scholar] [CrossRef]
- Hsiao, N.W.; Tseng, T.S.; Lee, Y.C.; Chen, W.C.; Lin, H.H.; Chen, Y.R.; Wang, Y.T.; Hsu, H.J.; Tsai, K.C. Serendipitous discovery of short peptides from natural products as tyrosinase inhibitors. J. Chem. Inf. Model. 2014, 54, 3099–3111. [Google Scholar] [CrossRef]
- Tseng, T.S.; Tsai, K.C.; Chen, W.C.; Wang, Y.T.; Lee, Y.C.; Lu, C.K.; Don, M.J.; Chang, C.Y.; Lee, C.H.; Lin, H.H.; et al. Discovery of potent cysteine-containing dipeptide inhibitors against tyrosinase: A comprehensive investigation of 20 × 20 dipeptides in inhibiting dopachrome formation. J. Agric. Food Chem. 2015, 63, 6181–6188. [Google Scholar] [CrossRef]
- Ochiai, A.; Tanaka, S.; Tanaka, T.; Taniguchi, M. Rice bran protein as a potent source of antimelanogenic peptides with tyrosinase inhibitory activity. J. Nat. Prod. 2016, 79, 2545–2551. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Liu, L.; Yang, H.; Guo, H.; Liu, X.; Tan, Y.; Wang, W.; Quan, J.; Zhu, L. A novel heptapeptide with tyrosinase inhibitory activity identified from a phage display library. Appl. Biochem. Biotechnol. 2017, 181, 219–232. [Google Scholar] [CrossRef]
- Morita, H.; Kayashita, T.; Kobata, H.; Gonda, A.; Takeya, K.; Itokawa, H.; Itokawa, H. Pseudostellarins A-C, new tyrosinase inhibitory cyclic peptides from Pseudostellaria heterophylla. Tetrahedron 1994, 50, 6797–6804. [Google Scholar] [CrossRef]
- Morita, H.; Kayashita, T.; Kobata, H.; Gonda, A.; Takeya, K.; Itokawa, H. Pseudostellarins D-F, new tyrosinase inhibitory cyclic peptides from Pseudostellaria heterophylla. Tetrahedron 1994, 50, 9975–9982. [Google Scholar] [CrossRef]
- Lien, C.Y.; Chen, C.Y.; Lai, S.T.; Chan, C.F. Kinetics of mushroom tyrosinase and melanogenesis inhibition by N-acetyl-pentapeptides. Sci. World J. 2014, 2014, 409783. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Chompoo, J.; Taira, N.; Fukuta, M.; Gima, S.; Tawata, S. Solid-phase synthesis of mimosine tetrapeptides and their inhibitory activities on neuraminidase and tyrosinase. J. Agric. Food Chem. 2011, 59, 12858–12863. [Google Scholar] [CrossRef]
- Kim, H.; Choi, J.; Cho, J.K.; Kim, S.Y.; Lee, Y.S. Solid-phase synthesis of kojic acid-tripeptides and their tyrosinase inhibitory activity, storage stability, and toxicity. Bioorg. Med. Chem. Lett. 2004, 14, 2843–2846. [Google Scholar] [CrossRef]
- Kahn, V. Effect of Proteins, Protein Hydrolyzates and Amino Acids on o-Dihydroxyphenolase Activity of Polyphenol Oxidase of Mushroom, Avocado, and Banana. J. Food Sci. 1985, 50, 111–115. [Google Scholar] [CrossRef]
- Iba, W. Nutrition Requirement of Cultured Abalone Post Larvae and Juveniles: A Review. Indones. Aquac. J. 2008, 3, 45–57. [Google Scholar] [CrossRef]
- Lou, Q.M.; Wang, Y.M.; Xue, C.H. Lipid and fatty acid composition of two species of abalone, Haliotis discus hannai Ino and Haliotis diversicolor Reeve. J. Food Biochem. 2013, 37, 296–301. [Google Scholar] [CrossRef]
- Latuihamallo, M.; Iriana, D.A.; Apituley, D. Amino acid and fatty acid of abalone Haliotis squamata cultured in different aquaculture systems. Procedia Food Sci. 2015, 3, 174–181. [Google Scholar] [CrossRef]
- Venugopal, V.; Gopakumar, K. Shellfish: Nutritive value, health benefits, and consumer safety. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1219–1242. [Google Scholar] [CrossRef]
- Nithitanakool, S.; Pithayanukul, P.; Bavovada, R.; Saparpakorn, P. Molecular docking studies and anti-tyrosinase activity of Thai mango seed kernel extract. Molecules 2009, 14, 257–265. [Google Scholar] [CrossRef]
- Manavalan, B.; Basith, S.; Shin, T.H.; Choi, S.; Kim, M.O.; Lee, G. MLACP: Machine-learning-based prediction of anticancer peptides. Oncotarget 2017, 8, 77121–77136. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. AIPpred: Sequence-based prediction of anti-inflammatory peptides using random forest. Front. Pharmacol. 2018, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Ansari, H.R.; Gautam, A.; Raghava, G.P. Identification of B-cell epitopes in an antigen for inducing specific class of antibodies. Biol. Direct. 2013, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci. Rep. 2018, 8, 1697. [Google Scholar] [CrossRef] [PubMed]
- Sanders, W.S.; Johnston, C.I.; Bridges, S.M.; Burgess, S.C.; Willeford, K.O. Prediction of cell penetrating peptides by support vector machines. PLoS Comp. Biol. 2011, 7, e1002101. [Google Scholar] [CrossRef]
- Singh, A.; Halgamuge, M.N.; Lakshmiganthan, R. Impact of Different Data Types on Classifier Performance of Random Forest, Naïve Bayes, and K-Nearest Neighbors Algorithms. Int. J. Adv. Comp. Sci. Appl. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Hassan Khan, M.T.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Wang, Y.; Guo, Z.; Tan, T.; Zhang, Y. Novel tyrosinase inhibitory peptide with free radical scavenging ability. J. Enzym. Inhib. Med. Chem. 2019, 34, 1633–1640. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin whitening agents: Medicinal chemistry perspective of tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 403–425. [Google Scholar] [CrossRef]
- Lata, S.; Sharma, B.K.; Raghava, G.P.S. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8, 263. [Google Scholar] [CrossRef]
- Sharma, A.K.; Gupta, A.; Kumar, S.; Dhakan, D.B.; Sharma, V.K. Woods: A fast and accurate functional annotator and classifier of genomic and metagenomic sequences. Genomics 2015, 106, 1–6. [Google Scholar] [CrossRef]
- Gupta, S.; Sharma, A.K.; Jaiswal, S.K.; Sharma, V.K. Prediction of biofilm inhibiting peptides: An in silico approach. Front. Microbiol. 2016, 7, 949. [Google Scholar] [CrossRef]
- Maillo, J.; Ramírez, S.; Triguero, I.; Herrera, F. kNN-IS: An Iterative Spark-based design of the k-Nearest Neighbors classifier for big data. Knowl.-Based Syst. 2017, 117, 3–15. [Google Scholar] [CrossRef]
- Thanh Noi, P.; Kappas, M. Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using Sentinel-2 imagery. Sensors 2018, 18, 18. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Beaufays, J.; Lins, L.; Thomas, A.; Brasseur, R. In silico predictions of 3D structures of linear and cyclic peptides with natural and non-proteinogenic residues. J. Pept. Sci. 2011, 18, 17–24. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 4, 1188–1190. [Google Scholar] [CrossRef]
- Ismaya, T.; Rozeboom, J.; Weijn, A.; Mes, J.; Fusetti, F.; Wichers, J.; Dijkstra, W. Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Heo, L.; Lee, M.S.; Seok, C. GalaxyPepDock: A protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 2015, 43, W431–W435. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, F.; Goddard, D.; Huang, C.; Couch, S.; Greenblatt, M.; Meng, C.; Ferrin, E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
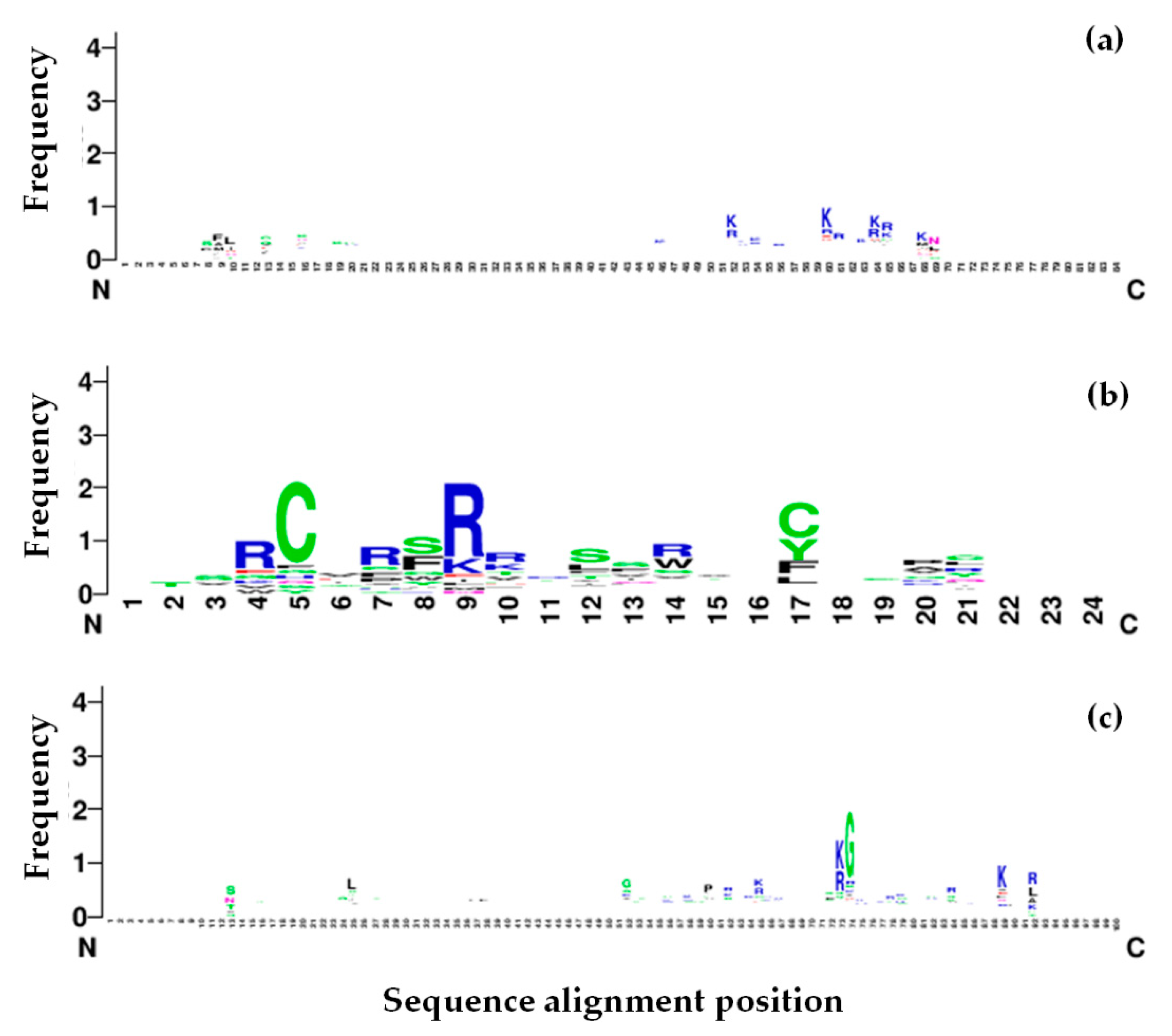
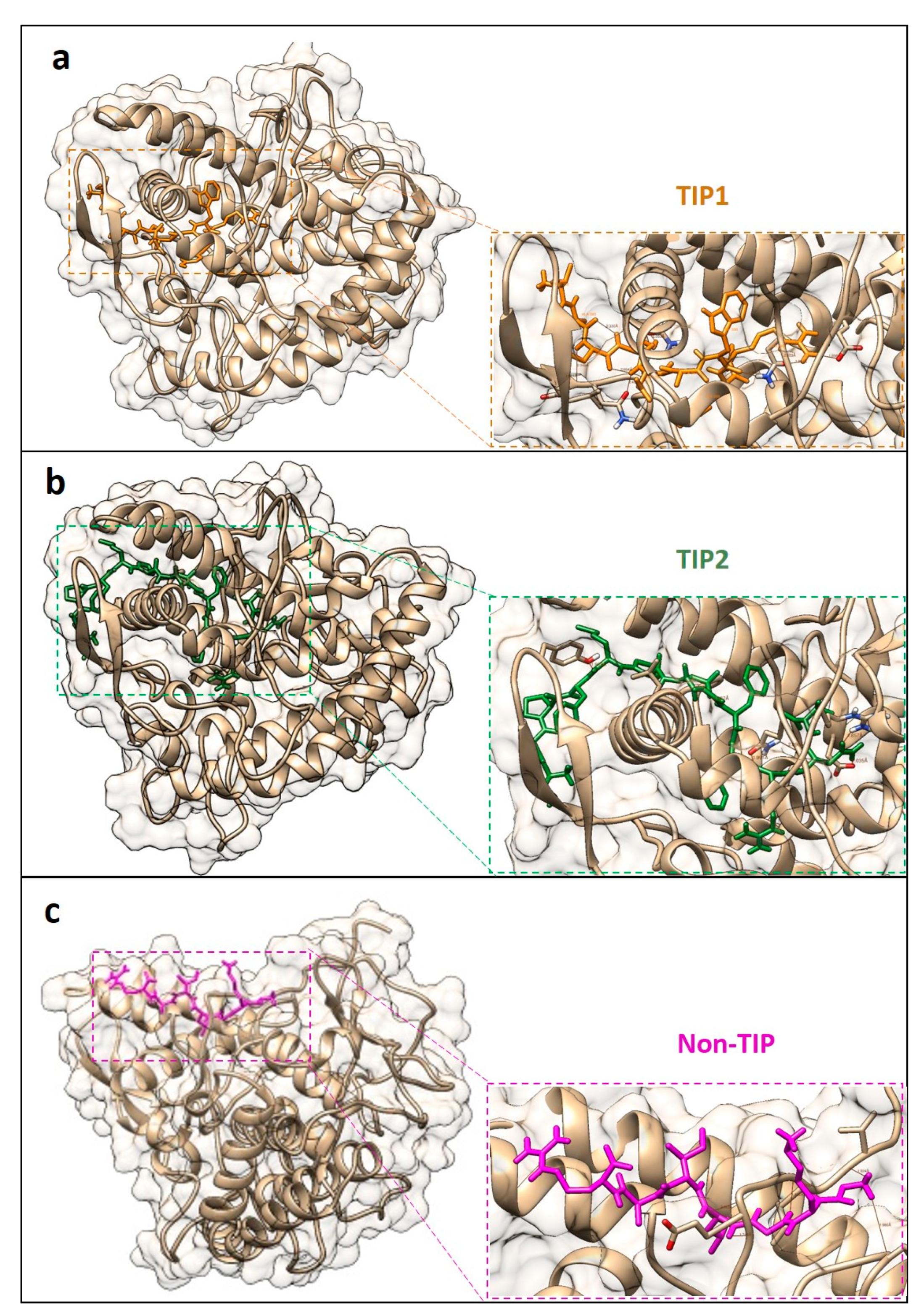
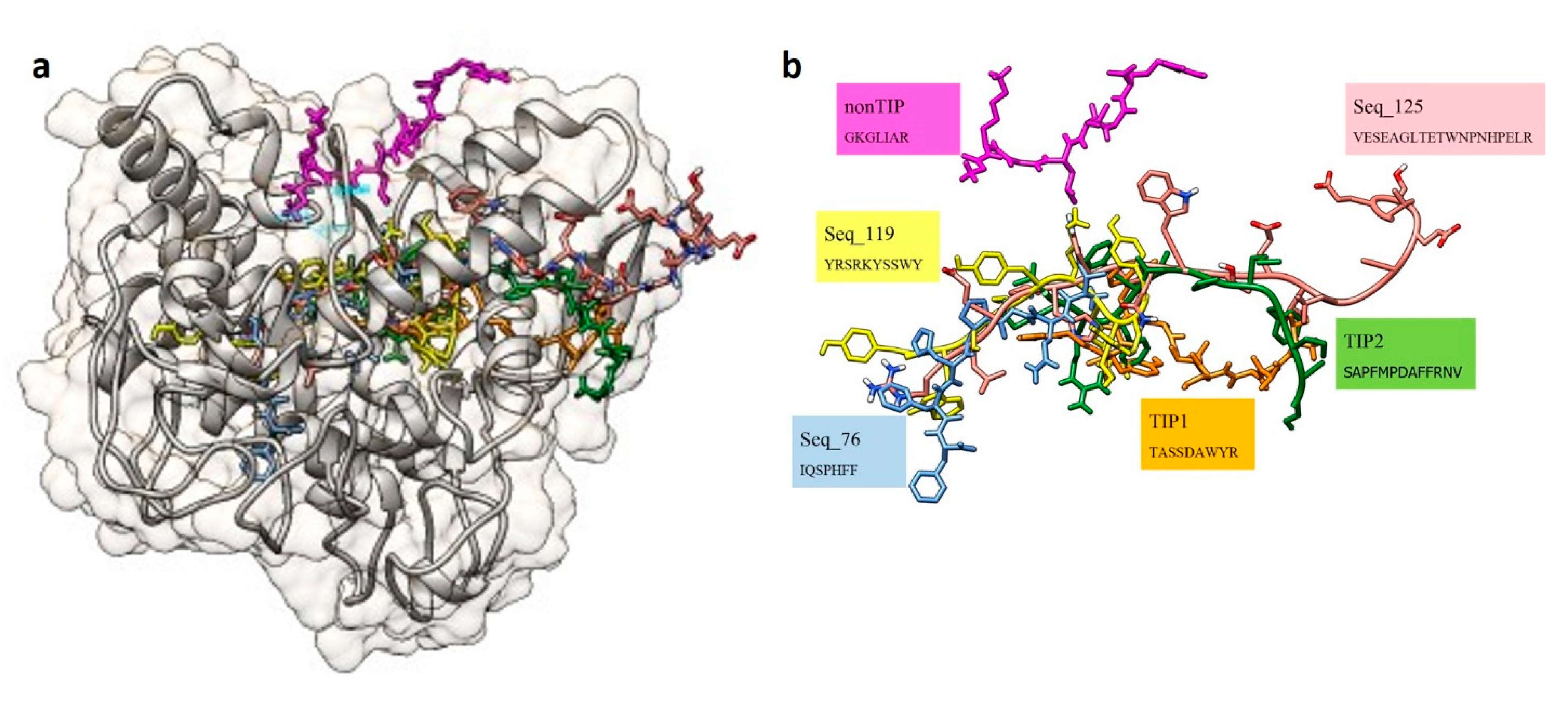

| Machine Learning Prediction Algorithms | Performance Measurement | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | Accuracy | Sensitivity | Specificity | ROC 1 | |
| kNN | 0.89 | 1.00 | 0.97 | 1.00 | 0.96 | 1.00 |
| RF | 0.97 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 |
| Peptides | Peptide Residues | Tyrosinase Residues | Distance (Å) |
|---|---|---|---|
| TIP1 | SER 3 | GLU 160 | 1.981 |
| SER 5 | ASN 173 | 2.402 | |
| SER 4 | GLN 43 | 2.053 | |
| TRP 7 | GLN 132 | 1.901 | |
| ARG 9 | GLN 132 | 2.023 | |
| ARG 9 | GLN 132 | 1.939 | |
| ARG 9 | GLU 97 | 1.898 | |
| TIP2 | ASP 7 | LEU 34 | 1.952 |
| ARG 11 | GLN 132 | 1.991 | |
| ASN 12 | GLN 132 | 2.096 | |
| ASN 12 | ARG 19 | 1.857 | |
| ASN 12 | GLU 97 | 2.035 | |
| Non-TIP | GLY 1 | ILE 12 | 1.832 |
| GLY 1 | GLY 11 | 1.924 | |
| GLY 1 | THR 359 | 1.980 | |
| LYS 2 | PRO 13 | 1.903 | |
| LEU 4 | ILE 16 | 1.746 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kongsompong, S.; E-kobon, T.; Chumnanpuen, P. K-Nearest Neighbor and Random Forest-Based Prediction of Putative Tyrosinase Inhibitory Peptides of Abalone Haliotis diversicolor. Molecules 2021, 26, 3671. https://doi.org/10.3390/molecules26123671
Kongsompong S, E-kobon T, Chumnanpuen P. K-Nearest Neighbor and Random Forest-Based Prediction of Putative Tyrosinase Inhibitory Peptides of Abalone Haliotis diversicolor. Molecules. 2021; 26(12):3671. https://doi.org/10.3390/molecules26123671
Chicago/Turabian StyleKongsompong, Sasikarn, Teerasak E-kobon, and Pramote Chumnanpuen. 2021. "K-Nearest Neighbor and Random Forest-Based Prediction of Putative Tyrosinase Inhibitory Peptides of Abalone Haliotis diversicolor" Molecules 26, no. 12: 3671. https://doi.org/10.3390/molecules26123671
APA StyleKongsompong, S., E-kobon, T., & Chumnanpuen, P. (2021). K-Nearest Neighbor and Random Forest-Based Prediction of Putative Tyrosinase Inhibitory Peptides of Abalone Haliotis diversicolor. Molecules, 26(12), 3671. https://doi.org/10.3390/molecules26123671







