Molecular Dynamics Simulations of a Cytochrome P450 from Tepidiphilus thermophilus (P450-TT) Reveal How Its Substrate-Binding Channel Opens
Abstract
1. Introduction
2. Results
3. Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sono, M.; Roach, M.P.; Coulter, E.D.; Dawson, J.H. Heme-containing oxygenases. Chem. Rev. 1996, 96, 2841–2888. [Google Scholar] [CrossRef] [PubMed]
- Groves, J.T. The bioinorganic chemistry of iron in oxygenases and supramolecular assemblies. Proc. Natl. Acad. Sci. USA 2003, 100, 3569–3574. [Google Scholar] [CrossRef]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005, 105, 2253–2278. [Google Scholar] [CrossRef] [PubMed]
- Ruppel, J.; Fields, K.; Snyder, N.; Zhang, X.; Kadish, K.; Smith, K.; Guilard, R. Handbook of Porphyrin Science; World Scientific Publishing Co.: Singapore, 2010. [Google Scholar]
- Ortiz de Montellano, P.R. Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem. Rev. 2010, 110, 932–948. [Google Scholar] [CrossRef] [PubMed]
- Meunier, B.; De Visser, S.P.; Shaik, S. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 2004, 104, 3947–3980. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Russell, D.W. Clinical importance of the cytochromes P450. Lancet 2002, 360, 1155–1162. [Google Scholar] [CrossRef]
- Fleming, I.; Busse, R. Endothelium-derived epoxyeicosatrienoic acids and vascular function. Hypertension 2006, 47, 629–633. [Google Scholar] [CrossRef]
- Johnson, E.F.; Stout, C.D. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 331–336. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Yadav, J.S. Microbial P450 enzymes in bioremediation and drug discovery: Emerging potentials and challenges. Curr. Protein Pept. Sci. 2018, 19, 75–86. [Google Scholar] [CrossRef]
- Andersen, J.F.; Tatsuta, K.; Gunji, H.; Ishiyama, T.; Hutchinson, C.R. Substrate specificity of 6-deoxyerythronolide B hydroxylase, a bacterial cytochrome P450 of erythromycin A biosynthesis. Biochemistry 1993, 32, 1905–1913. [Google Scholar] [CrossRef]
- Wang, B.; Yang, L.-P.; Zhang, X.-Z.; Huang, S.-Q.; Bartlam, M.; Zhou, S.-F. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab. Rev. 2009, 41, 573–643. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 2001, 14, 611–650. [Google Scholar] [CrossRef]
- Munro, A.W.; Lindsay, J.G. Bacterial cytochromes P-450. Molec. Microbiol. 1996, 20, 1115–1125. [Google Scholar] [CrossRef]
- Hanukoglu, I. Electron transfer proteins of cytochrome P450 systems. In Advances in Molecular and Cell Biology; Bittar, E.E., Ed.; Elsevier: Amsterdam, The Netherlands, 1996; Volume 14, pp. 29–56. [Google Scholar] [CrossRef]
- De Mot, R.; Parret, A.H. A novel class of self-sufficient cytochrome P450 monooxygenases in prokaryotes. Trends Microbiol. 2002, 10, 502–508. [Google Scholar] [CrossRef]
- Warman, A.J.; Robinson, J.W.; Luciakova, D.; Lawrence, A.D.; Marshall, K.R.; Warren, M.J.; Cheesman, M.R.; Rigby, S.E.; Munro, A.W.; McLean, K.J. Characterization of Cupriavidus metallidurans CYP116B1–A thiocarbamate herbicide oxygenating P450–phthalate dioxygenase reductase fusion protein. FEBS J. 2012, 279, 1675–1693. [Google Scholar] [CrossRef]
- Minerdi, D.; Sadeghi, S.J.; Di Nardo, G.; Rua, F.; Castrignanò, S.; Allegra, P.; Gilardi, G. CYP116B5: A new class VII catalytically self-sufficient cytochrome P 450 from A cinetobacter radioresistens that enables growth on alkanes. Molec. Microbiol. 2015, 95, 539–554. [Google Scholar] [CrossRef]
- Hrycay, E.G.; Bandiera, S.M. Monooxygenase, peroxidase and peroxygenase properties and reaction mechanisms of cytochrome P450 enzymes. In Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450; Hrycay, E.G., Bandiera, S.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 1–61. [Google Scholar] [CrossRef]
- Qiu, Y.; Tittiger, C.; Wicker-Thomas, C.; Le Goff, G.; Young, S.; Wajnberg, E.; Fricaux, T.; Taquet, N.; Blomquist, G.J.; Feyereisen, R. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 14858–14863. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ullrich, R.; Hofrichter, M.; Groves, J.T. Heme-thiolate ferryl of aromatic peroxygenase is basic and reactive. Proc. Natl. Acad. Sci. USA 2015, 112, 3686–3691. [Google Scholar] [CrossRef]
- Rittle, J.; Green, M.T. Cytochrome P450 compound I: Capture, characterization, and CH bond activation kinetics. Science 2010, 330, 933–937. [Google Scholar] [CrossRef]
- Nishida, C.R.; de Montellano, P.R.O. Thermophilic cytochrome P450 enzymes. Biochem. Biophys. Res. Commun. 2005, 338, 437–445. [Google Scholar] [CrossRef]
- Roberts, G.A.; Celik, A.; Hunter, D.J.; Ost, T.W.; White, J.H.; Chapman, S.K.; Turner, N.J.; Flitsch, S.L. A self-sufficient cytochrome P450 with a primary structural organization that includes a flavin domain and a [2Fe-2S] redox center. J. Biol. Chem. 2003, 278, 48914–48920. [Google Scholar] [CrossRef] [PubMed]
- Ciaramella, A.; Catucci, G.; Gilardi, G.; Di Nardo, G. Crystal structure of bacterial CYP116B5 heme domain: New insights on class VII P450s structural flexibility and peroxygenase activity. Int. J. Biol. Macromol. 2019, 140, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.; Tavanti, M.; Porter, J.L.; Kress, N.; De Visser, S.P.; Turner, N.J.; Flitsch, S.L. Regio- and Enantio-selective Chemo-enzymatic C-H-Lactonization of Decanoic Acid to (S)-delta-Decalactone. Angew. Chem. Int. Ed. Engl. 2019, 58, 5668–5671. [Google Scholar] [CrossRef]
- Tavanti, M.; Porter, J.L.; Levy, C.W.; Castellanos, J.R.G.; Flitsch, S.L.; Turner, N.J. The crystal structure of P450-TT heme-domain provides the first structural insights into the versatile class VII P450s. Biochem. Biophys. Res. Commun. 2018, 501, 846–850. [Google Scholar] [CrossRef] [PubMed]
- Pravda, L.; Berka, K.; Vařeková, R.S.; Sehnal, D.; Banáš, P.; Laskowski, R.A.; Koča, J.; Otyepka, M. Anatomy of enzyme channels. BMC Bioinform. 2014, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Urban, P.; Lautier, T.; Pompon, D.; Truan, G. Ligand access channels in cytochrome P450 enzymes: A review. Int. J. Mol. Sci. 2018, 19, 1617. [Google Scholar] [CrossRef]
- Gora, A.; Brezovsky, J.; Damborsky, J. Gates of enzymes. Chem. Rev. 2013, 113, 5871–5923. [Google Scholar] [CrossRef]
- Kingsley, L.J.; Lill, M.A. Substrate tunnels in enzymes: Structure–function relationships and computational methodology. Proteins 2015, 83, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Treado, J.D.; Grigas, A.T.; Levine, Z.A.; Regan, L.; O’Hern, C.S. Analyses of protein cores reveal fundamental differences between solution and crystal structures. Proteins 2020, 88, 1154–1161. [Google Scholar] [CrossRef]
- Tien, M.Z.; Meyer, A.G.; Sydykova, D.K.; Spielman, S.J.; Wilke, C.O. Maximum allowed solvent accessibilites of residues in proteins. PLoS ONE 2013, 8, e80635. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09 Gaussian; Gaussian 09 Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.; Grothaus, G.; Narayanan, S.; Onufriev, A. A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins 2006, 63, 928–938. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating p K as and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33 (Suppl. S2), W368–W371. [Google Scholar] [CrossRef]
- Becke, A. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785. [Google Scholar] [CrossRef] [PubMed]
- Hehre, W.J.; Ditchfield, R.; Pople, J.A. Self—Consistent molecular orbital methods. XII. Further extensions of Gaussian—Type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys. 1972, 56, 2257–2261. [Google Scholar] [CrossRef]
- Hariharan, P.C.; Pople, J.A. The influence of polarization functions on molecular orbital hydrogenation energies. Theor. Chim. Acta 1973, 28, 213–222. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Li, P.; Merz, K.M., Jr. MCPB. py: A python based metal center parameter builder. J. Chem. Inf. Model. 2016, 56, 599–604. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
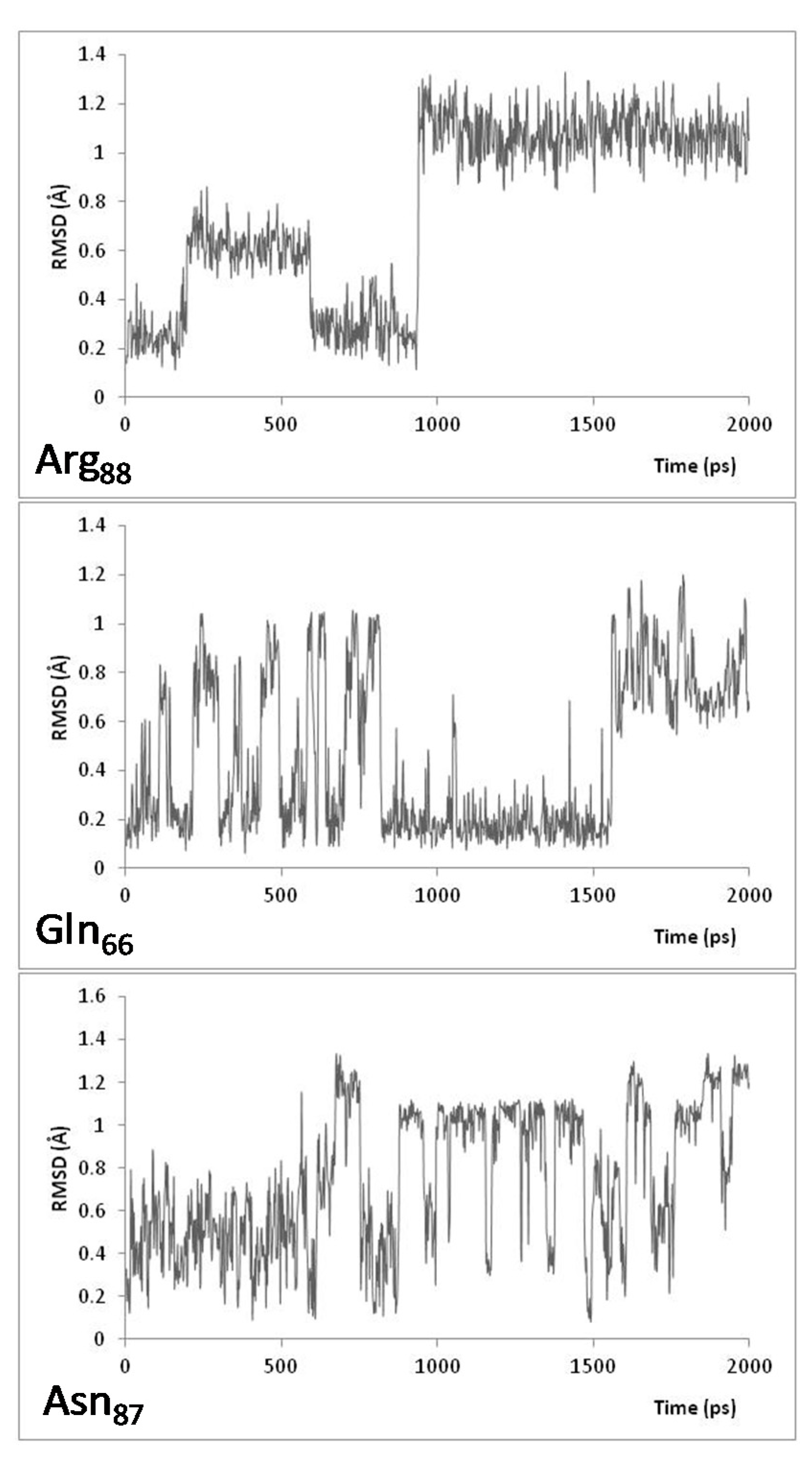
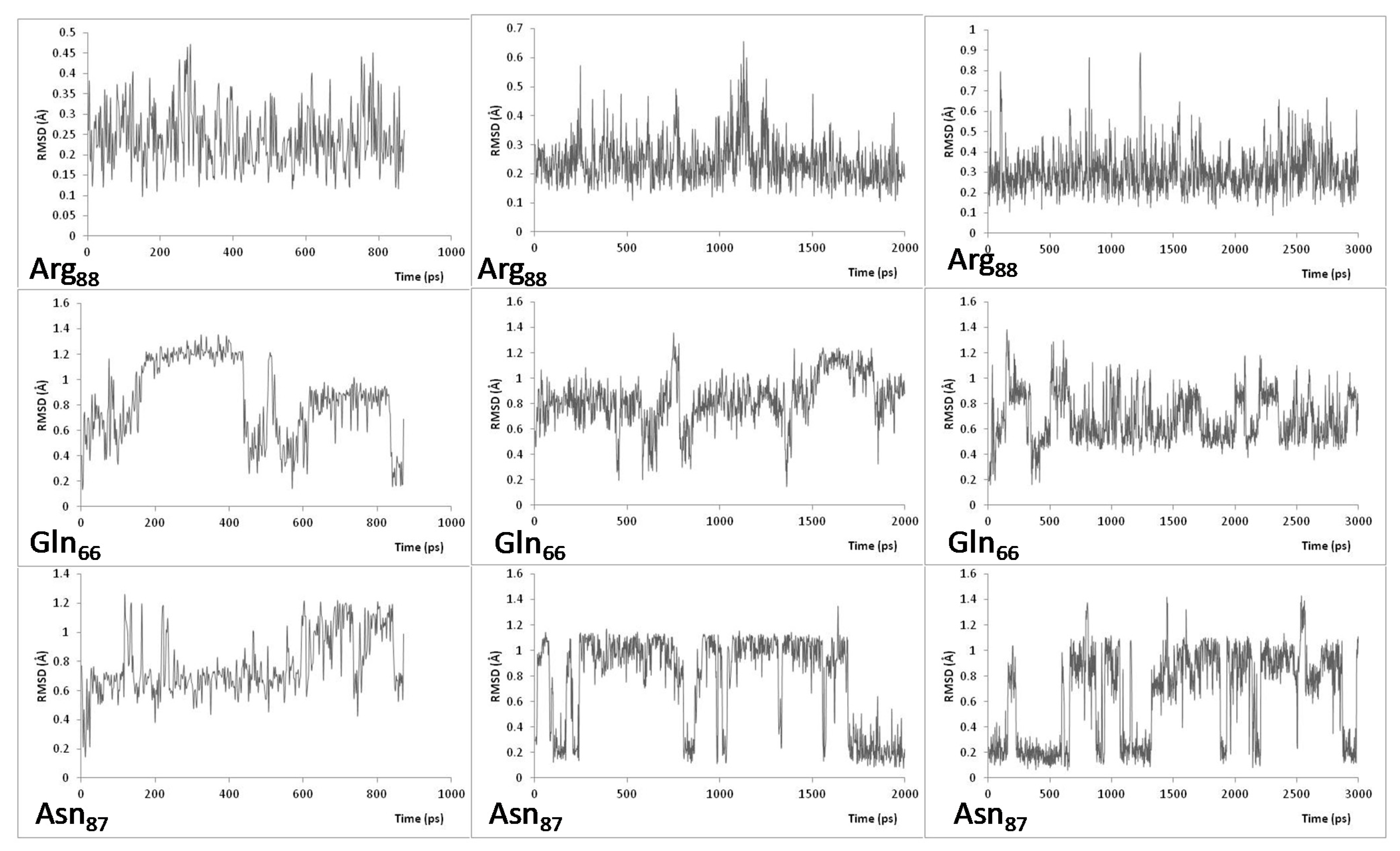
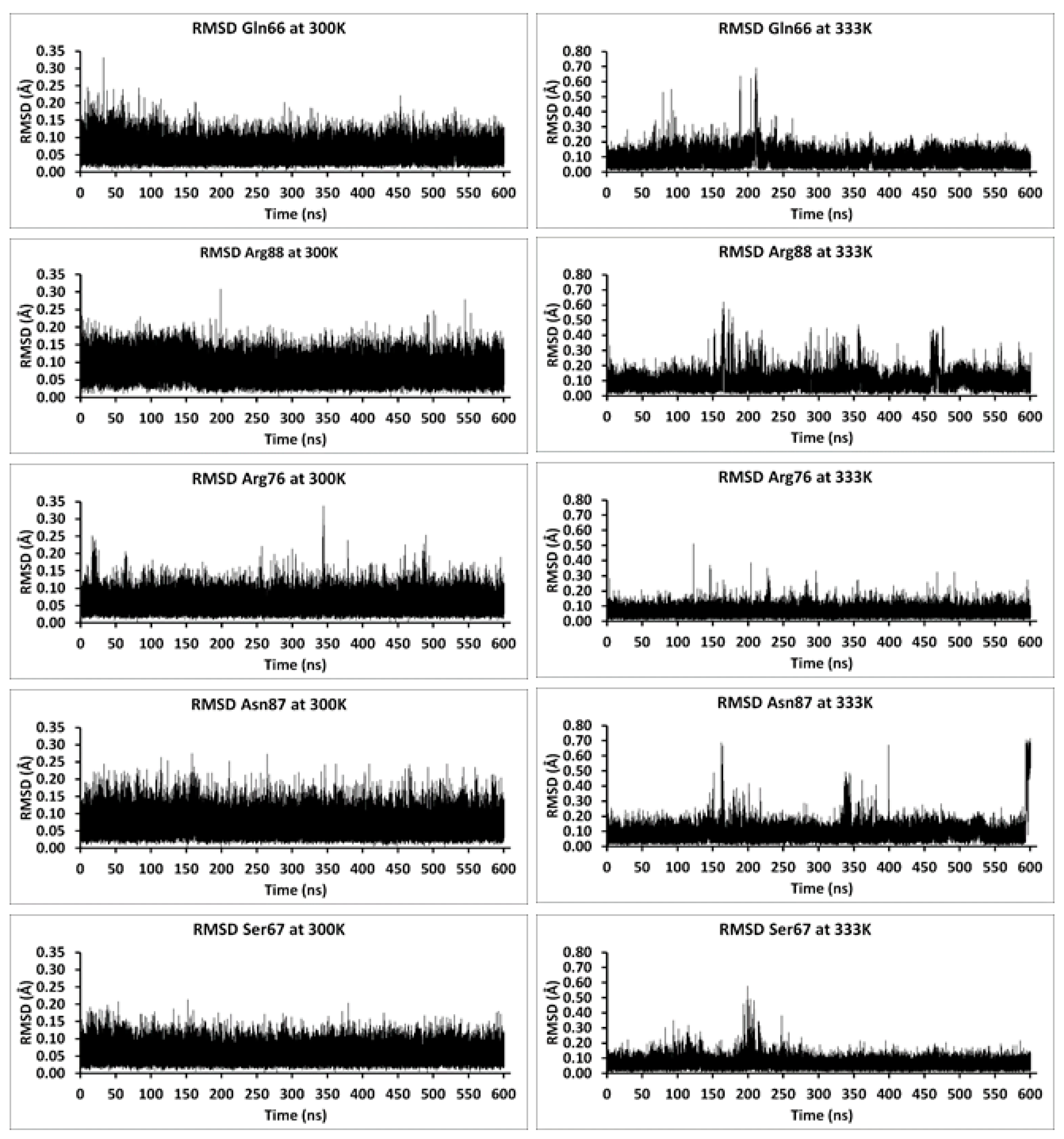
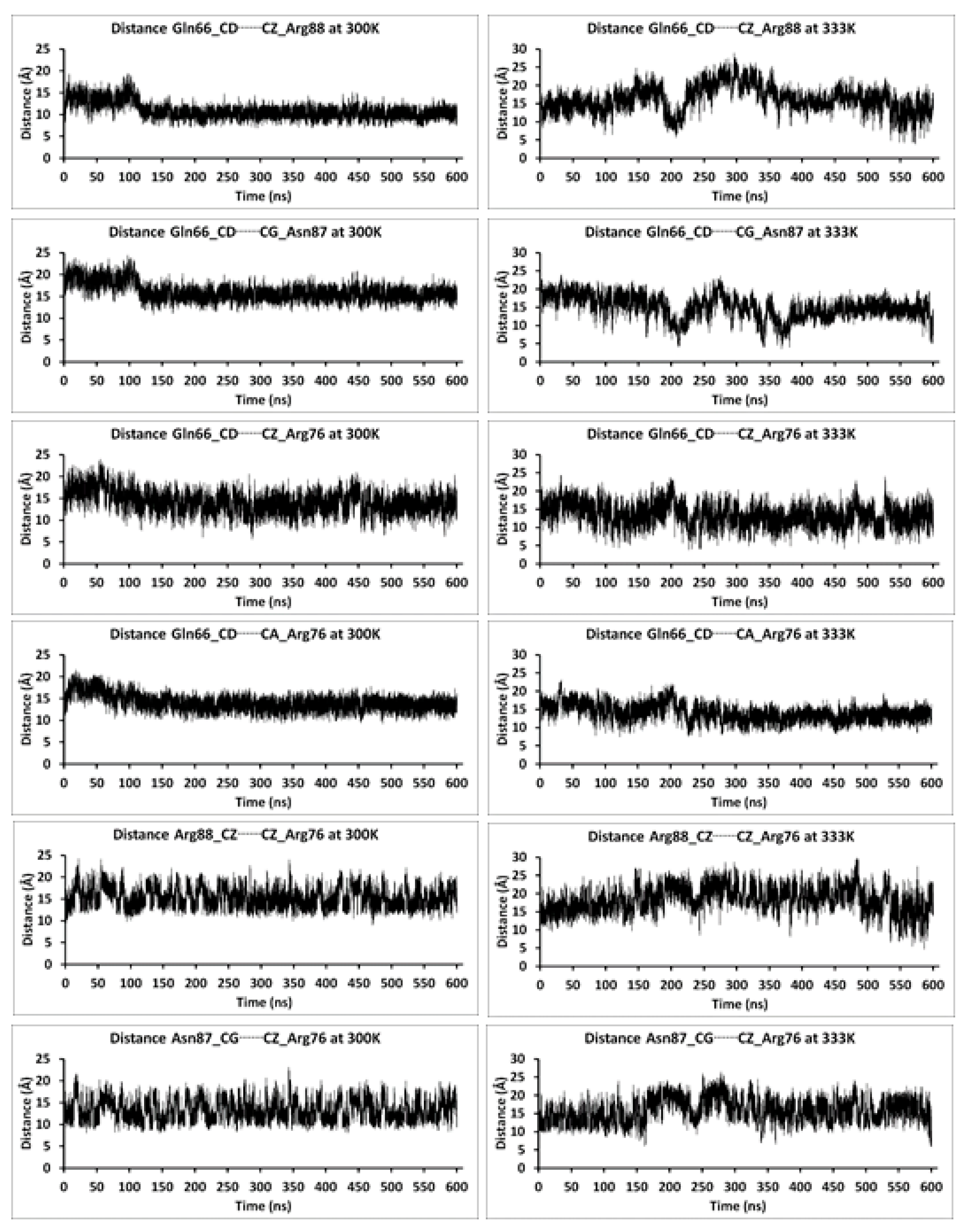
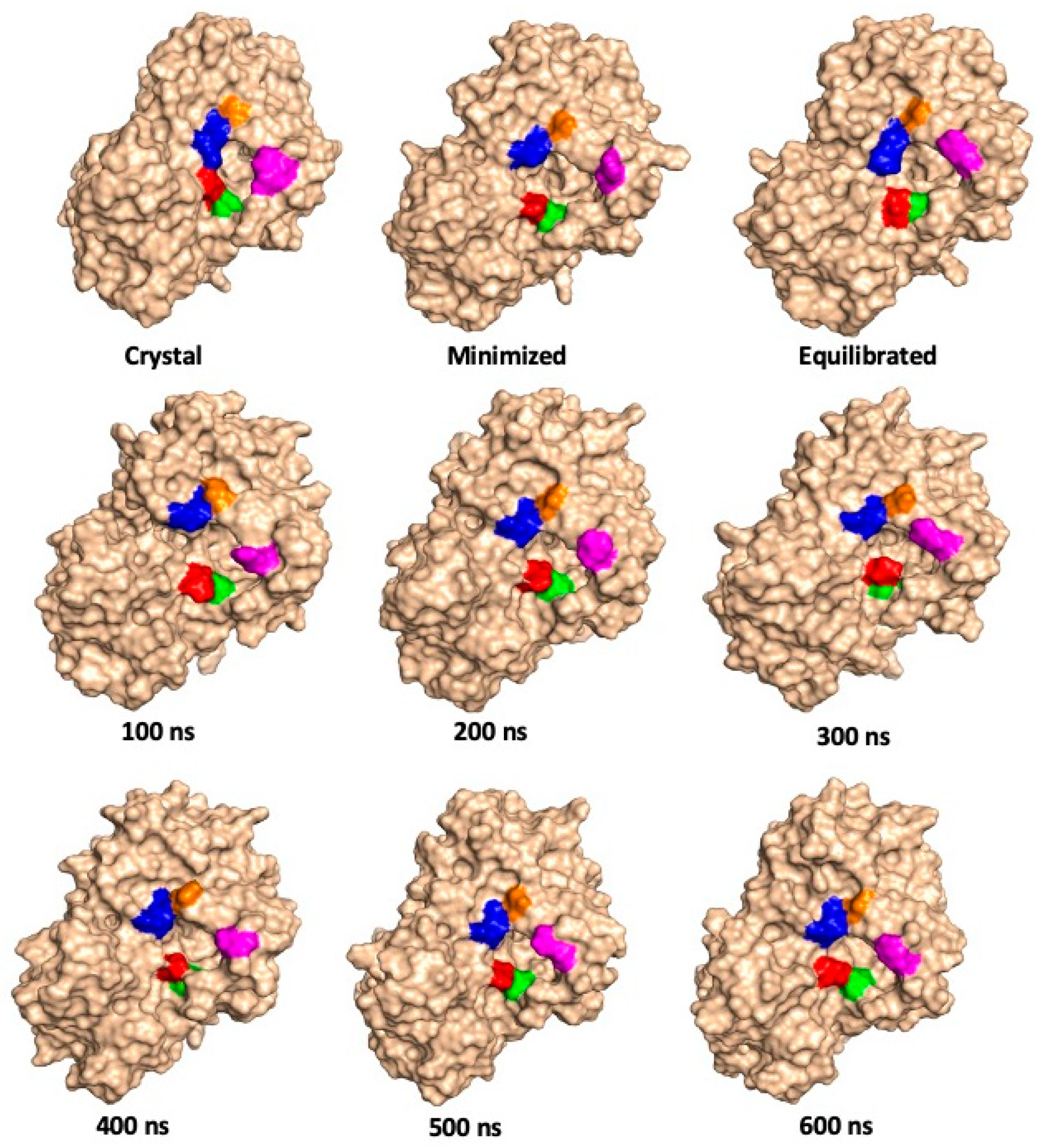
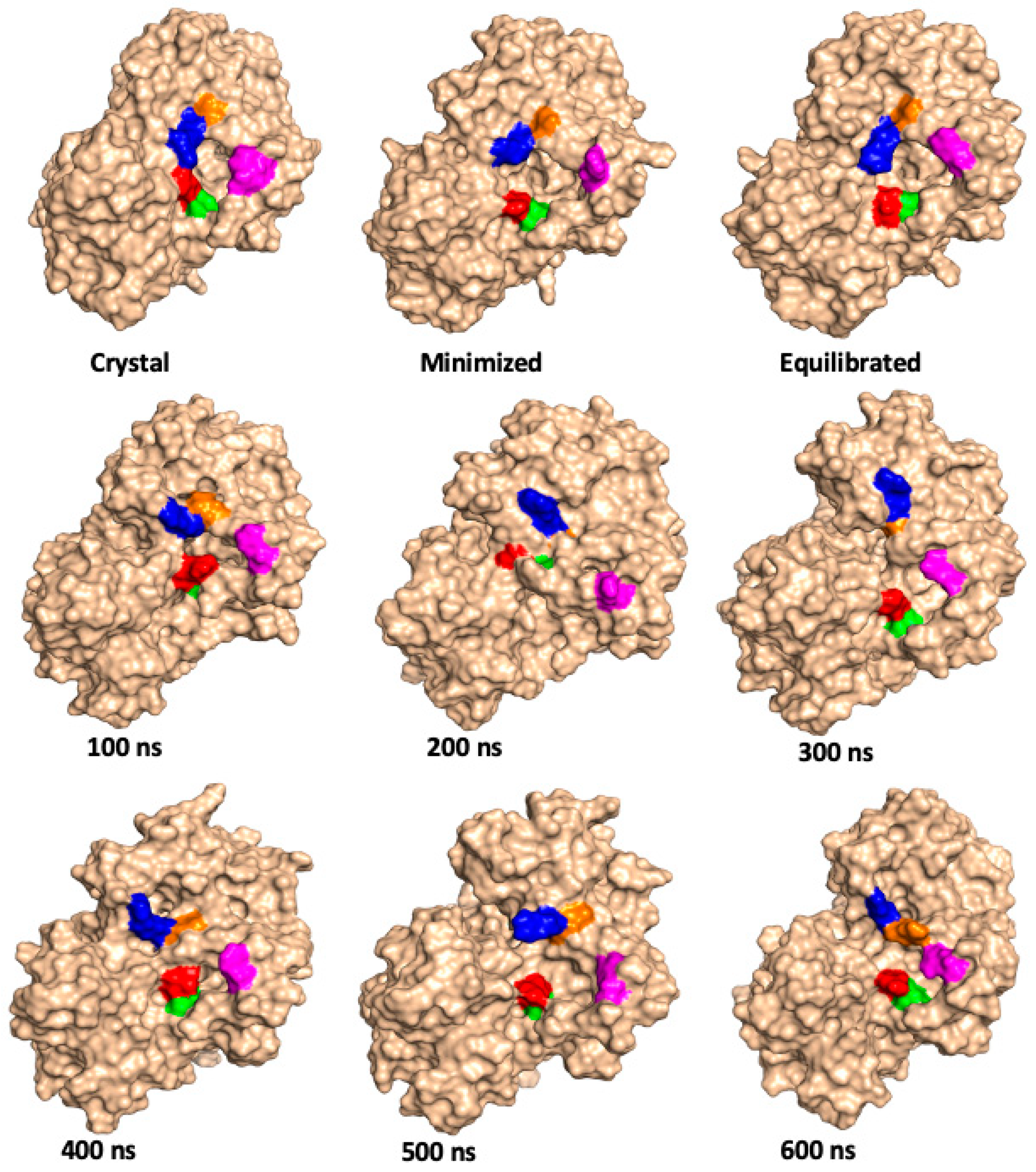
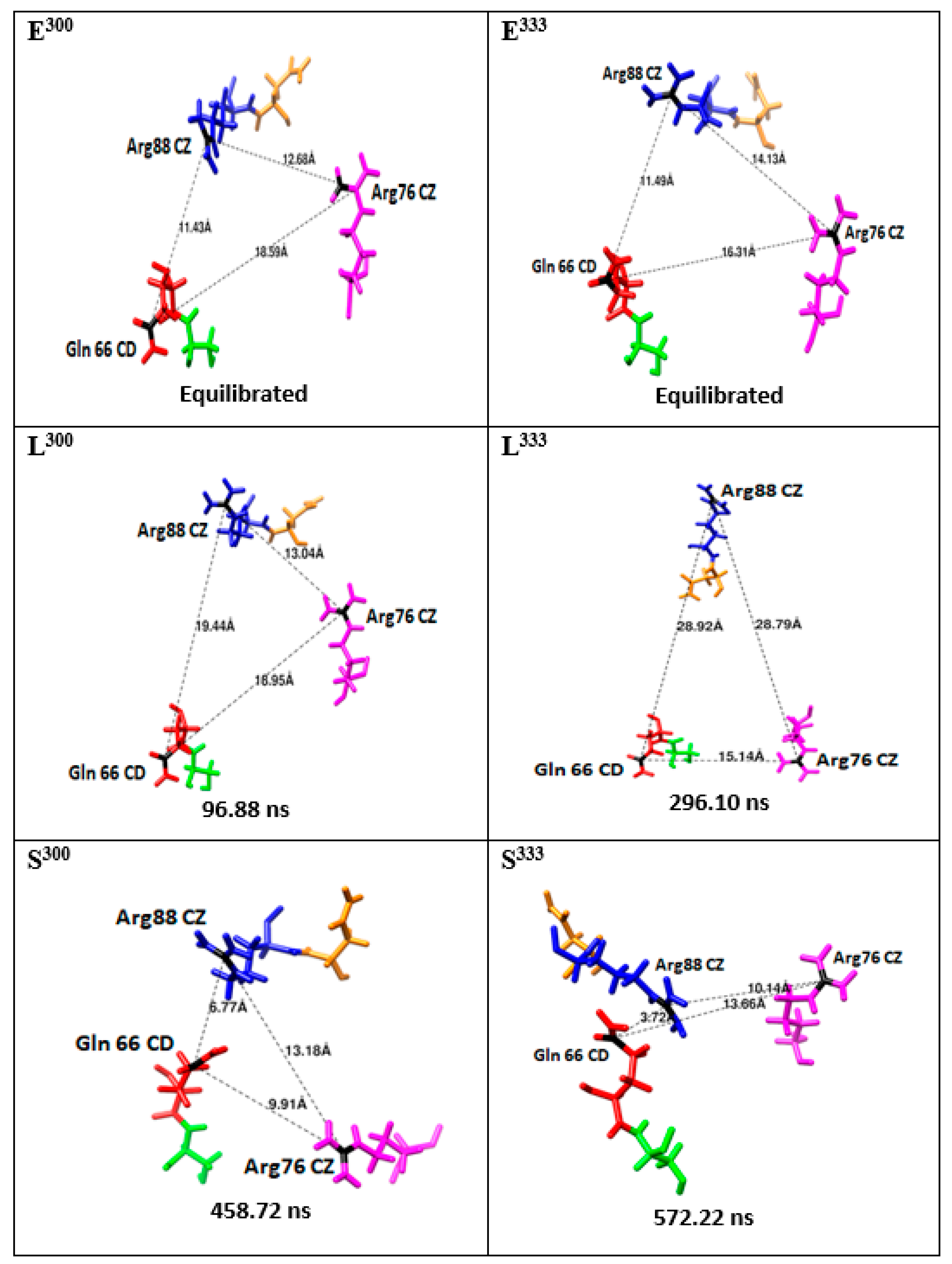
| Relative Solvent Accessibility (RAS) at 300 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residues | Crystal Structure | Minimized Structure | Equilibrated Structure | 100 ns | 200 ns | 300 ns | 400 ns | 500 ns | 600 ns |
| Leu65 | 0.31 | 0.32 | 0.36 | 0.38 | 0.23 | 0.25 | 0.26 | 0.23 | 0.24 |
| Gln66 | 0.27 | 0.40 | 0.26 | 0.50 | 0.46 | 0.48 | 0.35 | 0.41 | 0.38 |
| Ser67 | 0.26 | 0.20 | 0.21 | 0.20 | 0.14 | 0.15 | 0.16 | 0.22 | 0.27 |
| Phe68 | 0.15 | 0.16 | 0.20 | 0.20 | 0.09 | 0.08 | 0.11 | 0.13 | 0.14 |
| Arg76 | 0.58 | 0.55 | 0.40 | 0.62 | 0.51 | 0.56 | 0.40 | 0.49 | 0.49 |
| Asn87 | 0.22 | 0.27 | 0.26 | 0.47 | 0.35 | 0.42 | 0.57 | 0.34 | 0.35 |
| Arg88 | 0.47 | 0.39 | 0.55 | 0.38 | 0.36 | 0.36 | 0.36 | 0.37 | 0.36 |
| Val91 | 0.27 | 0.12 | 0.14 | 0.35 | 0.11 | 0.16 | 0.17 | 0.17 | 0.14 |
| Val246 | 0.25 | 0.26 | 0.21 | 0.11 | 0.03 | 0.02 | 0.03 | 0.03 | 0.01 |
| Phe397 | 0.05 | 0.03 | 0.04 | 0.06 | 0.07 | 0.04 | 0.09 | 0.06 | 0.08 |
| Relative Solvent Accessibility (RAS) at 333 K | |||||||||
| Residues | Crystal Structure | Minimized Structure | Equilibrated Structure | 100 ns | 200 ns | 300 ns | 400 ns | 500 ns | 600 ns |
| Leu65 | 0.31 | 0.32 | 0.32 | 0.39 | 0.11 | 0.27 | 0.20 | 0.24 | 0.22 |
| Gln66 | 0.27 | 0.40 | 0.42 | 0.60 | 0.13 | 0.42 | 0.26 | 0.38 | 0.54 |
| Ser67 | 0.26 | 0.20 | 0.23 | 0.30 | 0.30 | 0.16 | 0.20 | 0.13 | 0.16 |
| Phe68 | 0.15 | 0.16 | 0.23 | 0.42 | 0.56 | 0.17 | 0.14 | 0.11 | 0.22 |
| Arg76 | 0.58 | 0.55 | 0.48 | 0.38 | 0.38 | 0.59 | 0.57 | 0.31 | 0.46 |
| Phe86 | 0.32 | 0.45 | 0.32 | 0.34 | 0.32 | 0.59 | 0.38 | 0.31 | 0.17 |
| Asn87 | 0.22 | 0.27 | 0.23 | 0.56 | 0.05 | 0.15 | 0.18 | 0.20 | 0.65 |
| Arg88 | 0.47 | 0.39 | 0.35 | 0.40 | 0.41 | 0.32 | 0.52 | 0.80 | 0.46 |
| Val91 | 0.27 | 0.12 | 0.18 | 0.34 | 0.36 | 0.78 | 0.14 | 0.28 | 0.26 |
| Val246 | 0.25 | 0.26 | 0.14 | 0.32 | 0.11 | 0.27 | 0.04 | 0.02 | 0.04 |
| Phe397 | 0.05 | 0.03 | 0.03 | 0.14 | 0.02 | 0.04 | 0.05 | 0.05 | 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faponle, A.S.; Roy, A.; Adelegan, A.A.; Gauld, J.W. Molecular Dynamics Simulations of a Cytochrome P450 from Tepidiphilus thermophilus (P450-TT) Reveal How Its Substrate-Binding Channel Opens. Molecules 2021, 26, 3614. https://doi.org/10.3390/molecules26123614
Faponle AS, Roy A, Adelegan AA, Gauld JW. Molecular Dynamics Simulations of a Cytochrome P450 from Tepidiphilus thermophilus (P450-TT) Reveal How Its Substrate-Binding Channel Opens. Molecules. 2021; 26(12):3614. https://doi.org/10.3390/molecules26123614
Chicago/Turabian StyleFaponle, Abayomi S., Anupom Roy, Ayodeji A. Adelegan, and James W. Gauld. 2021. "Molecular Dynamics Simulations of a Cytochrome P450 from Tepidiphilus thermophilus (P450-TT) Reveal How Its Substrate-Binding Channel Opens" Molecules 26, no. 12: 3614. https://doi.org/10.3390/molecules26123614
APA StyleFaponle, A. S., Roy, A., Adelegan, A. A., & Gauld, J. W. (2021). Molecular Dynamics Simulations of a Cytochrome P450 from Tepidiphilus thermophilus (P450-TT) Reveal How Its Substrate-Binding Channel Opens. Molecules, 26(12), 3614. https://doi.org/10.3390/molecules26123614







