Classical Food Quality Attributes and the Metabolic Profile of Cambuci, a Native Brazilian Atlantic Rainforest Fruit
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Physicochemical Analyses
2.3. Metabolite Extraction for NMR Analyses
2.4. NMR Analyses
2.5. Data Processing
2.6. Statistical Analysis
3. Results and Discussion
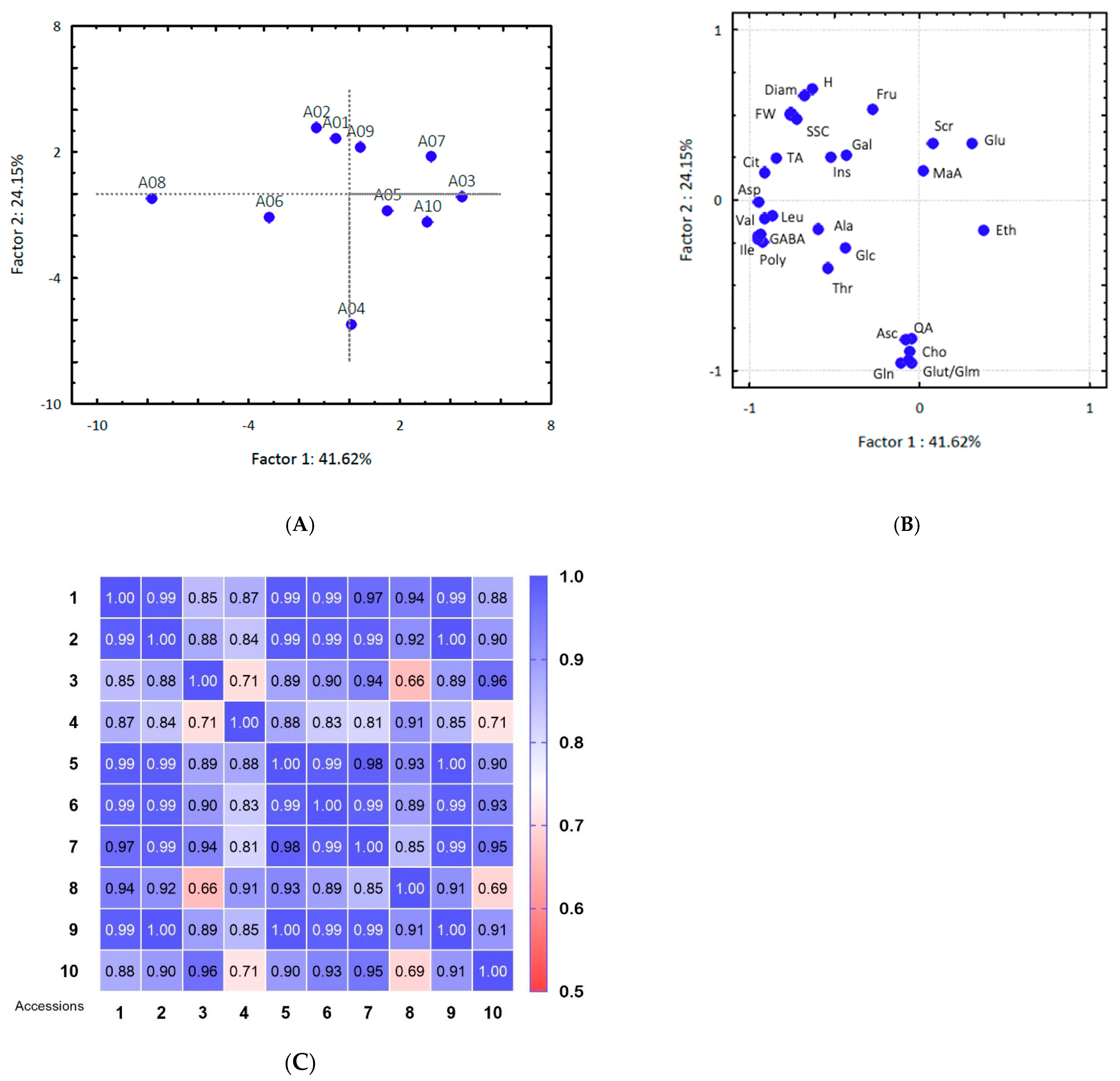
3.1. Physicochemical Parameters
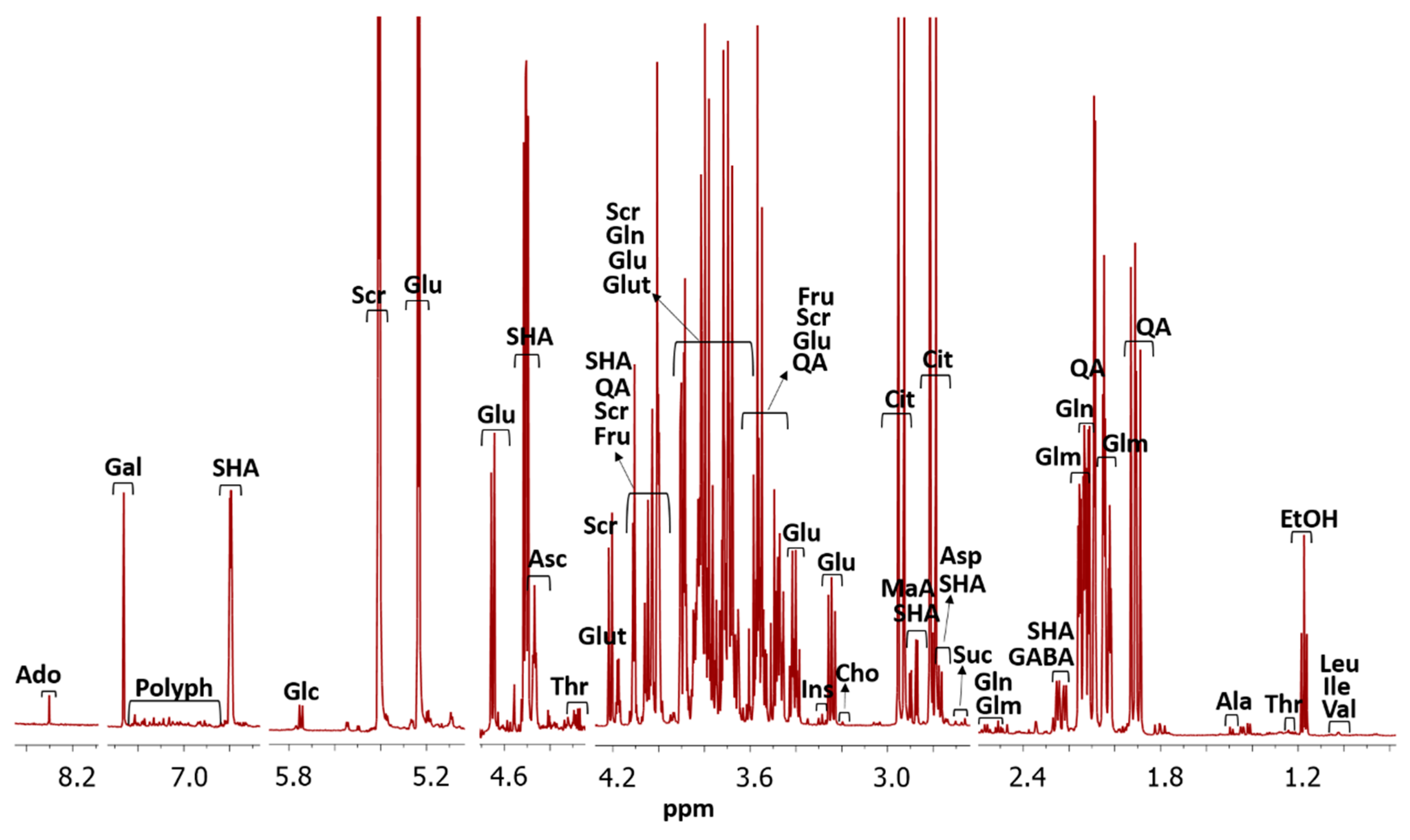
3.2. NMR Spectra Metabolite Assignment
3.3. Metabolite Amounts
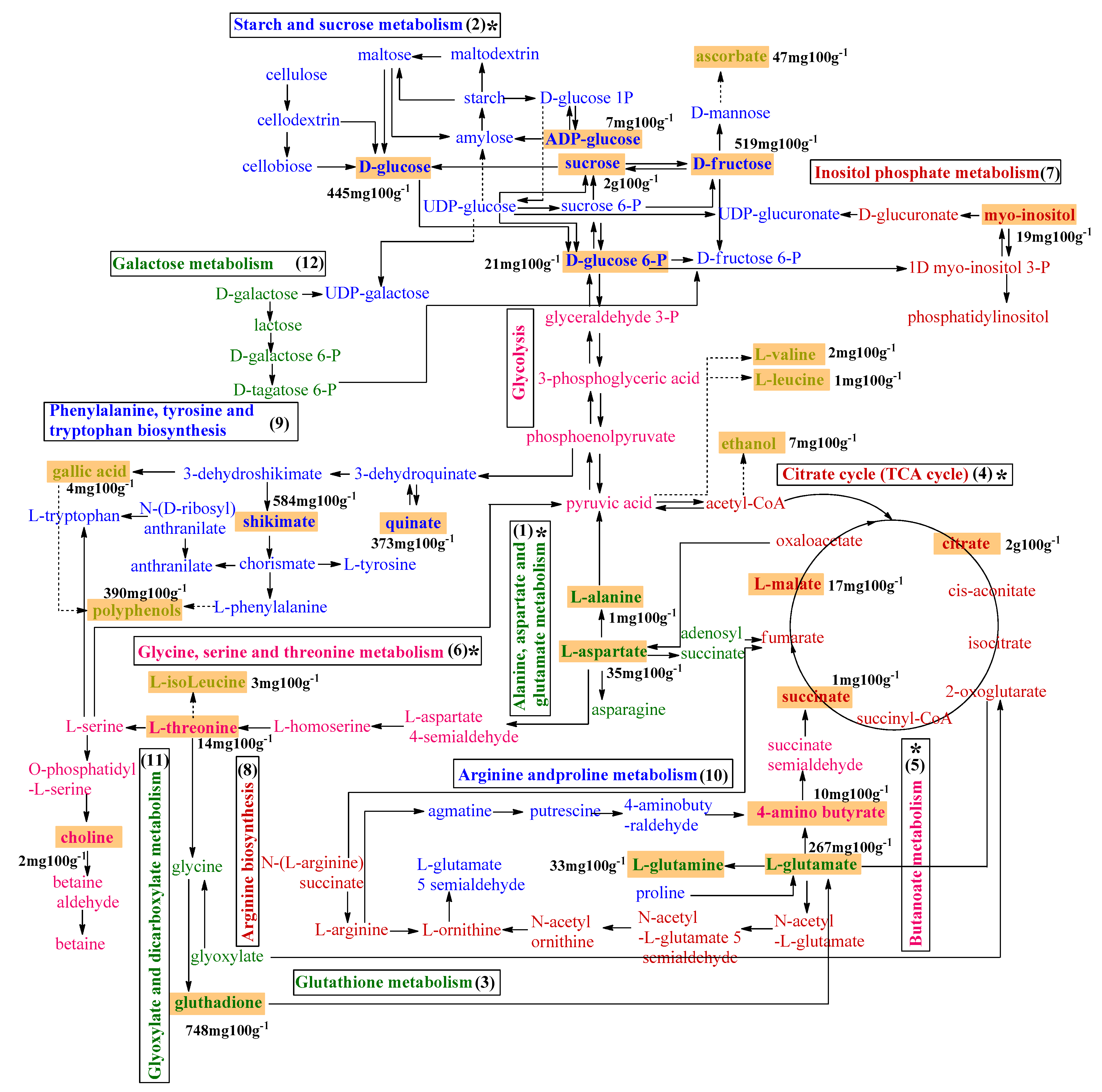
3.4. Metabolic Pathway of Common Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Tokairin, T.O.; Neto, H.B.; Jacomino, A.P. Cambuci—Campomanesia phaea (O. Berg.) Landrum. In Exotic Fruits; Academic Press: Cambridge, MA, USA, 2018; pp. 91–95. ISBN 9780128031384. [Google Scholar]
- Donado-Pestana, C.M.; Belchior, T.; Festuccia, W.T.; Genovese, M.I. Phenolic compounds from cambuci (Campomanesia phaea O. Berg) fruit attenuate glucose intolerance and adipose tissue inflammation induced by a high-fat, high-sucrose diet. Food Res. Int. 2015, 69, 170–178. [Google Scholar] [CrossRef]
- Tokairin, T.O.; Silva, A.P.G.; Spricigo, P.C.; Alencar, S.M.; Jacomino, A.P. Cambuci: A native fruit from the Brazilian Atlantic forest showed nutraceutical characteristics. Rev. Bras. Frutic. 2018, 40. [Google Scholar] [CrossRef]
- Lescano, C.H.; Freitas de Lima, F.; Caires, A.R.L.; de Oliveira, I.P. Chapter 25 - Polyphenols Present in Campomanesia Genus: Pharmacological and Nutraceutical Approach. In Polyphenols in Plants, 2nd ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 407–420. ISBN 978-0-12-813768-0. [Google Scholar]
- Sobolev, A.P.; Mannina, L.; Proietti, N.; Carradori, S.; Daglia, M.; Giusti, A.M.; Antiochia, R.; Capitani, D. Untargeted NMR-based methodology in the study of fruit metabolites. Molecules 2015, 20, 4088–4108. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhao, Y.; Yang, J.; Jiang, Y.; Lu, F.; Jia, Y.; Yang, B. Metabolomic analyses of banana during postharvest senescence by 1H-high resolution-NMR. Food Chem. 2017, 218, 406–412. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Ma, J.; Wang, H.; Li, F.; Qin, D.; Wu, J.; Zhu, G. NMR-based global metabolomics approach to decipher the metabolic effects of three plant growth regulators on strawberry maturation. Food Chem. 2018, 269, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Parijadi, A.A.R.; Putri, S.P.; Ridwani, S.; Dwivany, F.M.; Fukusaki, E. Metabolic profiling of Garcinia mangostana (mangosteen) based on ripening stages. J. Biosci. Bioeng. 2018, 125, 238–244. [Google Scholar] [CrossRef] [PubMed]
- AOAC Official Methods of Analysis of AOAC International. Assoc. Off. Anal. Chem. Int. 2000, 9, 471. [CrossRef]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef]
- Sousa, S.A.A.; Magalhães, A.; Ferreira, M.M.C. Optimized bucketing for NMR spectra: Three case studies. Chemom. Intell. Lab. Syst. 2013, 122, 93–102. [Google Scholar] [CrossRef]
- The MathWorks Inc. MATLAB 9.10 and Statistics Toolbox 9.10. 2021. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; Team, R.C.: Vienna, Austria, 2021; Available online: https://matlab.mathworks.com (accessed on 2 January 2021).
- Chong, J.; Wishart, D.S.; Xia, J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinforma. 2019, 68, e86. [Google Scholar] [CrossRef]
- Azevedo, M.C.S.; Silva, R.R.; Jacomino, A.P.; Genovese, M.P. Physicochemical variability of cambuci fruit (Campomanesia phaea) from a same orchard, from different locations and at different ripening stages. J. Food Sci. Agric. 2016, 39, 750–761. [Google Scholar] [CrossRef]
- Bianchini, F.G.; Balbi, R.V.; Pio, R.; Fernandes, D.; Pasqual, M.; Valério, E.; Vilas, D.B. Caracterização morfológica e química de frutos de cambucizeiro Morphological and chemical characterization of the fruits of cambuci fruit tree. Bragantia 2016, 10–18. [Google Scholar]
- dos Reis, L.C.R.; Facco, E.M.P.; Salvador, M.; Flôres, S.H.; de Oliveira Rios, A. Antioxidant potential and physicochemical characterization of yellow, purple and orange passion fruit. J. Food Sci. Technol. 2018, 55, 2679–2691. [Google Scholar] [CrossRef] [PubMed]
- Bassan, M.M.; Mourão Filho, F.D.A.A.; Caron, V.C.; do Couto, H.T.Z.; Jacomino, A.P. The harvesting system affects the quality and conservation of the ‘Tahiti’ acid lime. Sci. Hortic. 2013, 155, 72–77. [Google Scholar] [CrossRef]
- Su, M.; Zhang, B.; Ye, Z.; Chen, K.; Guo, J.; Gu, X.; Shen, J. Pulp volatiles measured by an electronic nose are related to harvest season, TSS concentration and TSS/TA ratio among 39 peaches and nectarines. Sci. Hortic. 2013, 150, 146–153. [Google Scholar] [CrossRef]
- Genovese, M.I.; Da Silva Pinto, M.; De Souza Schmidt Goncalves, a.E.; Lajolo, F.M. Bioactive Compounds and Antioxidant Capacity of Exotic Fruits and Commercial Frozen Pulps from Brazil. Food Sci. Technol. Int. 2008, 14, 207–214. [Google Scholar] [CrossRef]
- Bianchini, F.G.; Balbi, R.V.; Pio, R.; Bruzi, A.T.; Silva, D.F. da Parents choice and genetic divergence between cambuci fruit tree accessions. Crop Breed. Appl. Biotechnol. 2017, 17, 214–220. [Google Scholar] [CrossRef]
- Santucci, C.; Tenori, L.; Luchinat, C. NMR fingerprinting as a tool to evaluate post-harvest time-related changes of peaches, tomatoes and plums. Food Res. Int. 2015, 75, 106–114. [Google Scholar] [CrossRef]
- Chen, J.; Chan, P.H.; Lam, C.T.W.; Li, Z.; Lam, K.Y.C.; Yao, P.; Dong, T.T.X.; Lin, H.; Lam, H.H.N.; Tsim, K.W.K. Fruit of Ziziphus jujuba (Jujube) at two stages of maturity: Distinction by metabolic profiling and biological assessment. J. Agric. Food Chem. 2015, 63, 739–744. [Google Scholar] [CrossRef]
- Soininen, P. Quantitative 1H NMR Spectroscopy—Chemical and Biological Applications; Kuopio Uniersity Publications: Kupio, Finland, 2008; ISBN 9789512709786. [Google Scholar]
- Li, X.; Hu, K. Chapter Three-Quantitative NMR Studies of Multiple Compound Mixtures. In Annual Reports on NMR Spectroscopy; Academic Press: Cambridge, MA, USA, 2017; Volume 90, pp. 85–143. ISBN 0066-4103. [Google Scholar]
- Giraudeau, P. Challenges and perspectives in quantitative NMR. Magn. Reson. Chem. 2017, 55, 61–69. [Google Scholar] [CrossRef]
- Marquez, B.L.; Williamson, R.T. Quantitative applications of NMR spectroscopy. Chem. Eng. Pharm. Ind. 2019, 1, 133–149. [Google Scholar] [CrossRef]
- Navarro, Y.; Soengas, R.; Iglesias, M.J.; Ortiz, F.L. Use of NMR for the Analysis and Quantification of the Sugar Composition in Fresh and Store-Bought Fruit Juices. J. Chem. Educ. 2020, 97, 831–837. [Google Scholar] [CrossRef]
- Das, U.N. Sucrose, fructose, glucose, and their link to metabolic syndrome and cancer. Nutrition 2015, 31, 249–257. [Google Scholar] [CrossRef]
- Lu, X.-H.; Sun, D.-Q.; Wu, Q.-S.; Liu, S.-H.; Sun, G.-M. Physico-Chemical Properties, Antioxidant Activity and Mineral Contents of Pineapple Genotypes Grown in China. Molecules 2014, 19, 8518–8532. [Google Scholar] [CrossRef] [PubMed]
- Sturm, K.; Koron, D.; Stampar, F. The composition of fruit of different strawberry varieties depending on maturity stage. Food Chem. 2003, 83, 417–422. [Google Scholar] [CrossRef]
- Chen, Y.; Ge, Y.; Zhao, J.; Wei, M.; Li, C.; Hou, J.; Cheng, Y.; Chen, J. Postharvest sodium nitroprusside treatment maintains storage quality of apple fruit by regulating sucrose metabolism. Postharvest Biol. Technol. 2019, 154, 115–120. [Google Scholar] [CrossRef]
- Kou, J.; Wei, Y.; He, X.; Xu, J.; Xu, F.; Shao, X. Infection of post-harvest peaches by Monilinia fructicola accelerates sucrose decomposition and stimulates the Embden–Meyerhof–Parnas pathway. Hortic. Res. 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Li, M.; Feng, F.; Cheng, L. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE 2012, 7, e33055. [Google Scholar] [CrossRef]
- Botton, A.; Rasori, A.; Ziliotto, F.; Moing, A.; Maucourt, M.; Bernillon, S.; Deborde, C.; Petterle, A.; Varotto, S.; Bonghi, C. The peach HECATE3-like gene FLESHY plays a double role during fruit development. Plant Mol. Biol. 2016, 91, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Girelli, C.R.; Accogli, R.; Del Coco, L.; Angilè, F.; De Bellis, L.; Fanizzi, F.P. 1H-NMR-based metabolomic profiles of different sweet melon (Cucumis melo L.) Salento varieties: Analysis and comparison. Food Res. Int. 2018, 114, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.P.; Famiani, F. Organic acids in fruits: Metabolism, functions and contents. Hortic. Rev. (Am. Soc. Hortic. Sci). 2018, 45, 371–430. [Google Scholar] [CrossRef]
- Sieniawska, E.; Baj, T. Tannins. In Pharmacognosy: Fundamentals, Applications and Strategy; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Donado-Pestana, C.M.; Pessoa, É.V.M.; Rodrigues, L.; Rossi, R.; Moura, M.H.C.; dos Santos-Donado, P.R.; Castro, É.; Festuccia, W.T.; Genovese, M.I. Polyphenols of cambuci (Campomanesia phaea (O. Berg.) fruit ameliorate insulin resistance and hepatic steatosis in obese mice. Food Chem. 2021, 340, 128169. [Google Scholar] [CrossRef]
- Reynertson, K.A.; Yang, H.; Jiang, B.; Basile, M.J.; Kennelly, E.J. Quantitative analysis of antiradical phenolic constituents from fourteen edible Myrtaceae fruits. Food Chem. 2008, 109, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Donado-Pestana, C.M.; Moura, M.H.C.; de Araujo, R.L.; de Santiago, G.L.; de Moraes Barros, H.R.; Genovese, M.I. Polyphenols from Brazilian native Myrtaceae fruits and their potential health benefits against obesity and its associated complications. Curr. Opin. Food Sci. 2018. [Google Scholar] [CrossRef]
- Bontpart, T.; Marlin, T.; Vialet, S.; Guiraud, J.L.; Pinasseau, L.; Meudec, E.; Sommerer, N.; Cheynier, V.; Terrier, N. Two shikimate dehydrogenases, VvSDH3 and VvSDH4, are involved in gallic acid biosynthesis in grapevine. J. Exp. Bot. 2016. [Google Scholar] [CrossRef] [PubMed]
- Estevez, A.M.; Estévez, R.J. A short overview on the medicinal chemistry of (–)-shikimic acid. Mini Rev. Med. Chem. 2012, 12, 1443–1464. [Google Scholar] [CrossRef]
- Ghosh, S.; Chisti, Y.; Banerjee, U.C. Production of shikimic acid. Biotechnol. Adv. 2012, 30, 1425–1431. [Google Scholar] [CrossRef]
- Marchiosi, R.; Ferro, A.P.; Ramos, A.V.G.; Baldoqui, D.C.; Constantin, R.P.; Constantin, R.P.; dos Santos, W.D.; Ferrarese-Filho, O. Calophyllum brasiliense Cambess: An alternative and promising source of shikimic acid. Sustain. Chem. Pharm. 2019. [Google Scholar] [CrossRef]
- Rabelo, T.K.; Zeidán-Chuliá, F.; Caregnato, F.F.; Schnorr, C.E.; Gasparotto, J.; Serafini, M.R.; de Souza Araújo, A.A.; Quintans-Junior, L.J.; Moreira, J.C.F.; Gelain, D.P. In Vitro Neuroprotective Effect of Shikimic Acid Against Hydrogen Peroxide-Induced Oxidative Stress. J. Mol. Neurosci. 2015. [Google Scholar] [CrossRef]
- Wang, G.W.; Hu, W.T.; Huang, B.K.; Qin, L.P. Illicium verum: A review on its botany, traditional use, chemistry and pharmacology. J. Ethnopharmacol. 2011, 136, 10–20. [Google Scholar] [CrossRef]
- Magwaza, L.S.; Mditshwa, A.; Tesfay, S.Z.; Opara, U.L. An overview of preharvest factors affecting vitamin C content of citrus fruit. Sci. Hortic. (Amsterdam). 2017, 216, 12–21. [Google Scholar] [CrossRef]
- Granger, M.; Eck, P. Chapter Seven - Dietary Vitamin C in Human Health. In New Research and Developments of Water-Soluble Vitamins; Academic Press: Cambridge, MA, USA, 2018; Volume 83, ISBN 1043-4526. [Google Scholar]
- Kim, I.; Ku, K.H.; Jeong, M.C.; Kwon, S.I.; Lee, J. Metabolite profiling and antioxidant activity of 10 new early- to mid-season apple cultivars and 14 traditional cultivars. Antioxidants 2020, 9, 443. [Google Scholar] [CrossRef] [PubMed]
- Galili, S.; Amir, R.; Galili, G. Genetic Engineering of Amino Acid Metabolism in Plants. Adv. Plant Biochem. Mol. Biol. 2008, 1, 49–80. [Google Scholar]
- Batista-Silva, W.; Nascimento, V.L.; Medeiros, D.B.; Nunes-Nesi, A.; Ribeiro, D.M.; Zsögön, A.; Araújo, W.L. Modifications in Organic Acid Profiles During Fruit Development and Ripening: Correlation or Causation? Front. Plant Sci. 2018, 9, 1689. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, K. Umami the Fifth Basic Taste: History of Studies on Receptor Mechanisms and Role as a Food Flavor. Biomed Res. Int. 2015, 189402. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. Proc. J. Experimental Botany 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Y.; Luo, W.; Deng, L.; Yao, S.; Zeng, K. Primary metabolites analysis of induced citrus fruit disease resistance upon treatment with oligochitosan, salicylic acid and Pichia membranaefaciens. Biol. Control 2020, 148, 104289. [Google Scholar] [CrossRef]
- Byeon, S.-E.; Lee, J. Differential responses of fruit quality and major targeted metabolites in three different cultivars of cold-stored figs (Ficus carica L.). Sci. Hortic. 2020, 260, 108877. [Google Scholar] [CrossRef]
- Nafees, M.; Jafar Jaskani, M.; Ahmad, I.; Maryam; Ashraf, I.; Maqsood, A.; Ahmar, S.; Azam, M.; Hussain, S.; Hanif, A.; et al. Biochemical Analysis of Organic Acids and Soluble Sugars in Wild and Cultivated Pomegranate Germplasm Based in Pakistan. Plants 2020, 9, 493. [Google Scholar] [CrossRef]
- Hussain, S.B.; Guo, L.X.; Shi, C.Y.; Khan, M.A.; Bai, Y.X.; Du, W.; Liu, Y.Z. Assessment of sugar and sugar accumulation-related gene expression profiles reveal new insight into the formation of low sugar accumulation trait in a sweet orange (Citrus sinensis) bud mutant. Mol. Biol. Rep. 2020, 47, 2781–2791. [Google Scholar] [CrossRef]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 65, 33–67. [Google Scholar] [CrossRef]
- Etienne, A.; Génard, M.; Lobit, P.; Mbeguié-A-Mbéguié, D.; Bugaud, C. What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 2013, 64, 1451–1469. [Google Scholar] [CrossRef] [PubMed]
- Tretter, L.; Patocs, A.; Chinopoulos, C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1086–1101. [Google Scholar] [CrossRef] [PubMed]
- Kinnersley, A.M.; Turano, F.J. Gamma aminobutyric acid (GABA) and plant responses to stress. CRC. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Tarkowski, Ł.P.; Signorelli, S.; Höfte, M. γ-Aminobutyric acid and related amino acids in plant immune responses: Emerging mechanisms of action. Plant Cell Environ. 2020, 43, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Podlešáková, K.; Ugena, L.; Spíchal, L.; Doležal, K.; De Diego, N. Phytohormones and polyamines regulate plant stress responses by altering GABA pathway. N. Biotechnol. 2019, 48, 53–65. [Google Scholar] [CrossRef]
- Batushansky, A.; Kirma, M.; Grillich, N.; Toubiana, D.; Anh Pham, P.; Balbo, I.; Fromm, H.; Galili, G.; Fernie, A.R.; Fait, A. Combined transcriptomics and metabolomics of Arabidopsis thaliana seedlings exposed to exogenous gaba suggest its role in plants is predominantly metabolic. Mol. Plant 2014, 7, 1065–1068. [Google Scholar] [CrossRef]
- Oms-Oliu, G.; Hertog, M.L.A.T.M.; Van de Poel, B.; Ampofo-Asiama, J.; Geeraerd, A.H.; Nicolai, B.M. Metabolic characterization of tomato fruit during preharvest development, ripening, and postharvest shelf-life. Postharvest Biol. Technol. 2011, 62, 7–16. [Google Scholar] [CrossRef]
- Mack, C.; Wefers, D.; Schuster, P.; Weinert, C.H.; Egert, B.; Bliedung, S.; Trierweiler, B.; Muhle-Goll, C.; Bunzel, M.; Luy, B.; et al. Untargeted multi-platform analysis of the metabolome and the non-starch polysaccharides of kiwifruit during postharvest ripening. Postharvest Biol. Technol. 2017. [Google Scholar] [CrossRef]
| Accession | Fresh mass | Height | Diameter | SSC | TA | Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 78.3 | bc | 42.4 | cd | 60.7 | cd | 11.0 | cd | 3.0 | c | 3.7 | a |
| 2 | 88.3 | c | 45.4 | d | 65.4 | d | 11.6 | c | 2.4 | bc | 4.8 | ab |
| 3 | 36.3 | a | 34.9 | a | 48.2 | a | 8.7 | a | 1.1 | a | 8.1 | c |
| 4 | 41.7 | a | 35.7 | ab | 47.6 | a | 7.8 | a | 2.2 | bc | 3.5 | a |
| 5 | 44.5 | a | 37.1 | abc | 51.8 | ab | 9.0 | ab | 2.4 | bc | 3.8 | a |
| 6 | 57.6 | ab | 42.2 | cd | 52.0 | ab | 14.0 | f | 2.6 | c | 5.7 | abc |
| 7 | 44.4 | a | 38.6 | abc | 53.0 | ab | 10.7 | bcd | 2.4 | bc | 4.4 | ab |
| 8 | 86.0 | c | 41.9 | cde | 66.4 | d | 13.4 | ef | 4.3 | d | 3.2 | a |
| 9 | 53.7 | a | 42.2 | cd | 56.4 | bc | 12.2 | de | 3.2 | c | 3.9 | a |
| 10 | 44.9 | a | 36.2 | abc | 52.1 | ab | 9.3 | abc | 1.5 | ab | 6.6 | bc |
| Mean | 57.0 | 39.6 | 55.1 | 10.8 | 2.5 | 4.8 | ||||||
| CV (%) | 26.2 | 10.0 | 8.5 | 8.6 | 14.7 | 18.0 | ||||||
| Accession | SCR | GLU | FRU | GLC | ADO | INS | ETH | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1553.0 | ab | 460.2 | abc | 426.5 | abc | 17.1 | a | 6.4 | abc | 11.6 | a | 6.8 | ab |
| 2 | 2080.1 | bcd | 444.7 | abc | 611.9 | bc | 24.0 | a | 6.0 | ab | 40.2 | a | 6.5 | ab |
| 3 | 1756.8 | abc | 571.7 | bc | 624.0 | bc | 26.2 | a | 2.2 | a | 18.2 | a | 11.3 | b |
| 4 | 784.0 | A | 220.9 | a | 284.4 | a | 23.1 | a | 6.1 | ab | 22.6 | a | 5.5 | ab |
| 5 | 1908.1 | bc | 481.8 | abc | 481.0 | abc | 11.5 | a | 4.2 | ab | 6.6 | a | 8.8 | ab |
| 6 | 3098.2 | d | 693.5 | c | 695.5 | c | 51.2 | b | 11.1 | bc | 3.2 | a | 9.8 | ab |
| 7 | 2073.1 | bcd | 416.8 | ab | 489.3 | abc | 9.9 | a | 2.5 | ab | 19.7 | a | 5.5 | ab |
| 8 | 1482.6 | ab | 235.4 | a | 571.7 | abc | 25.2 | a | 15.1 | c | 57.1 | a | 4.9 | ab |
| 9 | 2108.2 | bcd | 470.7 | abc | 656.8 | c | 8.7 | a | 9.0 | abc | 44.5 | a | 3.5 | a |
| 10 | 2617.0 | cd | 451.8 | abc | 348.9 | ab | 17.6 | a | 3.4 | ab | 13.0 | a | 9.2 | ab |
| Mean | 1946.1 | 444.7 | 519.0 | 21.5 | 6.6 | 23.7 | 7.2 | |||||||
| CV (%) | 25.4 | 27.7 | 26.0 | 51.2 | 63.6 | 135.9 | 48.6 | |||||||
| Accession | CIT | SHA | QA | ASC | MAA | GAL | SUC | POLY | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1614.5 | bcd | 549.7 | a | 265.7 | a | 41.7 | a | 15.1 | ab | 5.3 | abcd | 0.6 | a | 482.1 | abc |
| 2 | 1966.2 | cd | 461.5 | a | 325.2 | ab | 25.9 | a | 18.5 | ab | 9.5 | d | 0.6 | a | 374.0 | abc |
| 3 | 617.0 | a | 370.7 | a | 346.4 | abc | 40.8 | a | 12.3 | ab | 2.0 | abc | 0.2 | a | 8.7 | a |
| 4 | 1400.3 | abc | 521.3 | a | 466.9 | d | 103.1 | b | 18.5 | ab | 5.9 | bcd | 0.7 | ab | 509.7 | abc |
| 5 | 1843.3 | bcd | 662.0 | ab | 383.8 | abc | 38.4 | a | 13.7 | ab | 2.0 | abc | 0.6 | a | 340.7 | abc |
| 6 | 2385.2 | de | 928.3 | bc | 464.0 | cd | 56.7 | a | 6.8 | a | 1.5 | ab | 0.8 | ab | 750.8 | bc |
| 7 | 1484.9 | abcd | 464.7 | a | 333.4 | ab | 29.0 | a | 14.4 | ab | 1.8 | abc | 0.2 | a | 143.9 | a |
| 8 | 2934.4 | e | 1012.8 | c | 355.6 | abcd | 40.5 | a | 18.5 | ab | 5.5 | abcd | 2.5 | b | 836.8 | c |
| 9 | 1969.0 | cd | 559.3 | a | 366.5 | abcd | 53.9 | a | 32.3 | bc | 6.2 | cd | 0.5 | a | 281.2 | ab |
| 10 | 934.0 | ab | 315.0 | a | 425.5 | bcd | 41.3 | a | 21.1 | bc | 1.1 | a | 0.3 | a | 314.6 | ab |
| Mean | 1714.9 | 584.5 | 373.3 | 47.1 | 17.1 | 4.1 | 0.7 | 412.1 | ||||||||
| CV (%) | 26.0 | 27.9 | 15.0 | 42.9 | 87.4 | 51.7 | 121.6 | 57.3 |
| Accession | GLUT | GLN | THR | GABA | ILE | VAL | ALA | LEU | CHO | GLM | ASP | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 509.1 | a | 24.5 | A | 13.4 | a | 12.6 | ab | 4.1 | ab | 2.5 | ab | 0.6 | a | 1.5 | ab | 1.2 | a | 179.9 | a | 34.2 | ab |
| 2 | 578.6 | ab | 28.8 | A | 14.9 | ab | 10.1 | ab | 3.5 | ab | 2.2 | ab | 2.5 | a | 1.3 | ab | 1.4 | a | 193.6 | a | 34.0 | ab |
| 3 | 711.3 | ab | 33.0 | A | 9.0 | a | 5.1 | a | 1.3 | a | 0.7 | a | 2.2 | a | 0.3 | a | 1.6 | ab | 265.1 | a | 13.4 | a |
| 4 | 1122.0 | c | 52.0 | B | 17.1 | ab | 12.6 | ab | 4.2 | ab | 2.5 | ab | 2.6 | a | 1.5 | ab | 2.7 | b | 429.9 | b | 35.9 | ab |
| 5 | 738.6 | ab | 32.3 | A | 11.9 | a | 8.9 | ab | 3.1 | ab | 1.8 | ab | 0.6 | a | 1.0 | ab | 2.2 | ab | 274.1 | a | 37.7 | ab |
| 6 | 847.1 | bc | 34.3 | A | 26.7 | b | 17.8 | ab | 5.7 | ab | 3.5 | ab | 0.5 | a | 2.2 | ab | 1.5 | a | 275.9 | a | 59.1 | bc |
| 7 | 655.2 | ab | 26.7 | A | 11.2 | a | 2.0 | a | 0.8 | a | 0.3 | a | 1.0 | a | 0.1 | a | 1.5 | a | 204.8 | a | 17.3 | a |
| 8 | 764.2 | ab | 34.9 | A | 16.8 | ab | 25.9 | b | 7.8 | b | 4.7 | b | 4.8 | a | 2.8 | b | 2.0 | ab | 285.6 | a | 80.0 | c |
| 9 | 704.4 | ab | 27.2 | A | 5.7 | a | 6.7 | a | 2.2 | ab | 1.3 | ab | 0.5 | a | 0.7 | ab | 1.4 | a | 272.2 | a | 20.0 | a |
| 10 | 849.9 | bc | 34.2 | A | 15.0 | ab | 7.5 | a | 2.4 | ab | 1.4 | ab | 0.0 | a | 0.8 | ab | 1.9 | ab | 289.2 | a | 25.6 | a |
| Mean | 748.0 | 32.8 | 14.2 | 10.9 | 3.5 | 2.1 | 1.4 | 1.2 | 1.7 | 267.0 | 35.7 | |||||||||||
| CV (%) | 17.4 | 19.7 | 39.6 | 74.5 | 73.7 | 81.5 | 212.2 | 86.1 | 31.1 | 19.5 | 43.5 | |||||||||||
| Accession | Sugar | Rel. Sugar | Org Acids | Am Acids | Poly | Sum | S/OA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2439.6 | ab | 41.9 | a | 2492.7 | abc | 783.7 | a | 482.1 | abc | 6240.0 | a | 1.0 |
| 2 | 3136.8 | bc | 76.7 | a | 2807.3 | bcd | 870.9 | abc | 374.0 | abc | 7265.6 | ab | 1.1 |
| 3 | 2952.5 | b | 58.0 | a | 1389.2 | a | 1043.2 | abc | 86.9 | a | 5529.7 | a | 2.1 |
| 4 | 1289.2 | ab | 57.4 | a | 2516.7 | abc | 1683.1 | d | 509.7 | abc | 6056.1 | a | 0.5 |
| 5 | 2870.8 | b | 31.1 | a | 2943.9 | cd | 1112.1 | abc | 340.7 | abc | 7298.6 | ab | 1.0 |
| 6 | 4487.2 | c | 75.4 | a | 3868.8 | de | 1273.3 | c | 750.8 | bc | 10455.5 | c | 1.2 |
| 7 | 2979.3 | b | 37.7 | a | 2328.3 | abc | 921.1 | abc | 143.9 | a | 6410.3 | ab | 1.3 |
| 8 | 2289.6 | ab | 102.3 | a | 4369.9 | e | 1229.6 | bc | 836.8 | c | 8828.1 | bc | 0.5 |
| 9 | 3235.6 | bc | 65.8 | a | 2962.2 | cd | 1042.2 | abc | 281.2 | ab | 7587.1 | ab | 1.1 |
| 10 | 3417.8 | bc | 43.2 | a | 1738.3 | ab | 1227.6 | bc | 314.6 | ab | 6741.6 | ab | 2.0 |
| Mean | 2909.9 | 58.9 | 2741.7 | 1118.7 | 412.1 | 7241.2 | 1.2 | ||||||
| CV (%) | 23.8 | 58.2 | 19.8 | 15.8 | 57.3 | 15.9 | 44.9 |
| Pathway | Total | Expected | Hits | Raw p | -log(p) | Holm adjust | FDR | Impact | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Alanine, aspartate and glutamate metabolism | 22 | 0.37087 | 6 | 7.56 × 10−7 | 14.095 | 7.18 × 10−5 | 3.63 × 10−5 | 0.77338 |
| 2 | Starch and sucrose metabolism | 22 | 0.37087 | 3 | 0.0052788 | 5.244 | 0.4751 | 0.072396 | 0.52723 |
| 3 | Glutathione metabolism | 26 | 0.4383 | 2 | 0.069283 | 26.696 | 1 | 0.44341 | 0.40159 |
| 4 | Citrate cycle (TCA cycle) | 20 | 0.33715 | 2 | 0.043066 | 3.145 | 1 | 0.31803 | 0.15581 |
| 5 | Butanoate metabolism | 17 | 0.28658 | 3 | 0.0024648 | 60.057 | 0.22676 | 0.046798 | 0.13636 |
| 6 | Glycine, serine and threonine metabolism | 33 | 0.5563 | 3 | 0.016545 | 41.017 | 1 | 0.15883 | 0.1204 |
| 7 | Inositol phosphate metabolism | 28 | 0.47202 | 2 | 0.078956 | 25.389 | 1 | 0.47374 | 0.10251 |
| 8 | Arginine biosynthesis | 18 | 0.30344 | 3 | 0.0029249 | 58.345 | 0.26616 | 0.046798 | 0.08544 |
| 9 | Phenylalanine, tyrosine and tryptophan biosynthesis | 22 | 0.37087 | 1 | 0.3139 | 11.587 | 1 | 1 | 0.08008 |
| 10 | Arginine and proline metabolism | 34 | 0.57316 | 2 | 0.11026 | 22.049 | 1 | 0.58806 | 0.07781 |
| 11 | Glyoxylate and dicarboxylate metabolism | 29 | 0.48887 | 4 | 0.0011247 | 67.902 | 0.1046 | 0.026993 | 0.06012 |
| 12 | Galactose metabolism | 27 | 0.45516 | 3 | 0.0094826 | 46.583 | 0.84395 | 0.11379 | 0.04805 |
| 13 | Sulfur metabolism | 15 | 0.25287 | 1 | 0.22605 | 1.487 | 1 | 0.9435 | 0.03315 |
| 14 | Phosphatidylinositol signaling system | 26 | 0.4383 | 1 | 0.35971 | 10.224 | 1 | 1 | 0.03285 |
| 15 | Glycerophospholipid metabolism | 37 | 0.62374 | 1 | 0.47106 | 0.75277 | 1 | 1 | 0.03075 |
| 16 | Purine metabolism | 63 | 1.062 | 2 | 0.28755 | 12.464 | 1 | 1 | 0.00126 |
| 17 | Aminoacyl-tRNA biosynthesis | 46 | 0.77546 | 8 | 3.34 × 10−7 | 14.912 | 3.21 × 10−5 | 3.21 × 10−5 | 0 |
| 18 | Valine, leucine and isoleucine biosynthesis | 22 | 0.37087 | 4 | 0.00037527 | 78.879 | 0.035276 | 0.012009 | 0 |
| 19 | Nitrogen metabolism | 12 | 0.20229 | 2 | 0.016243 | 41.201 | 1 | 0.15883 | 0 |
| 20 | Valine, leucine and isoleucine degradation | 37 | 0.62374 | 3 | 0.022537 | 37.926 | 1 | 0.19669 | 0 |
| 21 | Ascorbate and aldarate metabolism | 18 | 0.30344 | 2 | 0.035399 | 33.411 | 1 | 0.28319 | 0 |
| 22 | Carbon fixation in photosynthetic organisms | 21 | 0.35401 | 2 | 0.047114 | 30.552 | 1 | 0.32307 | 0 |
| 23 | Glucosinolate biosynthesis | 65 | 10.958 | 3 | 0.092973 | 23.754 | 1 | 0.52502 | 0 |
| 24 | Monobactam biosynthesis | 8 | 0.13486 | 1 | 0.12745 | 2.06 | 1 | 0.64396 | 0 |
| 25 | Lysine biosynthesis | 9 | 0.15172 | 1 | 0.14224 | 19.502 | 1 | 0.68275 | 0 |
| 26 | Selenocompound metabolism | 13 | 0.21915 | 1 | 0.19903 | 16.143 | 1 | 0.86847 | 0 |
| 27 | Nicotinate and nicotinamide metabolism | 13 | 0.21915 | 1 | 0.19903 | 16.143 | 1 | 0.86847 | 0 |
| 28 | beta-Alanine metabolism | 18 | 0.30344 | 1 | 0.26494 | 13.282 | 1 | 1 | 0 |
| 29 | Propanoate metabolism | 20 | 0.33715 | 1 | 0.28983 | 12.385 | 1 | 1 | 0 |
| 30 | Pantothenate and CoA biosynthesis | 23 | 0.38773 | 1 | 0.32564 | 1.122 | 1 | 1 | 0 |
| 31 | Glycolysis / Gluconeogenesis | 26 | 0.4383 | 1 | 0.35971 | 10.224 | 1 | 1 | 0 |
| 32 | Cyanoamino acid metabolism | 29 | 0.48887 | 1 | 0.39213 | 0.93616 | 1 | 1 | 0 |
| 33 | Pyrimidine metabolism | 38 | 0.64059 | 1 | 0.48021 | 0.73354 | 1 | 1 | 0 |
| 34 | Cysteine and methionine metabolism | 46 | 0.77546 | 1 | 0.5481 | 0.6013 | 1 | 1 | 0 |
| 35 | Porphyrin and chlorophyll metabolism | 48 | 0.80917 | 1 | 0.56369 | 0.57325 | 1 | 1 | 0 |
| 36 | Amino sugar and nucleotide sugar metabolism | 50 | 0.84289 | 1 | 0.57876 | 0.54686 | 1 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spricigo, P.C.; Correia, B.S.B.; Borba, K.R.; Taver, I.B.; Machado, G.d.O.; Wilhelms, R.Z.; Queiroz Junior, L.H.K.; Jacomino, A.P.; Colnago, L.A. Classical Food Quality Attributes and the Metabolic Profile of Cambuci, a Native Brazilian Atlantic Rainforest Fruit. Molecules 2021, 26, 3613. https://doi.org/10.3390/molecules26123613
Spricigo PC, Correia BSB, Borba KR, Taver IB, Machado GdO, Wilhelms RZ, Queiroz Junior LHK, Jacomino AP, Colnago LA. Classical Food Quality Attributes and the Metabolic Profile of Cambuci, a Native Brazilian Atlantic Rainforest Fruit. Molecules. 2021; 26(12):3613. https://doi.org/10.3390/molecules26123613
Chicago/Turabian StyleSpricigo, Poliana Cristina, Banny Silva Barbosa Correia, Karla Rodrigues Borba, Isabela Barroso Taver, Guilherme de Oliveira Machado, Renan Ziemann Wilhelms, Luiz Henrique Keng Queiroz Junior, Angelo Pedro Jacomino, and Luiz Alberto Colnago. 2021. "Classical Food Quality Attributes and the Metabolic Profile of Cambuci, a Native Brazilian Atlantic Rainforest Fruit" Molecules 26, no. 12: 3613. https://doi.org/10.3390/molecules26123613
APA StyleSpricigo, P. C., Correia, B. S. B., Borba, K. R., Taver, I. B., Machado, G. d. O., Wilhelms, R. Z., Queiroz Junior, L. H. K., Jacomino, A. P., & Colnago, L. A. (2021). Classical Food Quality Attributes and the Metabolic Profile of Cambuci, a Native Brazilian Atlantic Rainforest Fruit. Molecules, 26(12), 3613. https://doi.org/10.3390/molecules26123613