1,2,3-Triazoles as Biomimetics in Peptide Science
Abstract
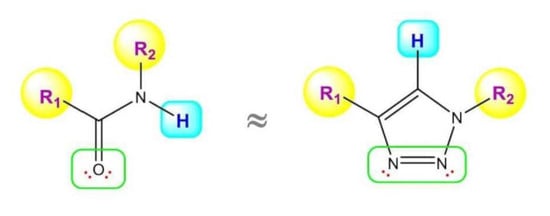
1. Introduction
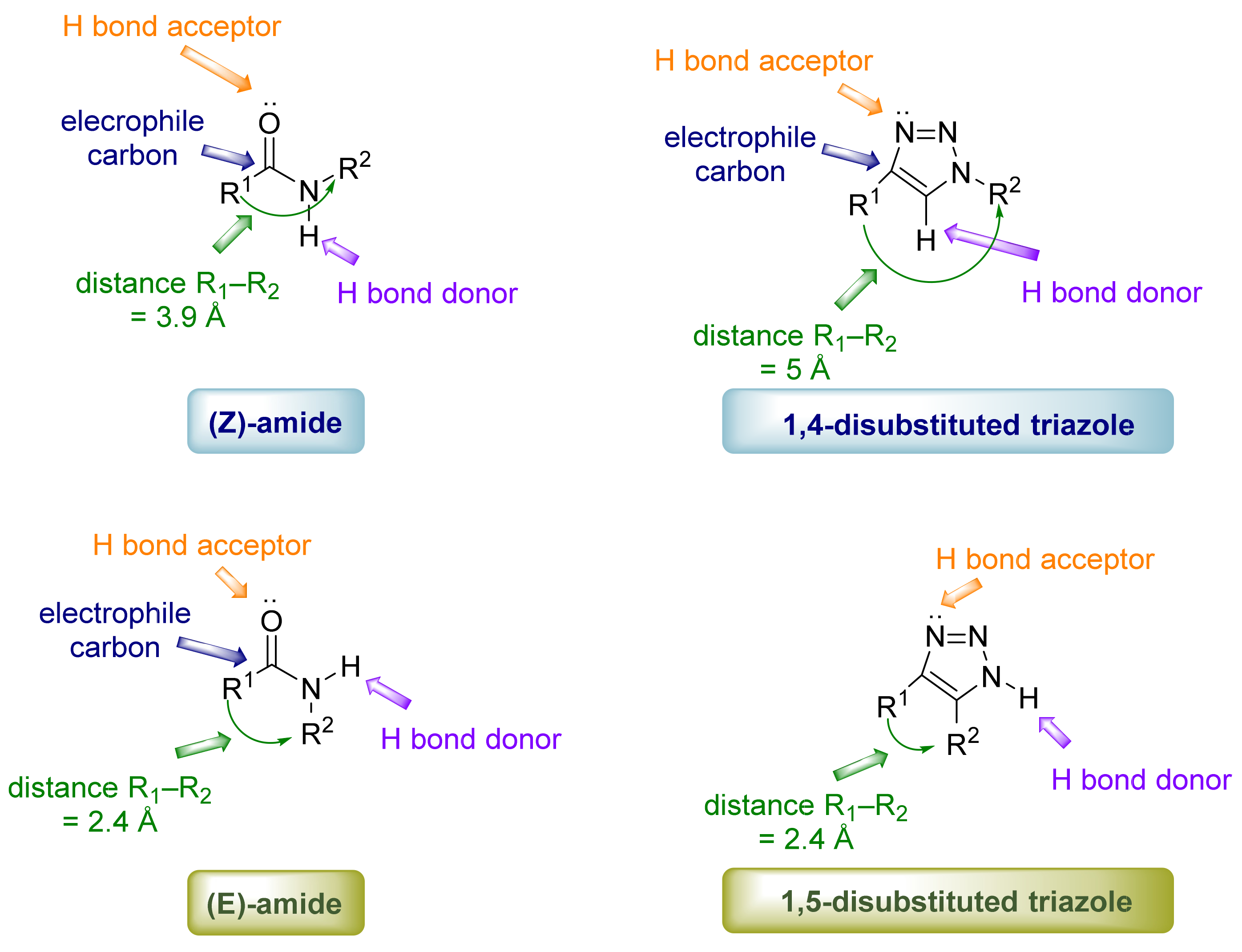
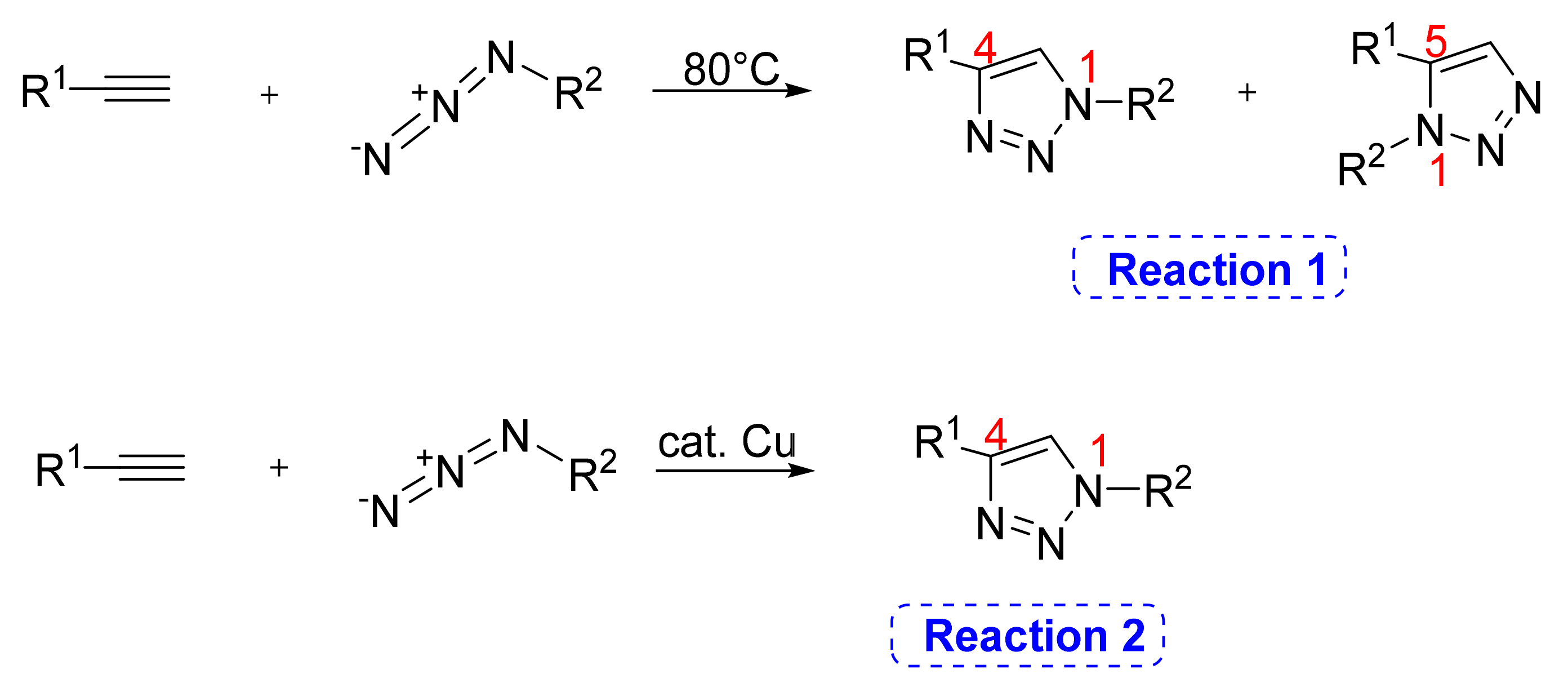
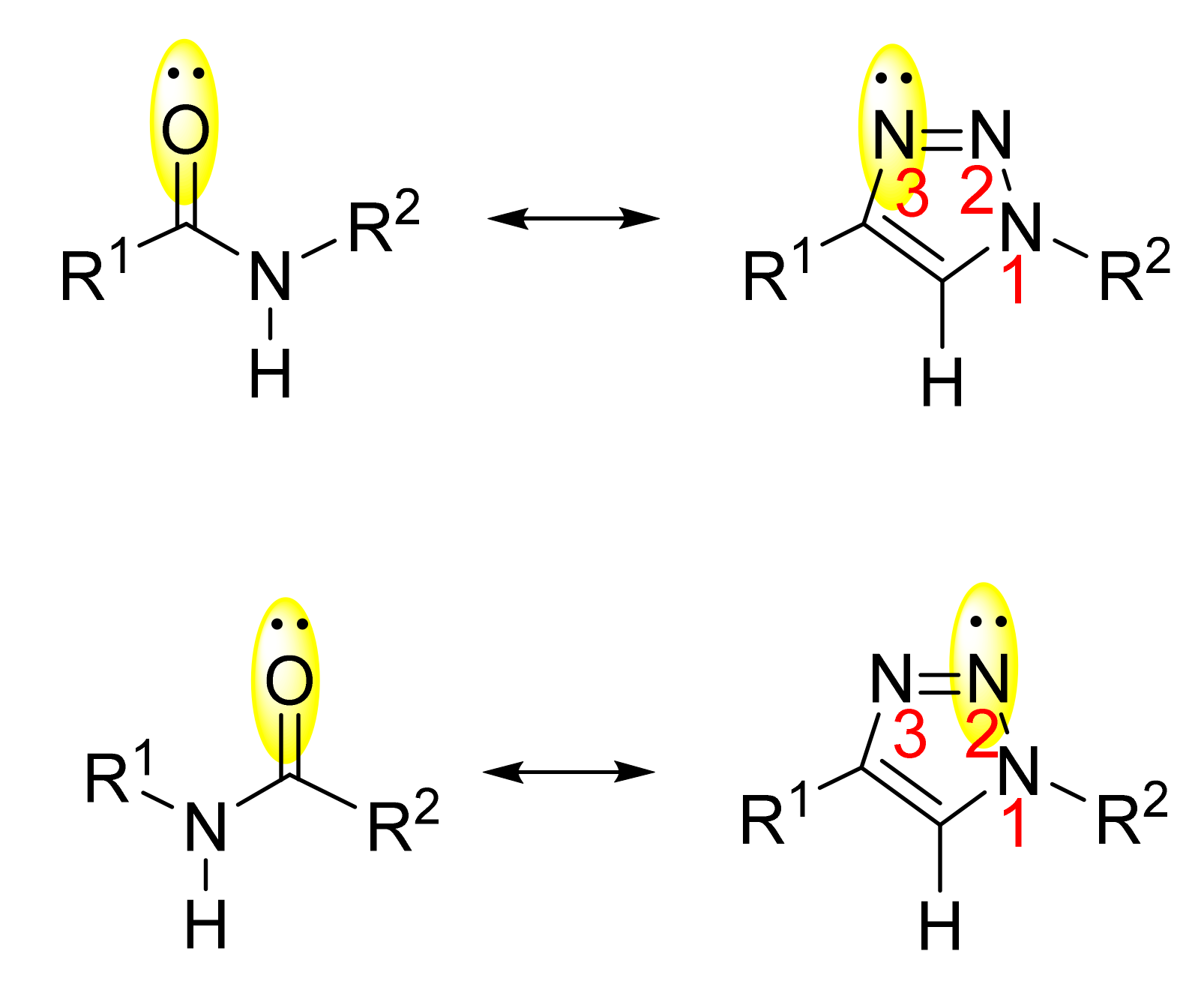
2. 1,2,3-. Triazole Moiety and Peptide Structure
2.1. Acyclic Analogues
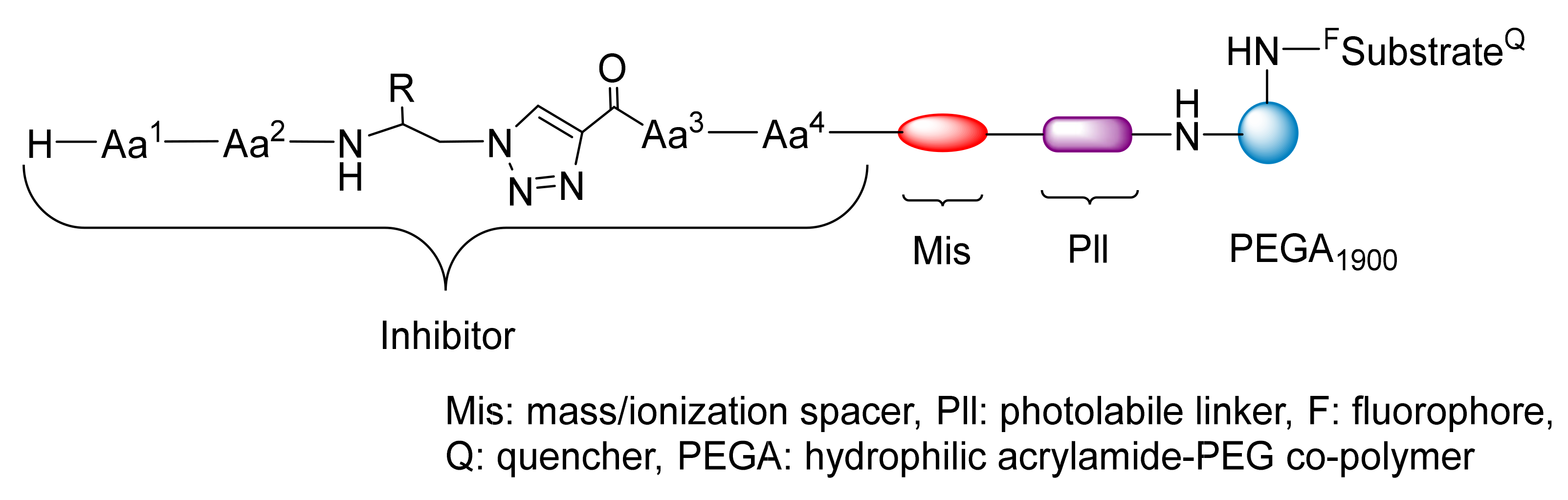
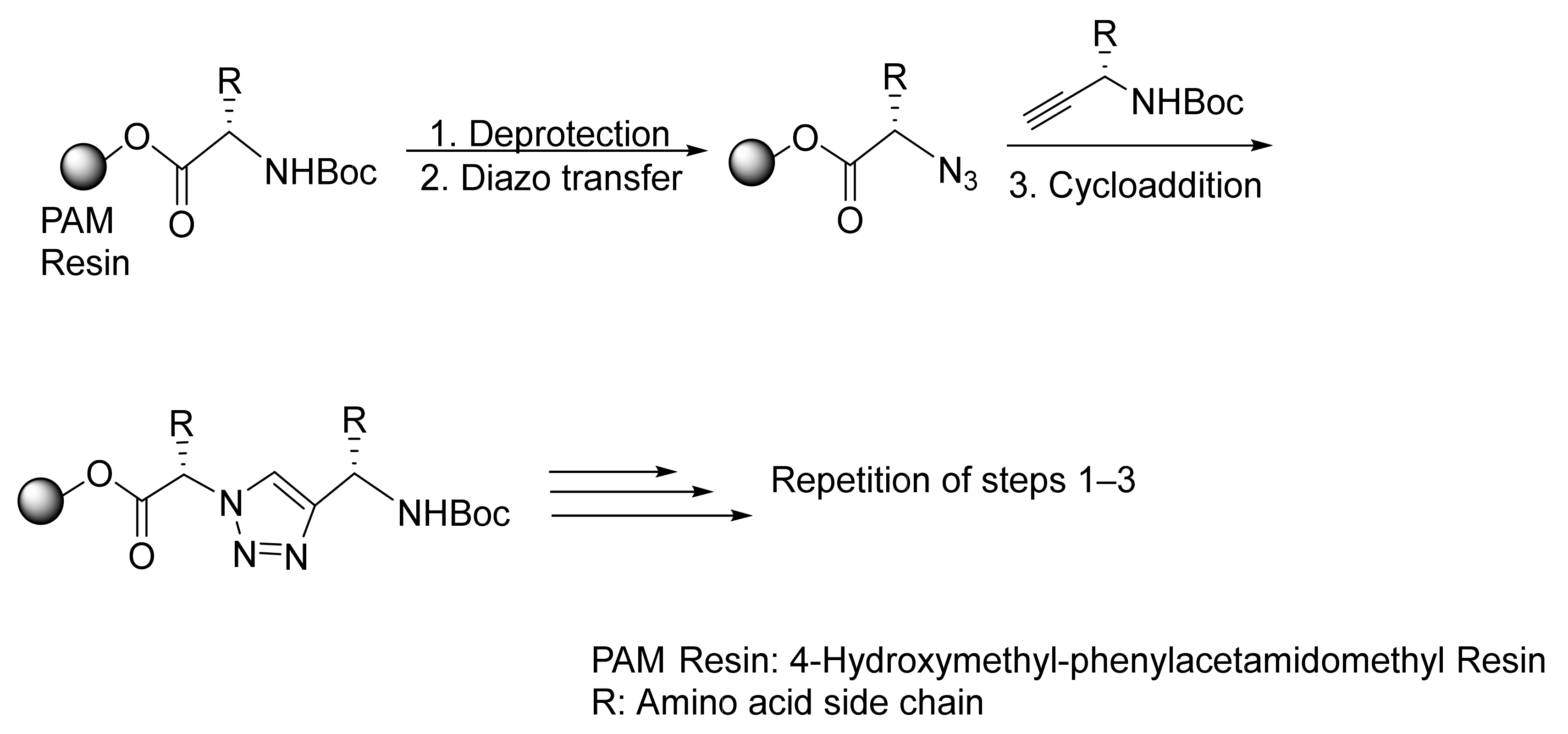
2.1.1. Peptidotriazole
2.1.2. β-Turn Mimetics
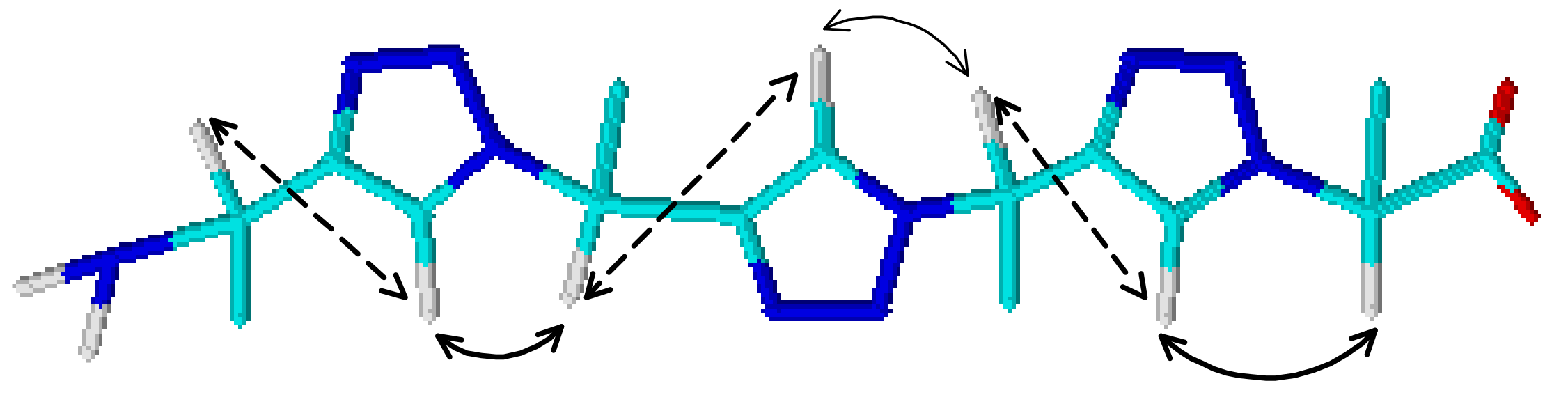
2.1.3. Triazolamer
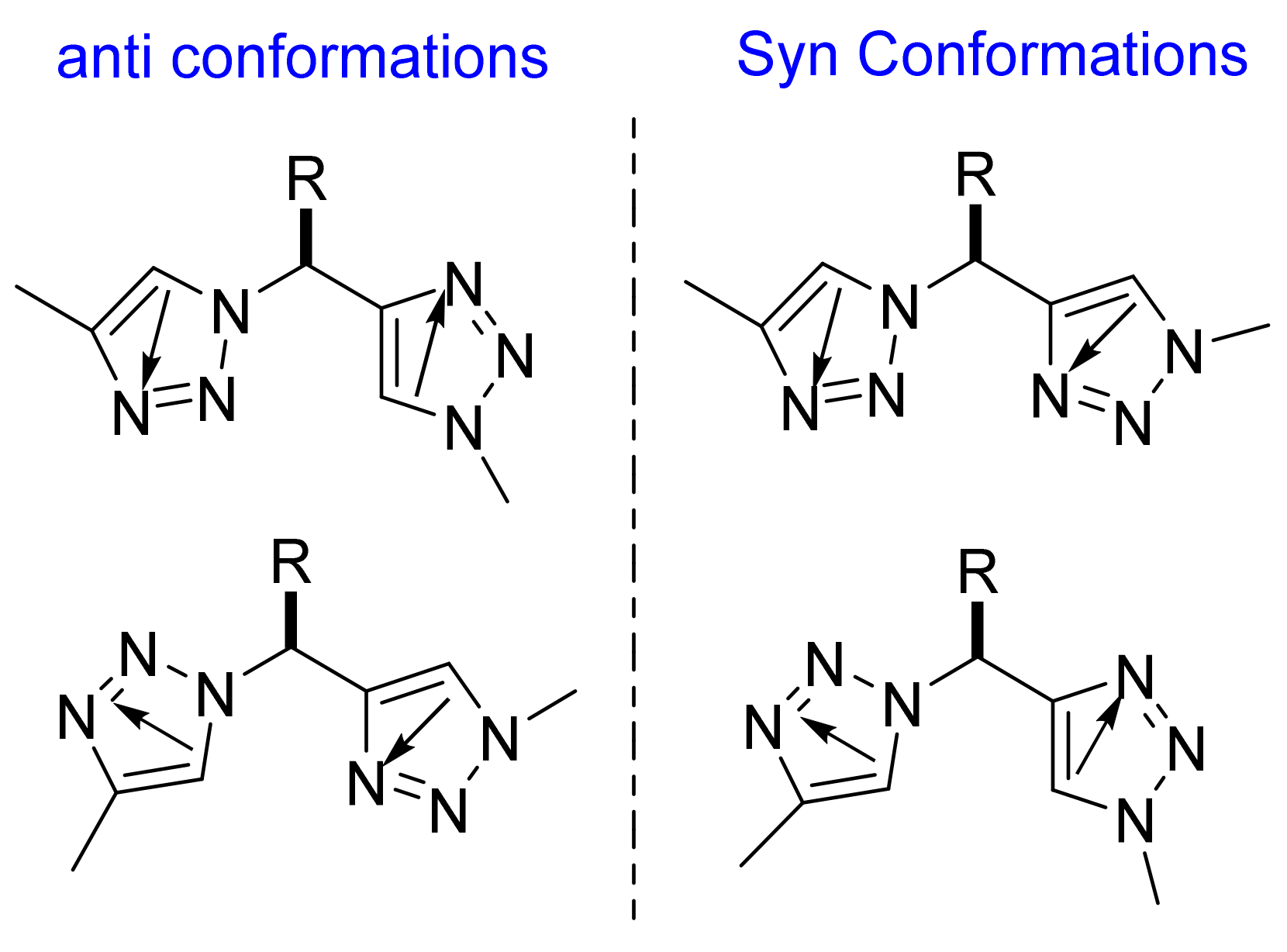
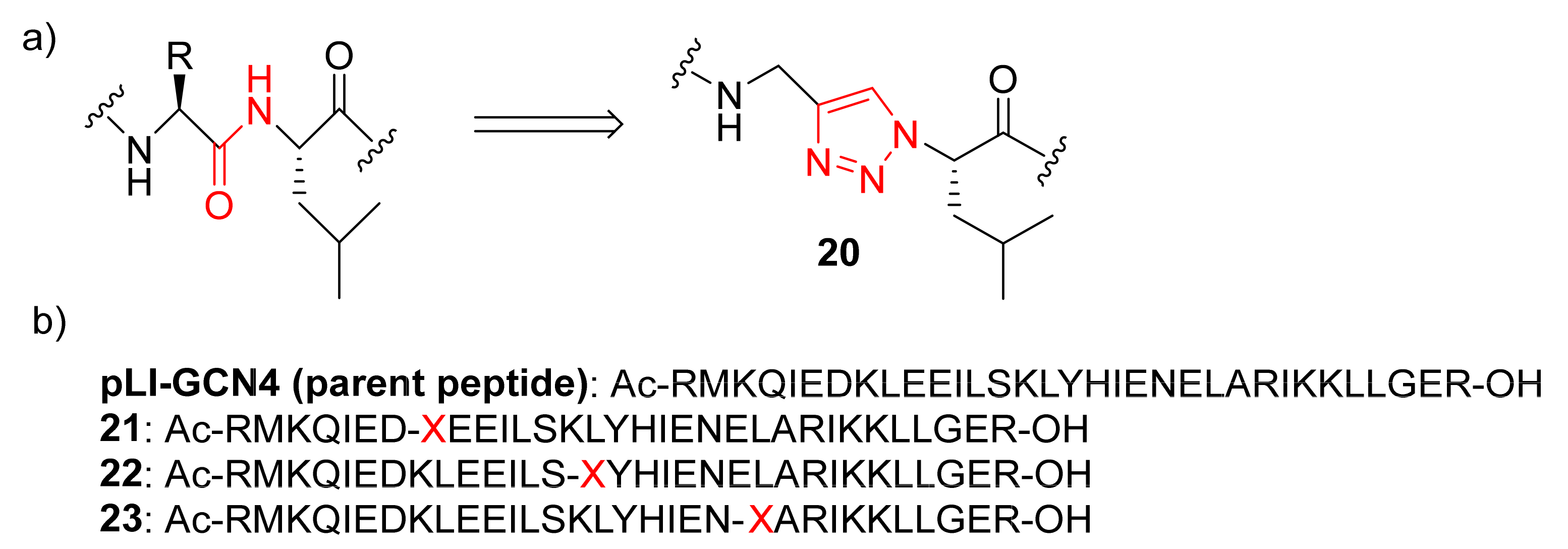
2.1.4. Coiled Coil Triazoles
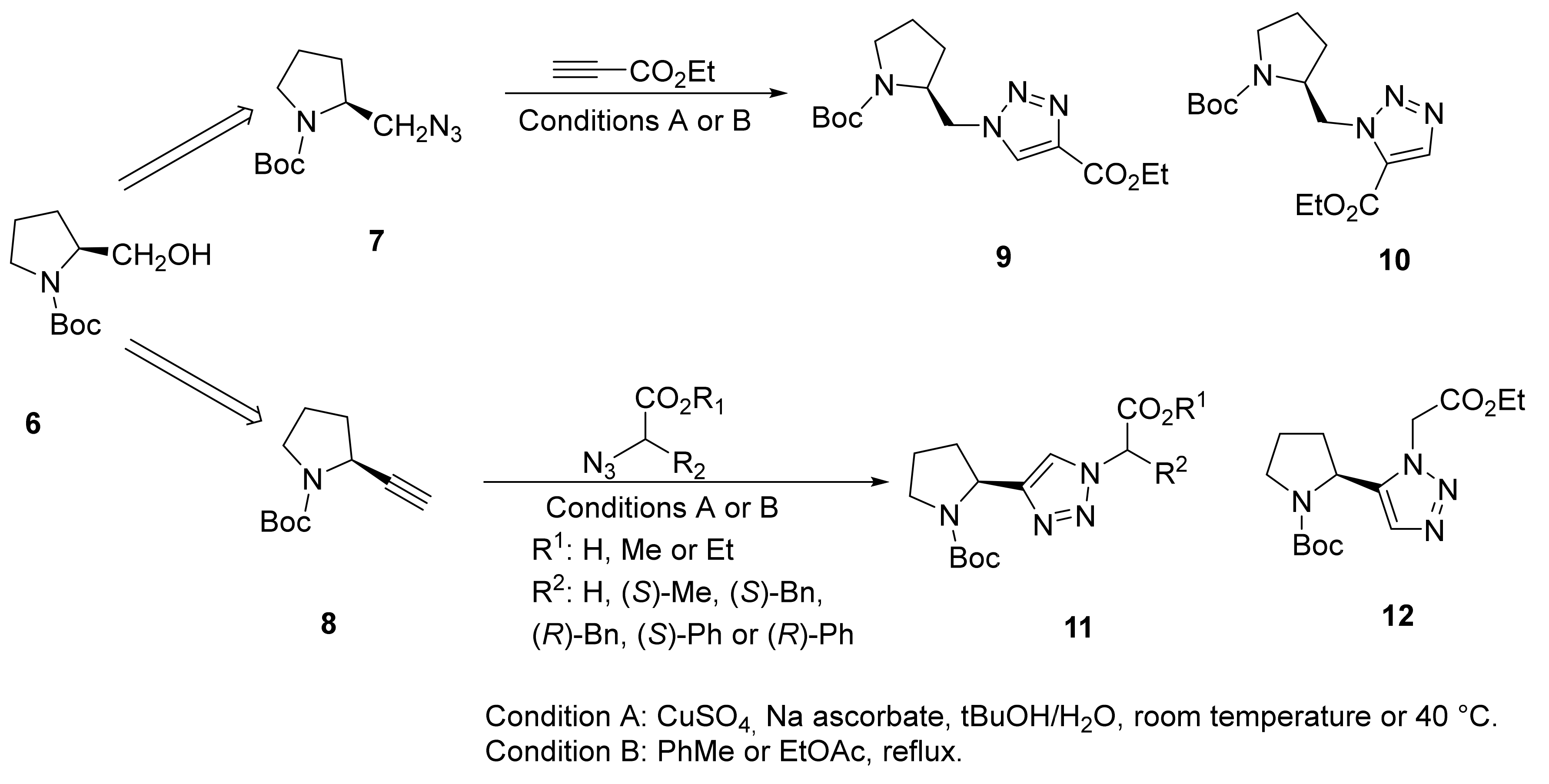
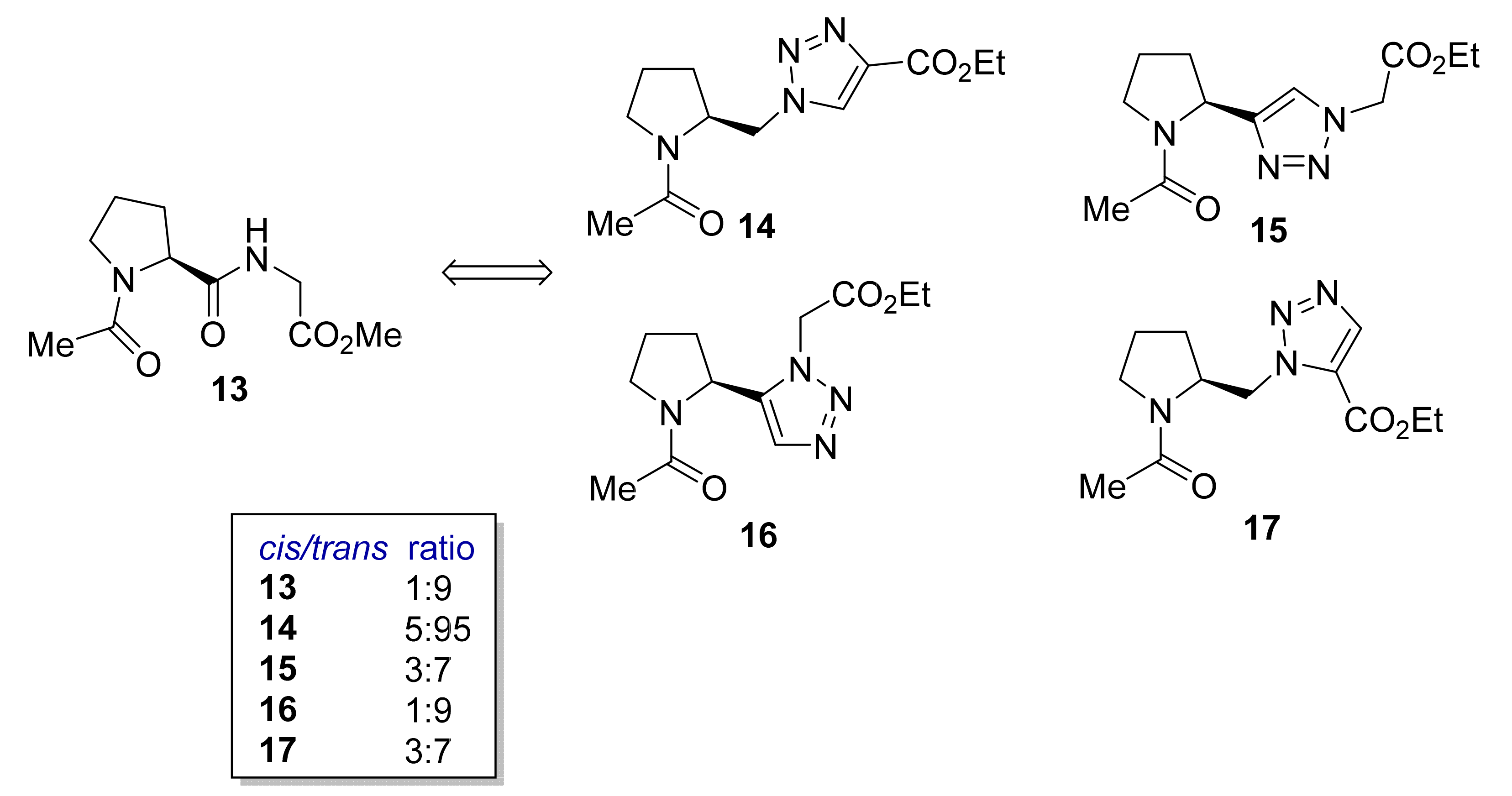
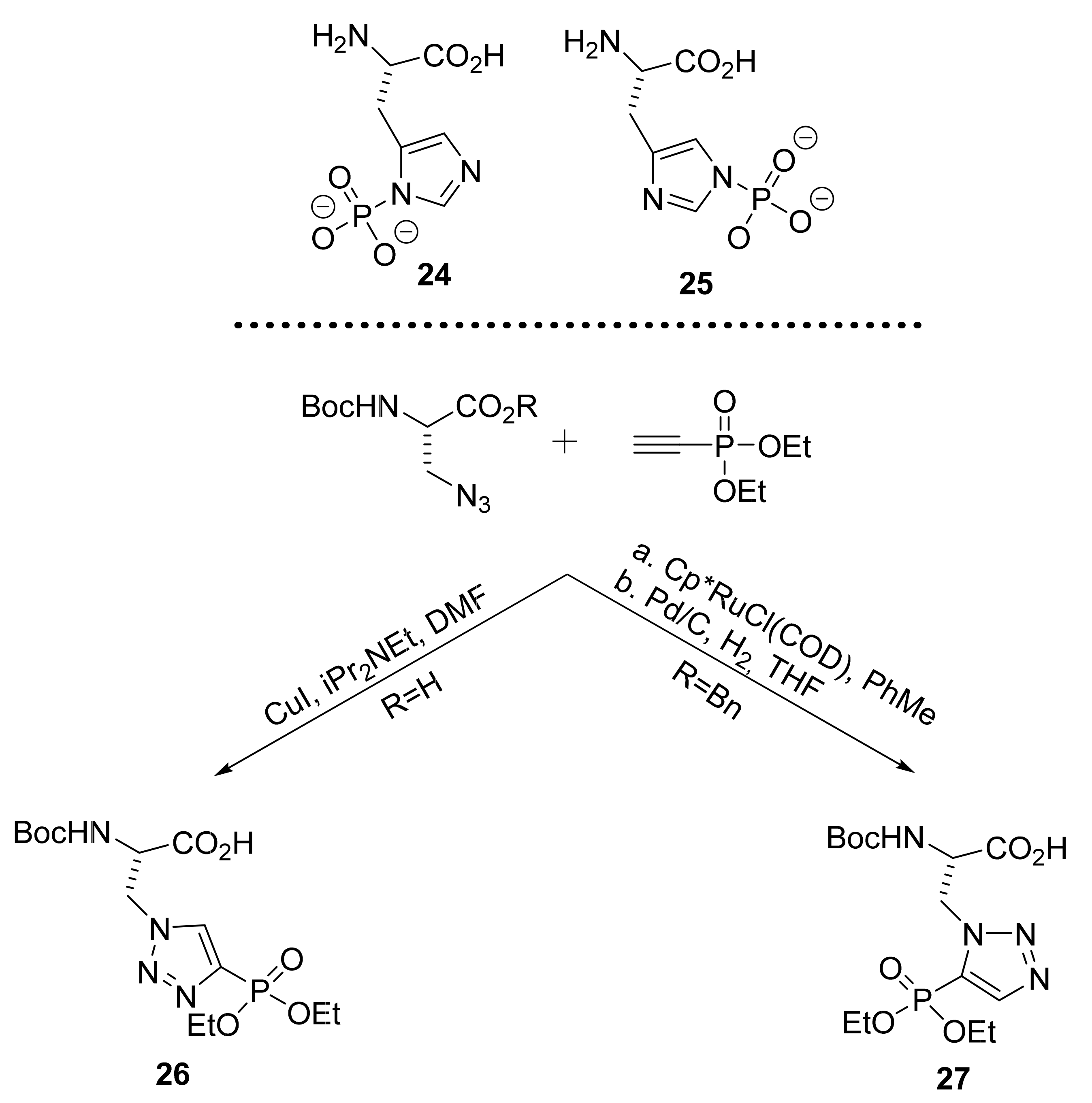
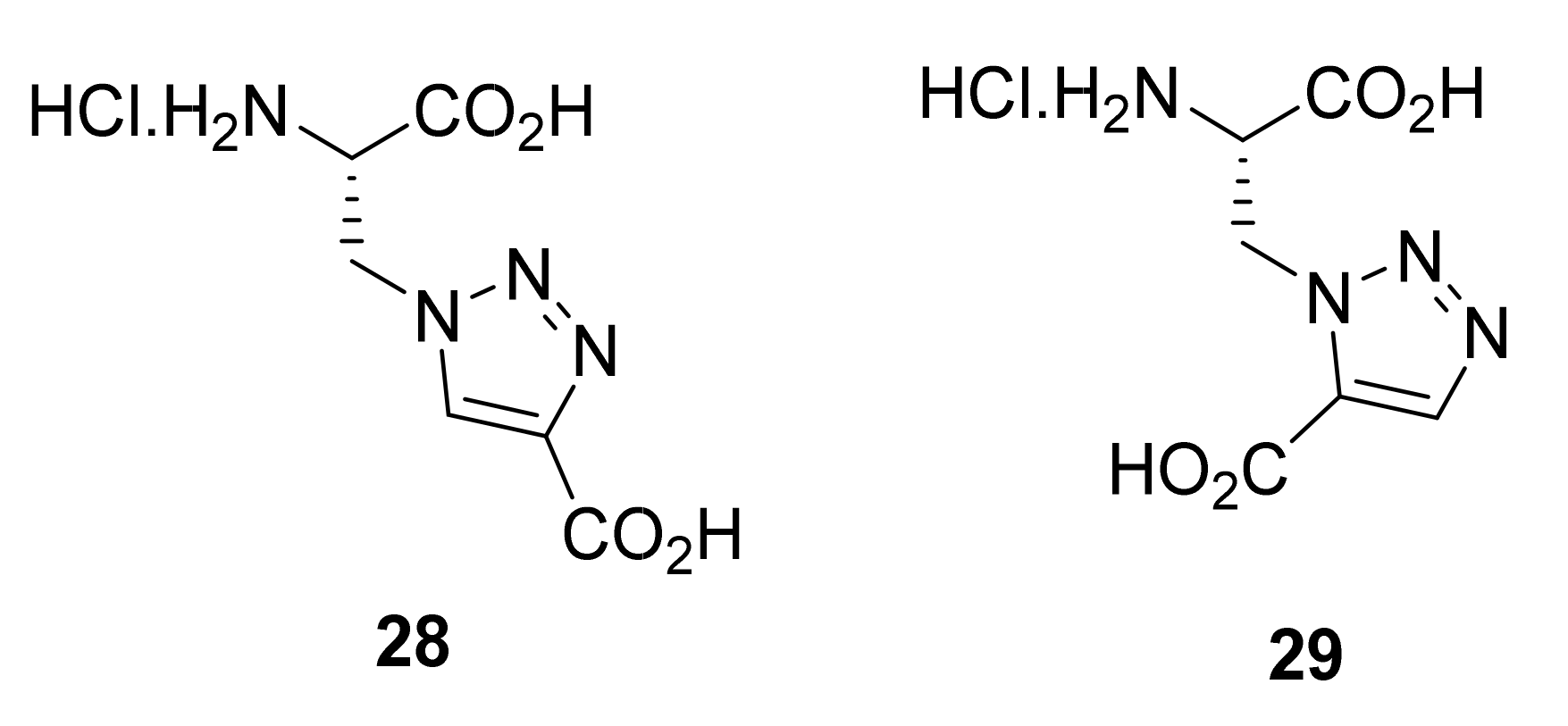
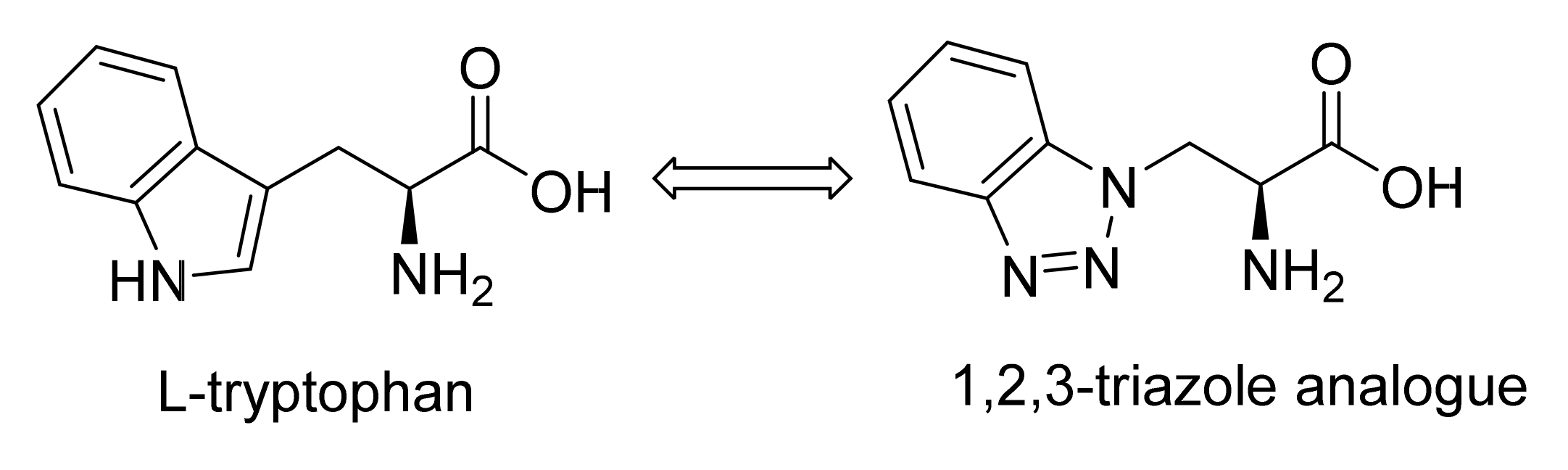
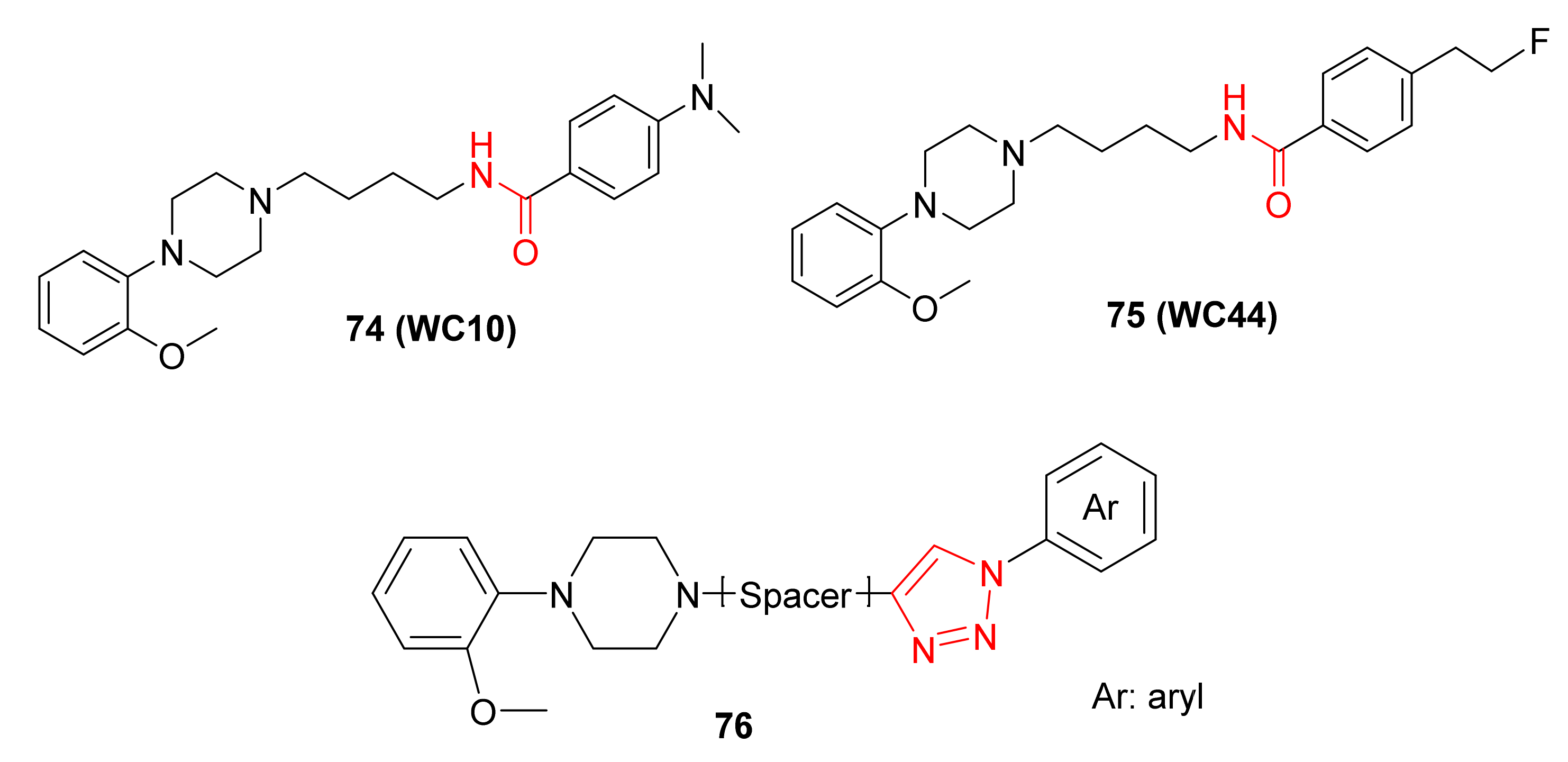
2.1.5. Amino Acid Mimics
2.2. Cyclic Compounds
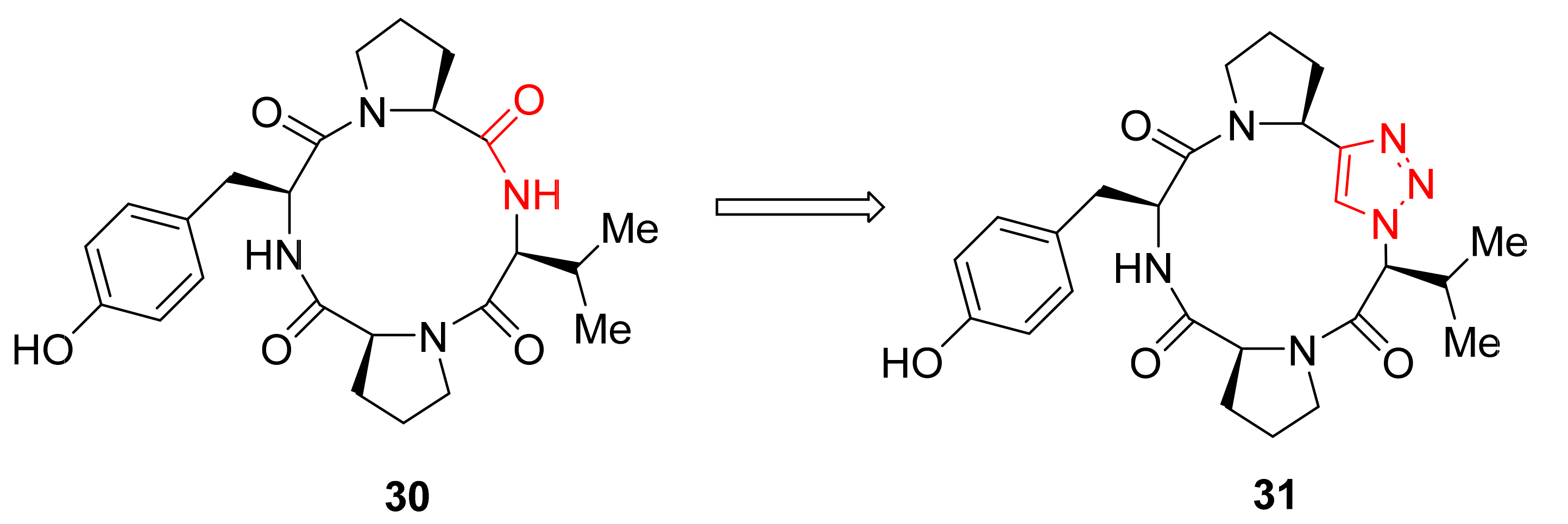
2.2.1. “Head-to-Tail” Cyclization
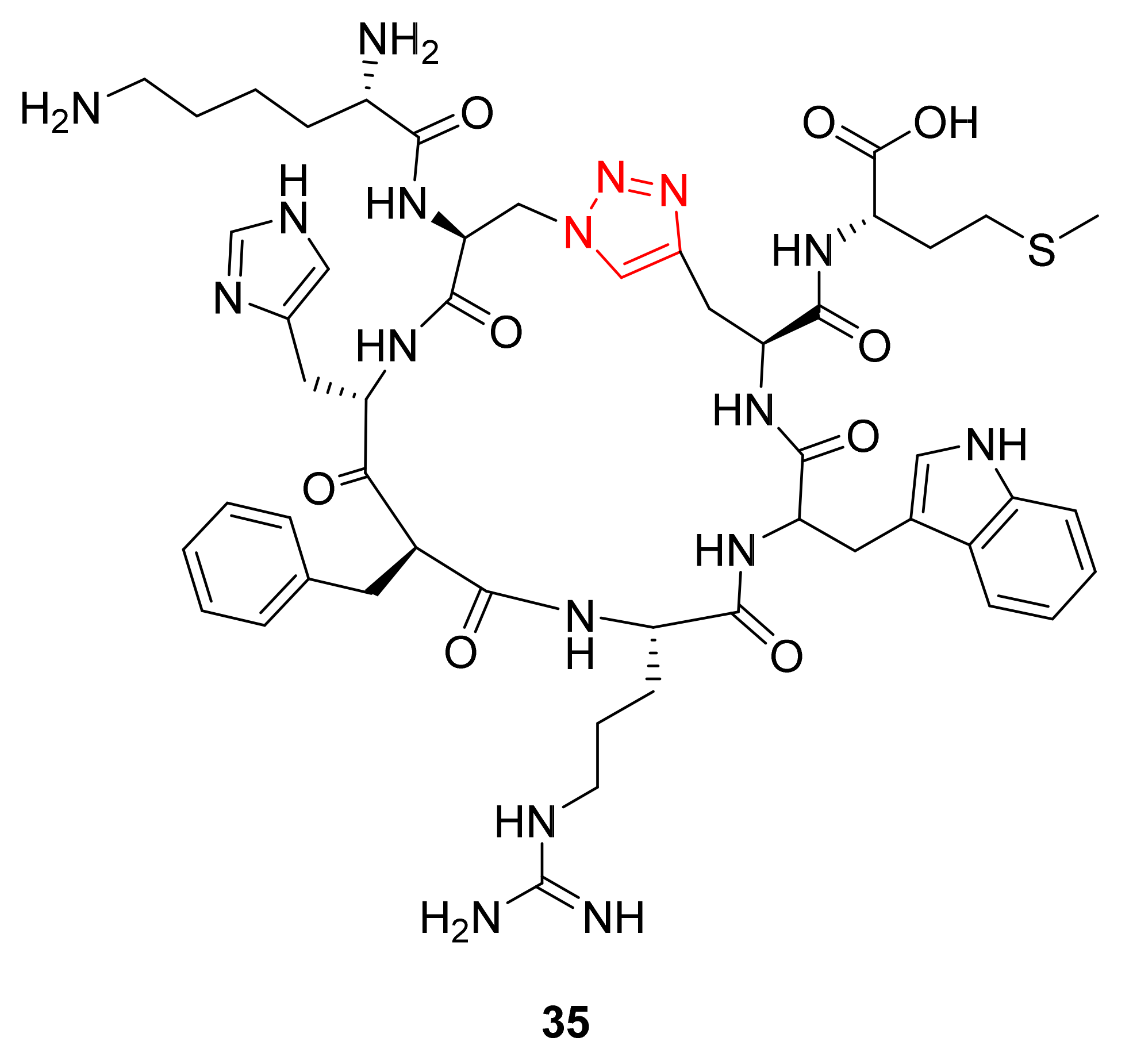
2.2.2. Cyclization Side-Chain–Side-Chain
- The synthesis of peptidotriazoles by the replacement of one or more amide bonds with triazole units;
- The preparation of β-turn secondary structures. Triazole-based compounds can mimic β-turn peptides, and triazole peptides can therefore be used for protein folding studies;
- The synthesis of triazolamer structures by replacing the amide bonds with 1,4-disubstituted 1,2,3-triazole;
- The replacement of a dipeptide within a multispiral α-helical secondary coiled coil structure;
- The modification of the amino acid side chain by the triazole motif to improve the stability or binding affinity through favorable interactions at the surface of the biological target;
- The macrocyclization of peptidomimetics in order to regulate the biological activity and stability of peptide macrocycles.
3. Compounds with Triazole Links to Other Features
3.1. Peptide–Carbohydrate
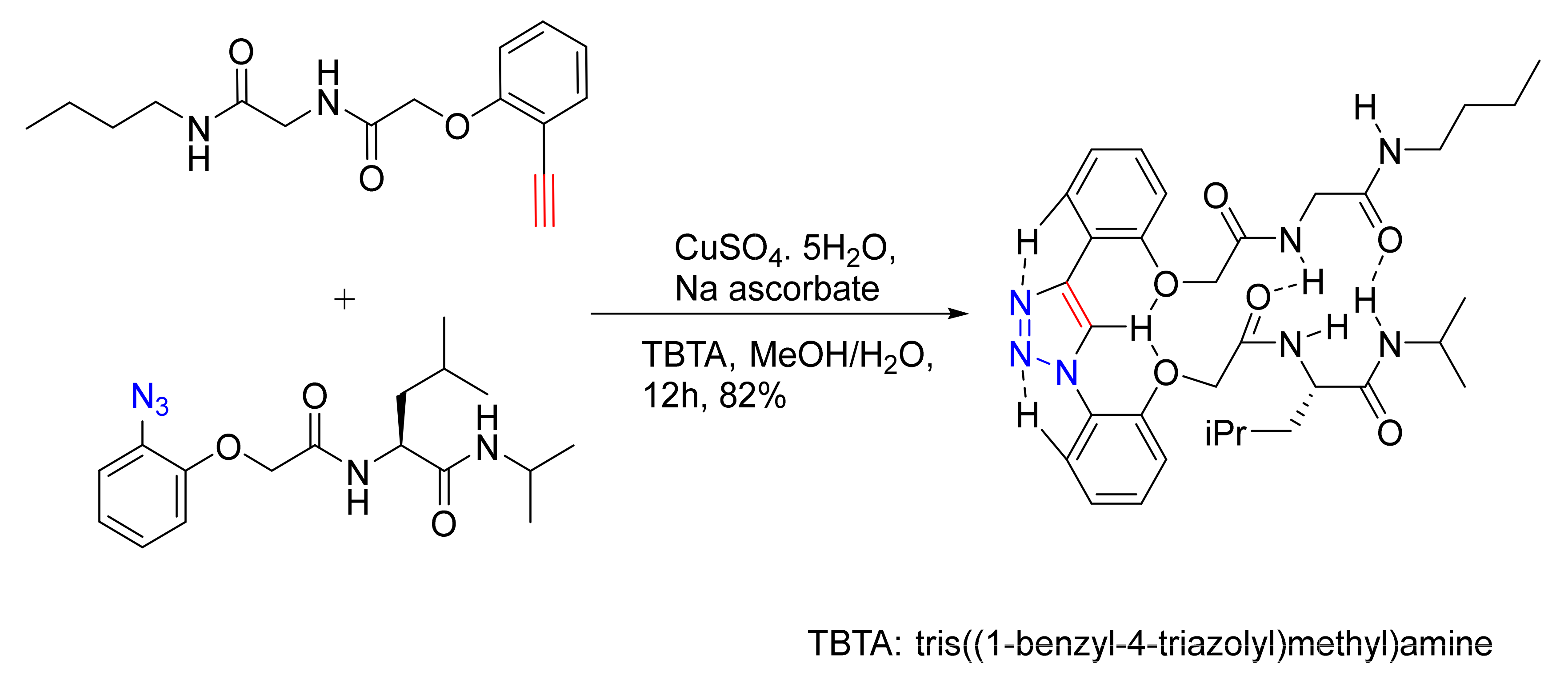
3.2. Peptide–Other Functions
3.3. Peptide–Polymer and Dendrimer
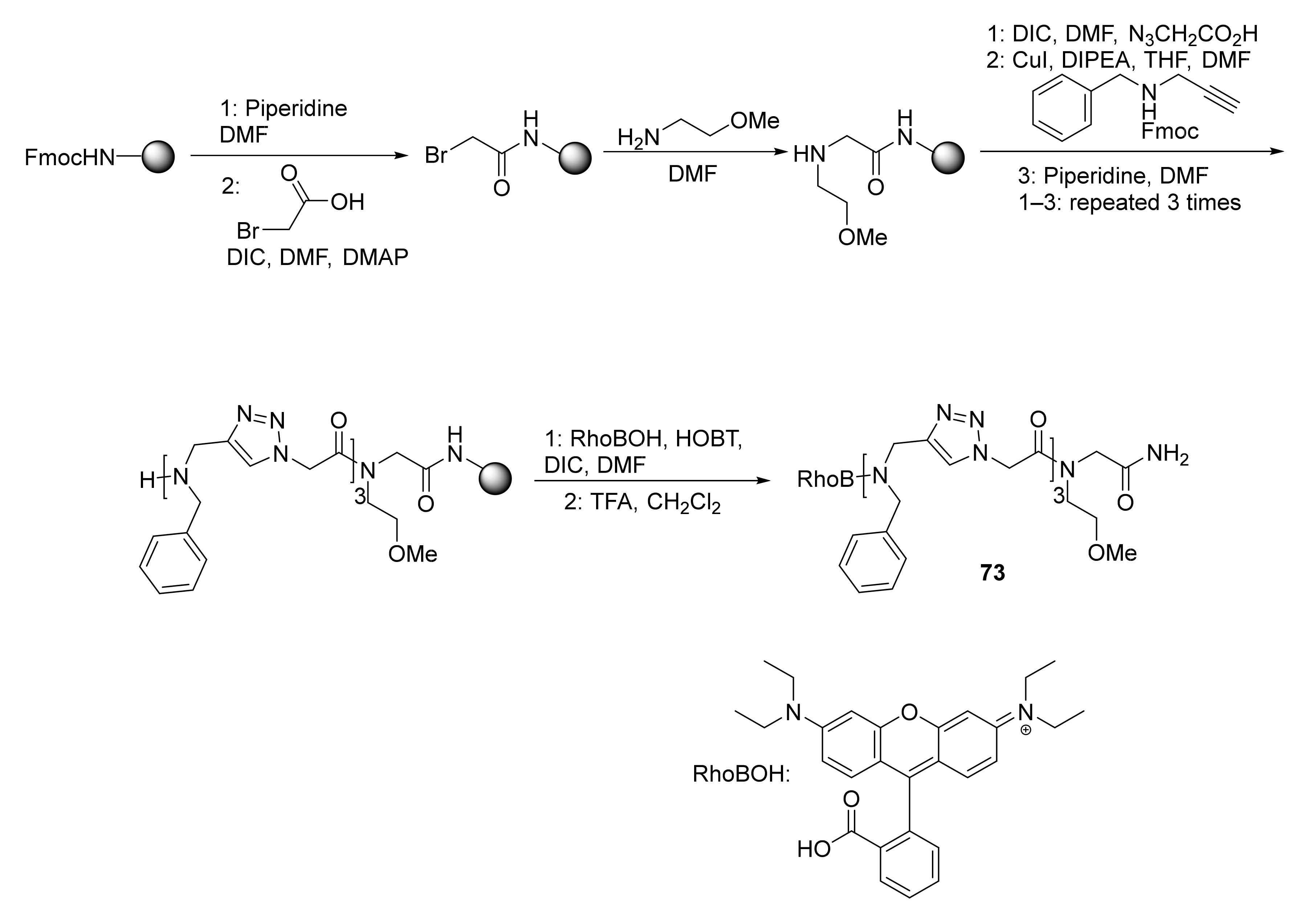
3.4. Conjugated Metal Complexes for Radiolabeling
- To conjugate carbohydrates into peptide sequences with more metabolically stable bonds;
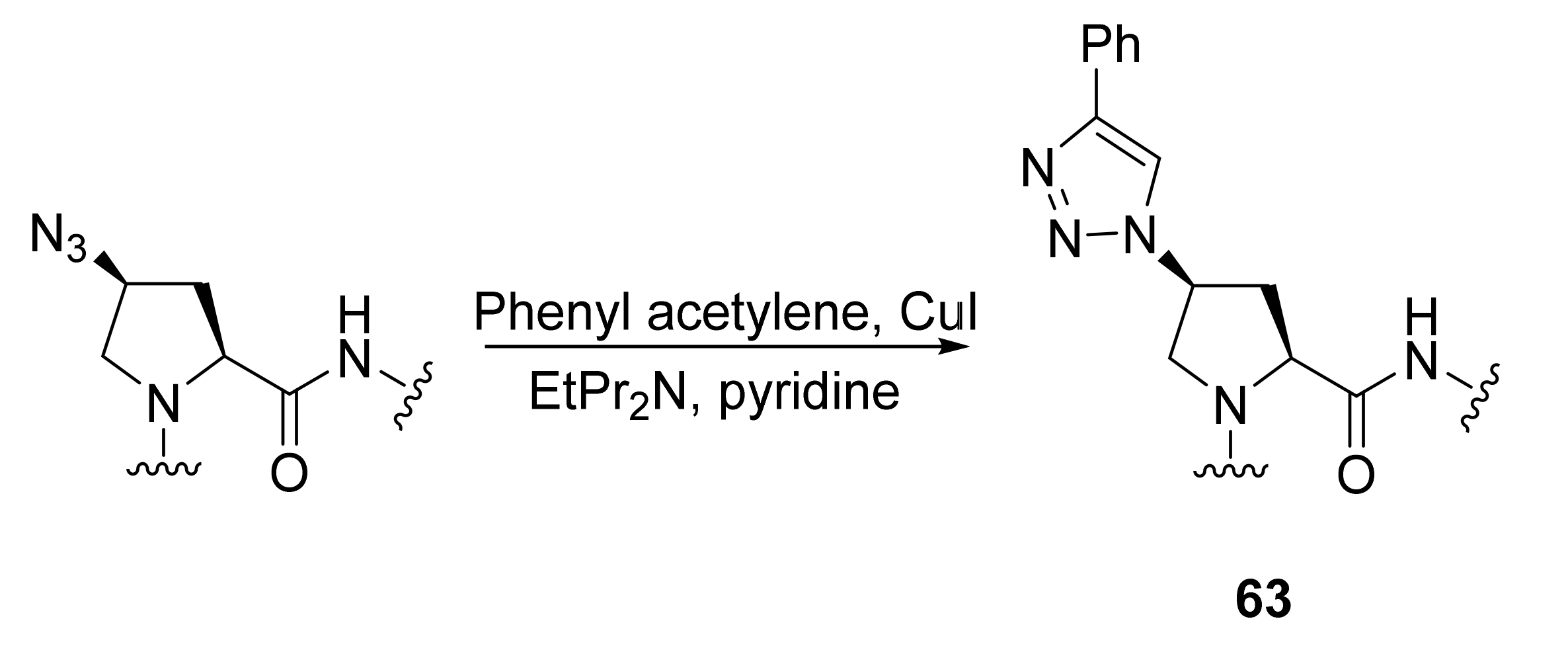
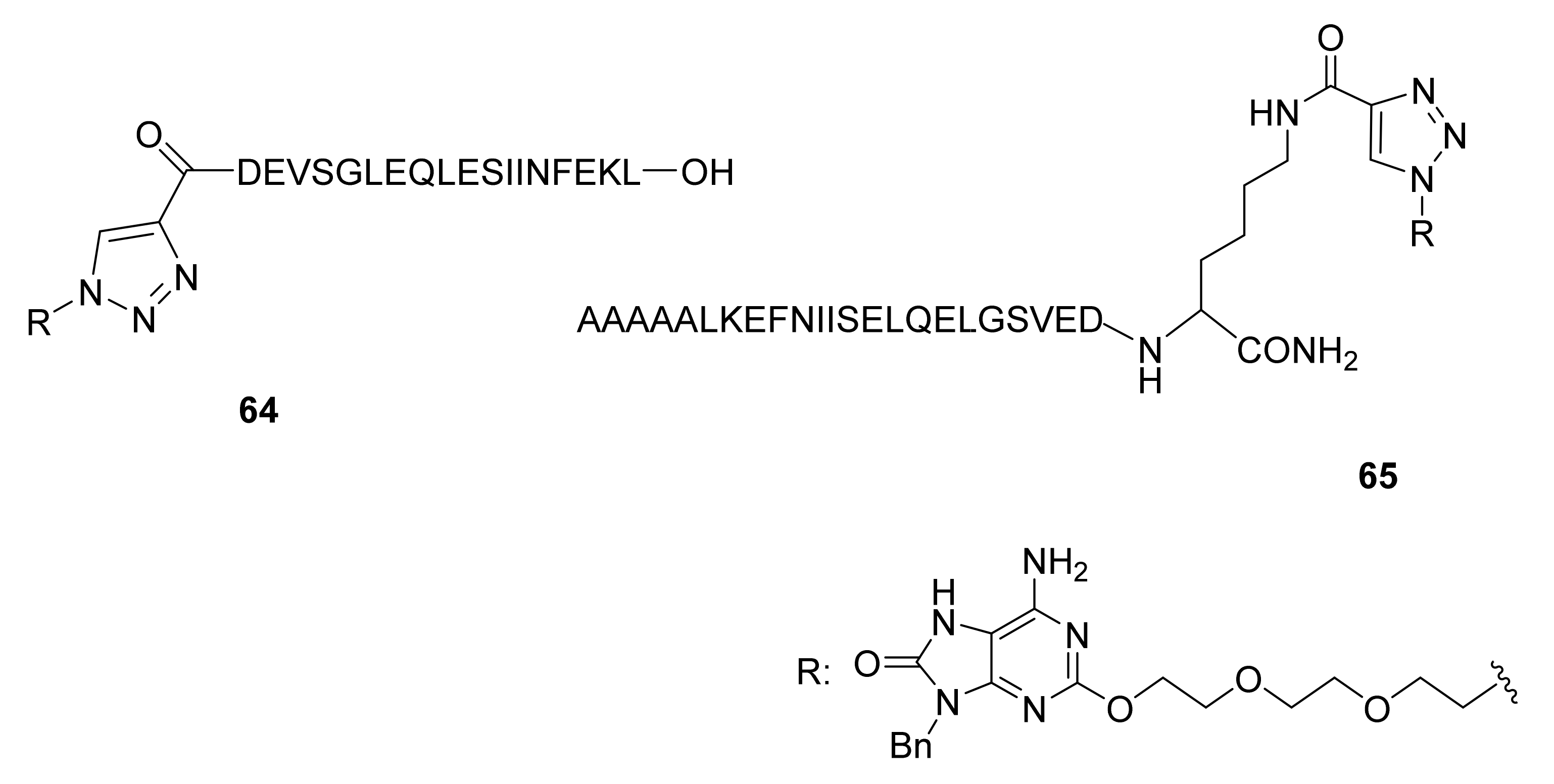
- To link peptides to functions that will be subsequently oriented in more rigid positions;
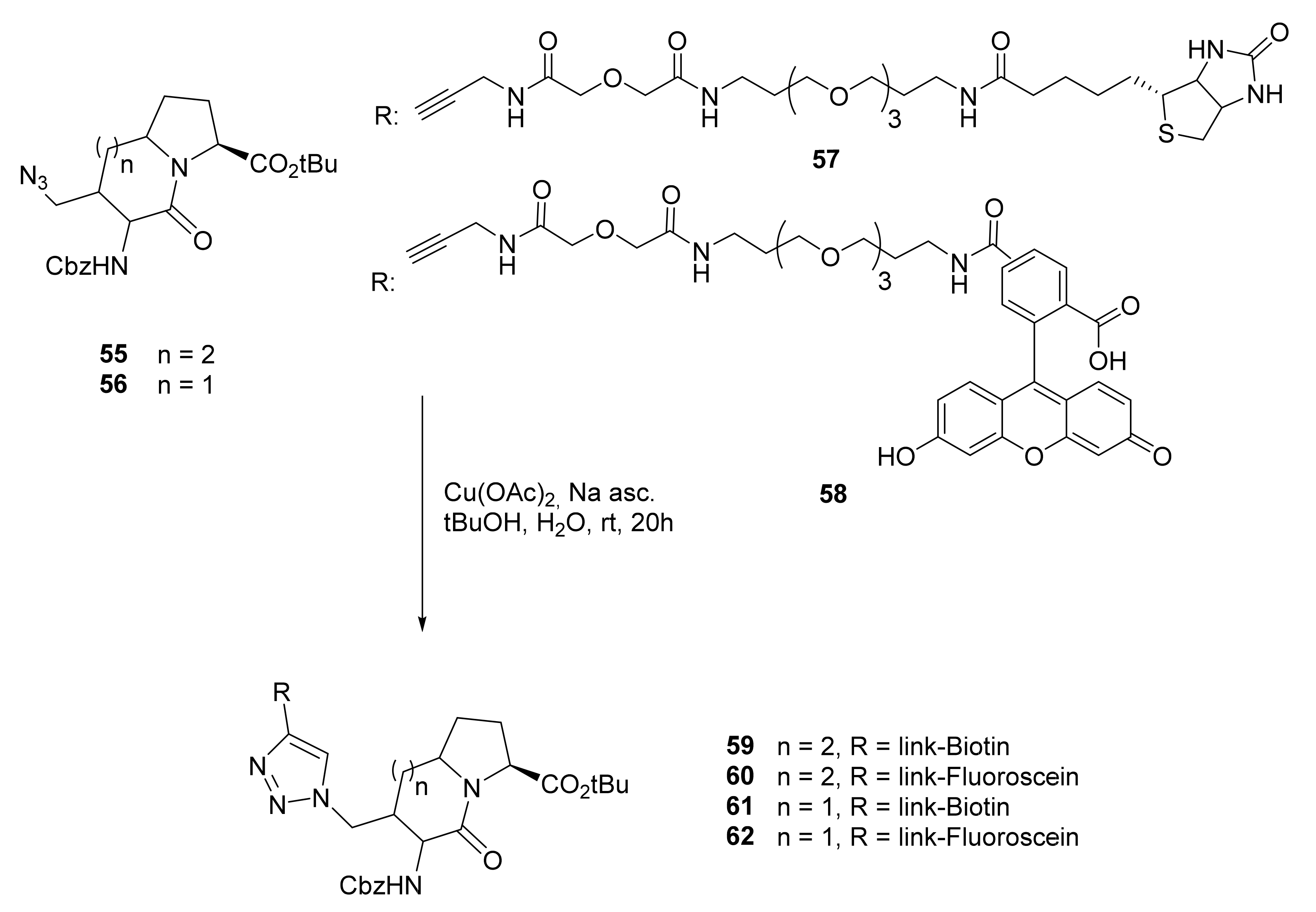
- To add biotin and fluorescein;
- In the synthesis of functional peptide polymer and dendrimer with improved properties;
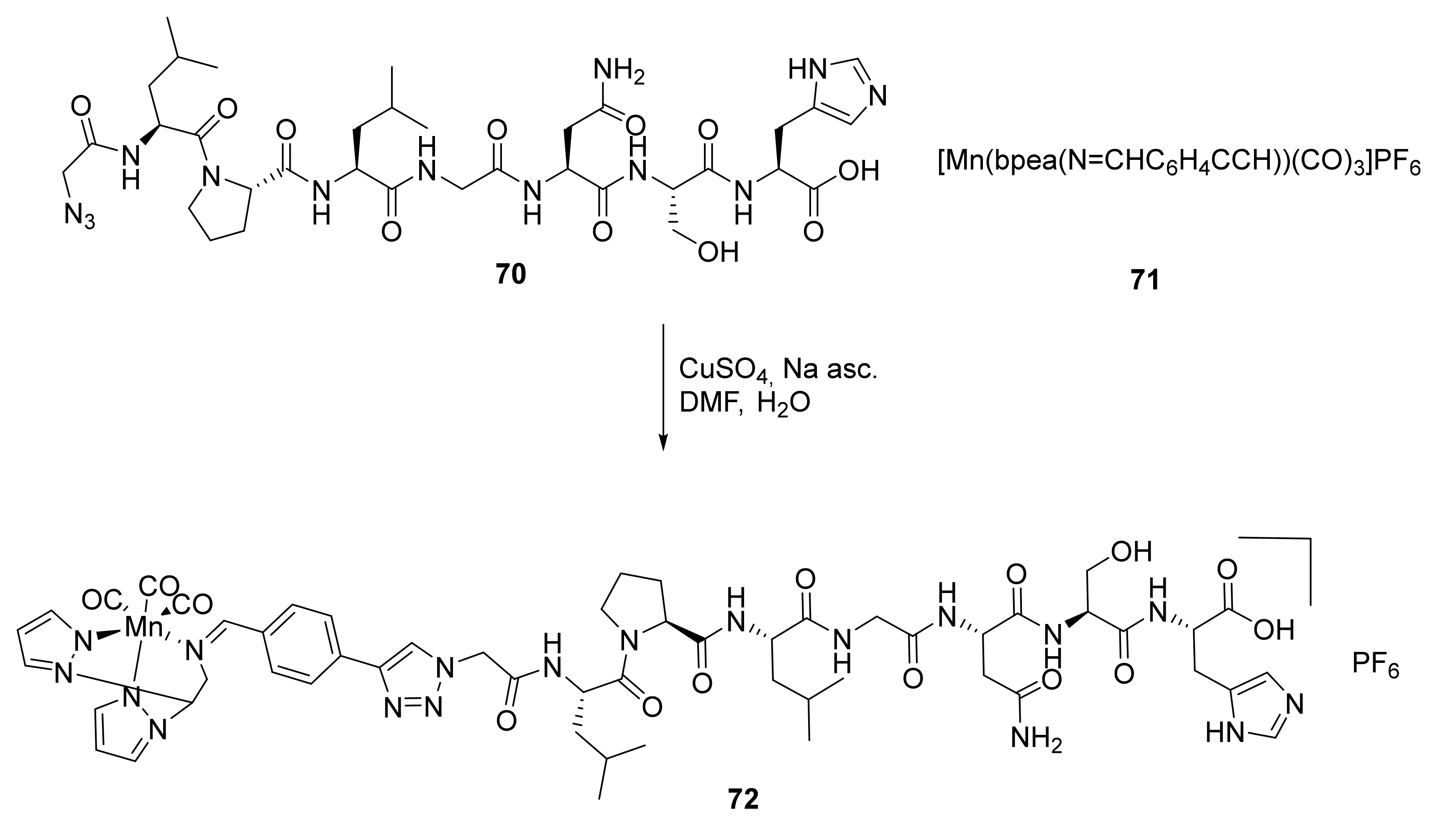
- In the preparation of conjugated metal complex peptidomimetics for radiolabeling, which is highly desirable for potential clinical applications using triazole as a bioisostere to replace the amide bond or as a tool to conjugate complex metal ions to peptides.
4. 1,2,3-Triazoles in Other Mimetics
4.1. Peptoids
4.2. Non-Peptide Mimetics in Which Triazoles Replace Amides
5. Conclusions
Funding
Conflicts of Interest
References
- Boas, L.C.P.V.; Campos, M.L.; Berlanda, R.L.A.; Neves, N.C.; Franco, O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542. [Google Scholar] [CrossRef] [PubMed]
- Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Janda, J.; Bastel, K.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of use of antiviral peptides against influenza virus. Viruses 2015, 7, 5428–5442. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.; Ardizzoni, A.; Ciociola, T.; Magliani, W.; Conti, S.; Blasi, E.; Cermelli, C. Antiviral activity of synthetic peptides derived from physiological proteins. Intervirology 2019, 61, 166–173. [Google Scholar] [CrossRef]
- Nyanguile, O. Peptide antiviral strategies as an alternative to treat lower respiratory viral infections. Front. Immunol. 2019, 10, 1366. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, A.S.K.; Lim, Y.S.; Fang, C.-M.; Loh, H.-S.; Le, C.F. The potential of antiviral peptides as COVID-19 therapeutics. Front. Pharmacol. 2020, 11, 575444. [Google Scholar] [CrossRef]
- Venegas-Rodriguez, R.; Santana-Sanchez, R.; Pena-Ruiz, R.; Bequet-Romero, M.; Hernandez-Cedeno, M.; Santiesteban-Licea, B.; Garcia, A.; Aroche, P.R.; Oliva-Perez, D.; Ortega-Gonzalez, L.M.; et al. CIGB-258 immunomodulatory peptide: A novel promising treatment for critical and severe COVID-19 patients. MedRxiv 2020, 20110601. [Google Scholar] [CrossRef]
- Kalita, P.; Padhi, A.K.; Zhang, K.Y.J.; Tripathi, T. Design of peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microbial. Pathog. 2020, 145, 104236. [Google Scholar] [CrossRef]
- Uddin, S. Chapter 25: Peptide Drug/Device Combinations. In Development of Biopharmaceutical Drug-Device Products; Jameel, F., Skoug, J., Nesbitt, R., Eds.; AAPS Advances in the Pharmaceutical Sciences Series; Springer: Cham, Switzerland, 2020; Volume 35, pp. 613–637. [Google Scholar]
- Fosgerau, K.; Hoffman, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Haggag, Y.A.; Donia, A.A.; Osman, M.A.; El-Gizawy, S.A. Peptides as drug candidates: Limitations and recent development perspectives. Biomed. J. Sci. Tech. Res. 2018, 8, 3. [Google Scholar] [CrossRef]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- D’Anessa, I.; Leva, F.S.D.; Teana, A.L.; Novellino, E.; Limongelli, V.; Marino, D.D. Bioinformatics and biosimulations as toolbox for peptides and peptidomimetics design: Where are we? Front. Mol. Biosci. 2020, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.D.; D’Annessa, I.; Tancredi, H.; Bagni, C.; Gallicchio, E. A unique binding mode of the eukaryotic translation initiation factor 4E for guiding the design of novel peptide inhibitors. Protein Sci. 2015, 24, 1370–1382. [Google Scholar] [CrossRef]
- Arumugam, A.C.; Agharbaoui, F.E.; Khazali, A.S.; Yusof, R.; Rahman, N.A.; Fuaad, A.A.H.A. Computational–aided design: Minimal peptide sequence to block dengue virus transmission into cells. J. Biomol. Struct. Dyn. 2020, 31, 1–10. [Google Scholar] [CrossRef]
- Marino, D.D.; Bruno, A.; Grimaldi, M.; Scrima, M.; Stillitano, I.; Amodio, G.; Sala, G.D.; Romagnoli, A.; Santis, A.D.; Moltedo, O.; et al. Binding of the Anti–FIV peptide C8 to differently charged membrane models: From first docking to membrane tubulation. Front. Chem. 2020, 8, 493. [Google Scholar] [CrossRef]
- Yao, P.; Zhang, J.; You, S.; Qi, W.; Su, R.; He, Z. Ferrocene-modified peptides as inhibitors against insulin amyloid aggregation based on molecular simulation. J. Mater. Chem. B 2020, 8, 3076–3086. [Google Scholar] [CrossRef]
- Watkins, A.M.; Bonneau, R.; Arora, P.S. Chapter 17: Modeling and design of peptidomimetics to modulate protein–protein interactions. In Modeling Peptide–Protein Interactions, Methods in Molecular Biology; Schueler-Furman, O., London, N., Eds.; Springer Protocols: Cham, Switzerland, 2017; Volume 1561, pp. 291–307. [Google Scholar]
- Hanold, L.E.; Oruganty, K.; Ton, N.T.; Beedle, A.M.; Kannan, N.; Kennedy, E.J. Inhibiting EGFR dimerization using triazolyl–bridged dimerization arm mimics. PLoS ONE 2015, 10, e0118796. [Google Scholar] [CrossRef]
- Emileh, A.; Tuzer, F.; Yeh, H.J.C.; Umashankara, M.; Moreira, D.R.M.; LaLonde, J.M.; Bewley, C.A.; Abrams, C.F.; Chaiken, I.M. A model of the peptide triazole entry inhibitor binding to HIV-1 gp120 and mechanism of bridging sheet disruption. Biochemical 2013, 52, 2245–2261. [Google Scholar] [CrossRef] [PubMed]
- Bonandi, E.; Christodoulou, M.S.; Fumagalli, G.; Perdicchia, D.; Rastelli, G.; Passarella, D. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov. Today 2017, 22, 1572–1581. [Google Scholar] [CrossRef]
- Recnik, L.-M.; Kandioller, W.; Mindt, T.L. 1,4-Disubstituted 1,2,3-triazoles as amide bond surrogates for the stabilisation of linear peptides with biological activity. Molecules 2020, 25, 3576. [Google Scholar] [CrossRef]
- Brik, A.; Alexandratos, J.; Lin, Y.C.; Elder, J.H.; Olson, A.J.; Wlodawer, A.; Goodsell, D.S.; Wong, C.H. 1,2,3-Triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. ChemBioChem 2005, 6, 1167–1169. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Meldal, M.; Diness, F. Recent fascinating aspects of the CuAAC click reaction. Trends Chem. 2020, 2, 569–584. [Google Scholar] [CrossRef]
- McKay, C.S.; Finn, M.G. Click chemistry in complex mixtures: Biorthogonal bioconjugation. Chem. Biol. 2014, 21, 1075–1101. [Google Scholar] [CrossRef]
- Bauer, M.; Wang, W.; Lorion, M.M.; Dong, C.; Ackermann, L. Internal peptide late–stage diversification: Peptide isosteric triazoles for primary and secondary C(sp3)–H activation. Angew. Chem. Int. Ed. 2017, 57, 203–207. [Google Scholar] [CrossRef]
- Segundo, M.S.; Correa, A. Pd–catalyzed site–selective C(sp2)–H radical acylation of phenylalanine containing peptides with aldehydes. Chem. Sci. 2019, 10, 8872–8879. [Google Scholar] [CrossRef]
- Wu, J.; Kaplaneris, N.; Ni, S.; Kaltenhauser, F.; Ackermann, L. Late–stage C(sp2)–H and C(sp3)–H glycosylation of C–aryl/alkyl glycopeptides: Mechanistic insights and fluorescence labeling. Chem. Sci. 2020, 11, 6521–6526. [Google Scholar] [CrossRef]
- Tornoe, C.W.; Sanderson, S.J.; Mottram, J.C.; Coombs, G.H.; Meldal, M. Combinatorial library of peptidotriazoles: Identification of [1,2,3]-triazole inhibitors against a recombinant leishmania mexicana cysteine protease. J. Comb. Chem. 2004, 6, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Güell, I.; Micaló, L.; Cano, L.; Badosa, E.; Ferre, R.; Montesinos, E.; Bardají, E.; Feliu, L.; Planas, M. Peptidotriazoles with antimicrobial activity against bacterial and fungal plant pathogens. Peptides 2012, 33, 9–17. [Google Scholar] [CrossRef]
- Valverde, I.E.; Lecaille, F.; Lalmanach, G.; Aucagne, V.; Delmas, A.F. Synthesis of a biologically active triazole-containing analogue of cystatin a through successive peptidmimetic Alkyne-Azide ligations. Angew. Chem. Int. Ed. 2012, 51, 718–722. [Google Scholar] [CrossRef]
- Tam, A.; Arnold, U.; Soellner, M.B.; Raines, R.T. Protein Prosthesis: 1,5-Disubstituted[1,2,3]triazoles as cis-peptide bond surrogates. J. Am. Chem. Soc. 2007, 129, 12670–12671. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Bittermann, H.; Gmeiner, P. Triazolopeptides: Chirospecific synthesis and cis/trans prolyl ratios of structural isomers. Tetrahedron 2006, 62, 8919–8927. [Google Scholar] [CrossRef]
- Paul, A.; Einsiedel, J.; Waibel, R.; Heinemann, F.W.; Meyer, K.; Gmeiner, P. Novel triazolopeptides: Chirospecific synthesis and conformational studies of proline derived analogs. Tetrahedron 2009, 65, 6156–6168. [Google Scholar] [CrossRef]
- Vrettos, E.I.; Valverde, I.E.; Mascarin, A.; Pallier, P.N.; Cerofolini, L.; Fragai, M.; Parigi, G.; Hirmiz, B.; Bekas, N.; Grob, N.M.; et al. Single peptide backbone surrogate mutations to regulate angiotensin GPCR subtype selectivity. Chem. Eur. J. 2020, 26, 10690–10694. [Google Scholar] [CrossRef]
- Oh, K.; Guan., Z. A convergent synthesis of new β-turn mimics by click chemistry. Chem. Commun. 2006, 3069– 3071. [Google Scholar] [CrossRef]
- Wu, C.-F.; Zhao, X.; Lan, W.-X.; Cao, C.; Liu, J.-T.; Jiang, X.-K.; Li, Z.-T. A 1,4-Diphenyl-1,2,3-Triazole-Based β-Turn mimic constructed by click chemistry. J. Org. Chem. 2012, 77, 4261–4270. [Google Scholar] [CrossRef] [PubMed]
- Hamley, A.W. Peptide fibrillization. Angew. Chem. Int. Ed. 2007, 46, 8128–8147. [Google Scholar] [CrossRef]
- Deike, S.; Rothemund, S.; Voigt, B.; Samantray, S.; Strodel, B.; Binder, W.H. β-turn mimetic synthetic peptides as amyloid-β aggregation inhibitors. Bioorg. Chem. 2020, 101, 104012. [Google Scholar] [CrossRef] [PubMed]
- Angelo, N.G.; Arora, P.S. Nonpeptidic foldamers from amino acids: Synthesis and characterization of 1,3-substituted triazole oligomers. J. Am. Chem. Soc. 2005, 127, 17134–17135. [Google Scholar] [CrossRef]
- Angelo, N.G.; Arora, P.S. Solution–and solid-phase synthesis of triazole oligomers that display protein-like functionality. J. Org. Chem. 2007, 72, 7963–7967. [Google Scholar] [CrossRef]
- Jochim, A.L.; Miller, S.E.; Angelo, N.G.; Arora, P.S. Evaluation of triazolamers as active site inhibitors of HIV-1 protease. Bioorg. Med. Chem. Lett. 2009, 19, 6023–6026. [Google Scholar] [CrossRef]
- Montagnat, O.D.; Lessene, G.; Hughes, A.B. Synthesis of Azide-Alkyne fragments for “Click” Chemical Applications. Part 2. formation of oligomers from orthogonally protected Chiral Trialkylsilylhomopropargyl Azides and Homopropargyl Alcohols. J. Org. Chem. 2010, 75, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Schröder, D.C.; Kracker, O.; Fröher, T.; Gora, J.; Jewginski, M.; NieB, A.; Antes, I.; Latajka, R.; Marion, A.; Sewald, N. 1,4-Disubstituted 1H-1,2,3-Triazole containing Peptidotriazolamers: A new class of peptidomimetics with interesting foldamer properties. Front. Chem. 2019, 7, 155. [Google Scholar] [CrossRef]
- Horne, W.S.; Yadav, M.K.; Stout, C.D.; Ghadiri, M.R. Heterocyclic peptide backbone modifications in an α-helical coiled coil. J. Am. Chem. Soc. 2004, 126, 15366–15367. [Google Scholar] [CrossRef] [PubMed]
- Ben Haj Salah, K.; Das, S.; Ruiz, N.; Andreu, V.; Martinez, J.; Wenger, E.; Amblard, M.; Didierjean, C.; Legrand, B.; Inguimbert, N. How are 1,2,3-triazoles accommodated in helical secondary structures? Org. Biomol. Chem. 2018, 16, 3576–3583. [Google Scholar] [CrossRef]
- Agouram, N.; El Hadrami, E.M.; Ben Tama, A.; Julve, M.; Anane, H.; Stiriba, S.E. Efficient synthesis of triazolic peptidomimetics via copper-catalyzed azide-alkyne [3+2] cycloaddition. Pharm. Chem. 2016, 8, 499–506. [Google Scholar]
- Maugard, M.; Vigneron, P.A.; Bolanos, J.P.; Bonvento, G. L-Serine links metabolism with neurotransmission. Prog. Neurobiol. 2021, 197, 101896. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.S.; Dunlop, R.A.; Powell, J.T.; Banack, S.A.; Cox, P.A. L-Serine: A naturally-occurring amino acid with theapeutic potential. Neurotox. Res. 2018, 33, 213–221. [Google Scholar] [CrossRef]
- Kee, J.M.; Villani, B.; Carpenter, L.R.; Muir, T.W. Development of stable phosphohistidine analogues. J. Am. Chem. Soc. 2010, 132, 14327–14329. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, M.; Seaver, B.; Esslinger, C.S. Design, synthesis, and biological activity of novel triazole amino acids used to probe binding interactions between ligand and neutral amino acid transport protein SN1. Bioorg. Med. Chem. Lett. 2007, 17, 4163–4166. [Google Scholar] [CrossRef]
- Stanley, N.J.; Sejer Pedersen, D.; Nielsen, B.; Kvist, T.; Mathiesen, J.M.; Bräuner-Osborne, H.; Taylor, D.K.; Abell, A.D. 1,2,3-Triazolyl amino acids a AMPA receptor ligands. Bioorg. Med. Chem. Lett. 2010, 20, 7512–7515. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Wolfe, H.; Wells, A.; Chien, H.C.; Colas, C.; Schlessinger, A.; Giacomini, K.M.; Thomas, A.A. L-Type amino acid transporter 1 activity of 1,2,3-triazolyl analogs of L-histidine and L-tryptophan. Bioorg. Med. Chem. Lett. 2019, 29, 2254–2258. [Google Scholar] [CrossRef]
- Buckton, L.K.; Rahimi, M.N.; McAlpine, S.R. Cyclic peptides as drugs for intracellular targets: The next frontier in peptide therapeutic development. Chem. A Eur. J. 2021, 27, 1487–1513. [Google Scholar] [CrossRef] [PubMed]
- Sohbari, C.; Foster, A.; Tavassoli, A. Methods for generating and screening libraries of genetically encoded cyclic peptides in drug discovery. Nat. Rev. Chem. 2020, 4, 90–101. [Google Scholar]
- Horne, W.S.; Stout, C.D.; Ghadiri, M.R. A heterocyclic peptide nanotube. J. Am. Chem. Soc. 2003, 125, 9372–9376. [Google Scholar] [CrossRef]
- Bock, V.D.; Perciaccante, R.; Jansen, T.P.; Hiemstra, H.; van Maarseveen, J.H. Click chemistry as a route to cyclic tetrapeptide analogues: Synthesis of cyclo-[Pro-Val-Ψ(triazole)-Pro-Tyr]. Org. Lett. 2006, 8, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, L.; Lu, J.; Qiu, S.; Yang, C.-Y.; Yi, H.; Wang, S. Cyclopeptide Smac mimetics as antagonists of IAP proteins. Bioorg. Med. Chem. Lett. 2010, 20, 3043–3046. [Google Scholar] [CrossRef]
- Roice, M.; Johannsen, I.; Meldal, M. High capacity poly(ethylene glycol) based amino polymers for peptide and organic synthesis. QSAR Comb. Sci. 2004, 23, 662–673. [Google Scholar] [CrossRef]
- Beierle, J.M.; Horne, W.S.; van Maarseveen, J.H.; Waser, B.; Reubi, J.C.; Ghadiri, M.R. Conformationally homogeneous heterocyclic pseudotetrapeptides as three-dimensional scaffolds for rational drug design: Receptor-selective somatostatin analogues. Angew. Chem. Int. Ed. Engl. 2009, 48, 4725–4729. [Google Scholar] [CrossRef]
- Isidro-Llobet, A.; Georgiou, K.H.; Galloway, W.R.J.D.; Giacomini, E.; Hansen, M.R.; Méndez-Abt, G.; Sing Tan, Y.; Carro, L.; Sore, H.F.; Spring, D.R. A diversity-oriented synthesis strategy enabling the combinatorial-type variation of macrocyclic peptidomimetic scaffolds. Org. Biomol. Chem. 2015, 13, 4570–4580. [Google Scholar] [CrossRef]
- Kidd, S.L.; Osberger, T.J.; Mateu, N.; Sore, H.F.; Spring, D.R. Recent applications of Diversity-Oriented Synthesis toward novel, 3-dimensional fragment collections. Front. Chem. 2018, 6, 460. [Google Scholar] [CrossRef] [PubMed]
- Pathoor, R.; Puthiyedath, T.; Bahulayan, D. An efficient green Diversity-Oriented Synthesis of pyrimidinone and indole appended macrocyclic peptidomimetics. Tetrahedron Lett. 2019, 60, 191–196. [Google Scholar] [CrossRef]
- Gabba, A.; Robakiewicz, S.; Taciak, B.; Ulewicz, K.; Broggini, G.; Rastelli, G.; Krol, M.; Murphy, P.V.; Passarella, D. Synthesis and biological evaluation of migrastatin macrotriazoles. Eur. J. Org. Chem. 2017, 2017, 60–69. [Google Scholar] [CrossRef]
- Davis, M.R.; Singh, E.K.; Wahyudi, H.; Alexander, L.D.; Kunicki, J.B.; Nazarova, L.A.; Fairweather, K.A.; Giltrap, A.M.; Jolliffe, K.A.; McAlpine, S.R. Synthesis of sansalvamide a peptidomimetics: Triazole, oxazole, thiazole, and pseudoproline containing compounds. Tetrahedron 2012, 68, 1029–1051. [Google Scholar] [CrossRef] [PubMed]
- Haney, C.M.; Werner, H.M.; McKay, J.J.; Horne, W.S. Thermodynamic origin of α-helix stabilization by side-chain cross-links in a small protein. Org. Biomol. Chem. 2016, 14, 5768–5773. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, S.A.; Coleska, A.; Ran, X.; Yi, H.; Yang, C.-Y.; Wang, S. Design of triazole-stapled BCL9 α-helical peptides to target the β-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction. J. Med. Chem. 2012, 55, 1137–1146. [Google Scholar] [CrossRef]
- Testa, C.; Scrima, M.; Grimaldi, L.; D’Ursi, A.M.; Dirain, M.L.S.; Germain, N.L.; Singh, A.; Luevano, C.H.; Chorev, M.; Rovero, P.; et al. 1,4-Disubstituted-[1,2,3]triazolyl-containing analogs of MT-II—Design, synthesis, conformational analysis, and biological activity. J. Med. Chem. 2014, 22, 9424–9434. [Google Scholar] [CrossRef]
- Lau, Y.H.; de Andrade, P.; Quah, S.-T.; Rossmann, M.; Laraia, L.; SkÖld, N.; Sum, T.J.; Rowling, P.J.E.; Joseph, T.L.; Verma, C.; et al. Functionalised staple linkages for modulating the cellular activity of stapled peptides. Chem. Sci. 2014, 5, 1804–1809. [Google Scholar] [CrossRef]
- Oueis, E.; Jaspars, M.; Westwood, N.J.; Naismith, J.H. Enzymatic macrocyclization of 1,2,3-triazole peptide mimetics. Angew. Chem. 2016, 128, 5936–5939. [Google Scholar] [CrossRef]
- Van der Wal, S.; Capicciotti, C.J.; Rontogianni, S.; Ben, R.N.; Liskamp, R.M.J. Synthesis and evaluation of linear CuAAC-oligomerized antifreeze neo-glycopeptides. Med. Chem. Commun. 2014, 5, 1159–1165. [Google Scholar] [CrossRef]
- Junior, E.F.C.; Guimaraes, C.F.R.C.; Franco, L.L.; Alves, R.J.; Kato, K.C.; Martins, H.R.; de Souza Filho, J.D.; Bemquerer, M.P.; Munhoz, V.H.O.; Resende, J.M.; et al. Glycotriazole-peptides derived from the peptide HSP1: Synergitic effect of triazole and saccharide rings on the antifungal activity. Amino Acids 2017, 49, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Brmble, M.A.; Kowalczyk, R.; Watson, A.J.A.W.; Fairbanks, A.J. Protecting-group-free one-pot synthesis of glycocomjugates directly from reducing sugars. Angew. Chem. 2014, 126, 12101–12105. [Google Scholar] [CrossRef]
- Yule, L.R.; Bower, R.L.; Kaur, H.; Kowalczyk, R.; Hay, D.L.; Brimble, M.A. Synthesis and amylin receptor of glycomimetics of Pramlintide using click chemistry. Org. Biomol. Chem. 2016, 14, 5238–5245. [Google Scholar] [CrossRef]
- Bera, S.; Zhanel, G.G.; Schweizer, F. Evaluation of amphiphilic aminoglycoside–peptide triazole conjugates as antibacterial agents. Bioorg. Med. Chem. Lett. 2010, 20, 3031–3035. [Google Scholar] [CrossRef]
- Kuijpers, B.H.M.; Groothuys, S.; Soede, A.C.; Laverman, P.; Boerman, O.C.; van Delft, F.L.; Rutjes, F.P.J.T. Preparation and evaluation of glycosylated arginine-glycine-aspartate (RGD) derivatives for integrin targeting. Bioconjug. Chem. 2007, 18, 1847–1854. [Google Scholar] [CrossRef]
- Arosio, D.; Bertoli, M.; Manzoni, L.; Scolastico, C. Click chemistry to functionalise peptidomimetics. Tetrahedron Lett. 2006, 47, 3697–3700. [Google Scholar] [CrossRef]
- Gopi, H.N.; Tirupula, K.C.; Baxter, S.; Ajith, S.; Chaiken, I.M. Click chemistry on azidoproline: High-affinity dual antagonist for HIV-1 envelope glycoprotein gp120. ChemMedChem 2006, 1, 54–57. [Google Scholar] [CrossRef]
- Weterings, J.J.; Khan, S.; van der Heden, G.J.; Drijfhout, J.W.; Melief, C.M.J.; Overkleeft, H.S.; van der Burg, S.H.; Ossendorp, F.; van der Marela, G.A.; Filippov, D.V. Synthesis of 2-alkoxy-8-hydroxyadenylpeptides: Towards synthetic epitope-based vaccines. Bioorg. Med. Chem. Lett. 2006, 16, 3258–3261. [Google Scholar] [CrossRef] [PubMed]
- Sapra, R.; Verma, R.P.; Maurya, G.P.; Dhawan, S.; Babu, J.; Haridas, V. Designer peptide and protein dendrimers: A cross-sectional analysis. Chem. Rev. 2019, 119, 11391–11441. [Google Scholar] [CrossRef]
- Mirakabad, F.S.T.; Khoramgah, M.S.; Keshavarz, F.K.; Tabarzad, M.; Ranjbari, J. Peptide dendrimers as valuable biomaterials in medical sciences. Life Sci. 2019, 233, 116754. [Google Scholar] [CrossRef]
- Haridas, V.; Sharma, Y.K.; Sahu, S.; Verma, R.P.; Sadanandan, S.; Kacheshwar, B.G. Designer peptide dendrimers using click reaction. Tetrahedron 2011, 67, 1873–1884. [Google Scholar] [CrossRef]
- Verma, R.P.; Shandilya, A.; Haridas, V. Peptide dendrimers with designer core for directed self-assembly. Tetrahedron 2015, 71, 8758–8765. [Google Scholar] [CrossRef]
- Haridas, V.; Kumar, P.P.P.; Dhawan, S.; Devaki, S.J. Designer peptide dendrons and dendrimers based soft materials through self-assembly. ChemistrySelect 2016, 1, 4582–4590. [Google Scholar] [CrossRef]
- Agut, W.; Agnaou, R.; Lecommandoux, S.; Taton, D. Synthesis of block copolypeptides by click chemistry. Macromol. Rapid Commun. 2008, 29, 1147–1155. [Google Scholar] [CrossRef]
- Krishnan, B.P.; Rai, R.; Asokan, A.; Sureshan, K.M. Crystal-to-crystal synthesis of triazole-linked pseudo-proteins via Topochemical Azide-Alkyne Cycloaddition reaction. J. Am. Chem. Soc. 2016, 138, 14824–14827. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, B.P.; Sureshan, K.M. Topochecical Azide-Alkyne Cycloaddition reaction in gels: Size-tunable synthesis of triazole-linked polypeptides. J. Am. Chem. Soc. 2017, 139, 1584–1589. [Google Scholar] [CrossRef]
- Hema, K.; Sureshan, K.M. β-Sheet to helical-sheet evolution induced by topochemical polymerization: Cross-α-amyloid-like packing in a pseudoprotein with Gly-Phe-Gly repeats. Angew. Chem. Int. Ed. 2020, 59, 8854–8859. [Google Scholar] [CrossRef]
- Athiyarath, V.; Sureshan, K.M. Designed synthesis of a 1D polymer in twist-stacked topology via single-crystal-to-single-crystal polymerization. Angew. Chem. 2020, 132, 15710–15715. [Google Scholar] [CrossRef]
- Mindt, T.L.; Struthers, H.; Brans, L.; Anguelov, T.; Schweinsberg, C.; Maes, V.; Tourwe’, D.; Schibli, R. “Click to Chelate”: Synthesis and installation of metal chelates into biomolecules in a single step. J. Am. Chem. Soc. 2006, 128, 15096–15397. [Google Scholar] [CrossRef]
- Vats, K.; Satpati, D.; Sharma, R.; Sarma, H.D.; Banerjee, S. Synthesis and comparative in vivo evaluation of 99mTc(CO)3-labeled PEGylated and non-PEGylated cRGDfK peptide monomers. Chem. Biol. Drug Des. 2016, 89, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Price, J.R.; Jensen, P.; Lovitt, C.J.; Shelper, T.; Duffy, S.; Windus, L.C.; Avery, V.M.; Rutledge, P.J.; Todd, M.H. Copper, nickel, and zinc cyclam-amino acid and cyclam-peptide complexes may be synthesized with “click” chemistry and are noncytotoxic. Inorg. Chem. 2011, 50, 12823–12935. [Google Scholar] [CrossRef]
- Pai, S.; Radacki, K.; Schatzschneider, U. Sonogashira, CuAAC, and oxime ligations for the synthesis of MnI tricarbonyl photoCORM peptide conjugates. Eur. J. Inorg. Chem. 2014, 2014, 2886–2895. [Google Scholar] [CrossRef]
- Grob, N.M.; Schmid, S.; Schibli, R.; Behe, M.; Mindt, T.L. Design of radiolabeled analogs of minigastrin by multiple amide-to-triazole substitutions. J. Med. Chem. 2020, 63, 4496–4505. [Google Scholar] [CrossRef]
- Grob, N.M.; Haussinger, D.; Deupi, X.; Schibli, R.; Behe, M.; Mindt, T.L. Triazolo-peptidomimetics: Novel radiolabeled minigastrin analogs for improved tumor targeting. J. Med. Chem. 2020, 63, 4484–4495. [Google Scholar] [CrossRef] [PubMed]
- Mascarin, A.; Valverde, I.E.; Mindt, T.L. Radiolabeled analogs of neurotensin (8–13) containing multiple 1,2,3-triazoles as stable amide bond mimics in the backbone. MedChemComm 2016, 7, 1640–1646. [Google Scholar] [CrossRef]
- Sun, J.; Li, Z. Peptoid applications in biomedicine and nanotechnology. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Woodhead Publishing: Cambridge, UK, 2018; pp. 183–213. [Google Scholar]
- Saini, A.; Verma, G. Peptoids: Tomorrow’s therapeutics. Nanostruct. Novel Ther. 2017, 251–280. [Google Scholar]
- Althuon, D.; Rönicke, F.; Fürniss, D.; Quan, J.; Wellhöfer, I.; Jung, N.; Schepers, U.; Bräse, S. Functionalized triazolopeptoids—A novel class for mitochondrial targeted delivery. Org. Biomol. Chem. 2015, 13, 4226–4230. [Google Scholar] [CrossRef]
- Shin, M.-K.; Hyun, Y.-J.; Lee, J.H.; Lim, H.-S. Comparison of cell permeability of cyclic peptoids and linear peptoids. ACS Comb. Sci. 2018, 20, 237–242. [Google Scholar] [CrossRef]
- D’Aamato, A.; Pierri, G.; Tedesco, C.; Sala, G.D.; Izzo, I.; Costabile, C.; Riccardis, F. reverse turn and loop secondary structures in stereodefined cyclic peptoid scaffolds. J. Org. Chem. 2019, 84, 10911–10928. [Google Scholar] [CrossRef]
- Jiang, L.; Kirshenbaum, K. A modular approach for organizing dimeric coiled coils on peptoid oligomer scaffolds. Org. Biomol. Chem. 2020, 18, 2312–2320. [Google Scholar] [CrossRef]
- Lam, H.; Schwochert, J.; Lao, Y.; Lau, T.; Lloyd, C.; Luu, J.; Kooner, O.; Morgan, J.; Lokey, S.; Auerbuch, V. Synthetic cyclic peptomers as type III secretion system inhibitors. Antimicrob. Agents Chemother. 2017, 61, e00060-17. [Google Scholar] [CrossRef] [PubMed]
- Barreto, A.F.S.; Santos, V.A.D.; Andrade, C.K.Z. Synthesis of acylhydrazino-peptomers, a new class of peptidomimetics, by consecutive Ugi and hydrazine-Ugi reactions. Beilstein J. Org. Chem. 2016, 12, 2865–2872. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.H.B.; Emery, F.S.; Ponte, G.D.; Donate, P.M. Synthesis of some functionalized peptomers via Ugi four-component reaction. Synth. Commun. 2015, 45, 1761–1767. [Google Scholar] [CrossRef]
- Furukawa, A.; Townsend, C.E.; Schwochert, J.; Pye, C.R.; Bednarek, M.A.; Lokey, R.S. Passive membrane permeability in cyclic peptomer scaffolds is robust to extensive variation in side chain functionality and backbone geometry. J. Med. Chem. 2016, 59, 9503–9512. [Google Scholar] [CrossRef]
- Samarasimhareddy, M.; Shamir, M.; Shalev, D.E.; Hurevich, M.; Friedler, A. A rapid and efficient building block approach for click cyclization of peptoids. Front. Chem. 2020, 8, 405. [Google Scholar] [CrossRef]
- Salvador, C.E.M.; Pieber, B.; Neu, P.M.; Torvisco, A.; Andrade, C.K.Z.; Kappe, C.O. A Sequential Ugi multicomponent-Cu-catalyzed azide-alkyne cycloaddition approach for the continuous flow generation of cyclic peptoids. J. Org. Chem. 2015, 80, 4590–4602. [Google Scholar] [CrossRef] [PubMed]
- Assuncao, E.L.F.; Carvalho, D.B.; Neves, A.R.d.; Shiguemotto, C.Y.K.; Portapilla, G.B.; Albuquerque, S.; Baroni, A.C.M. Synthesis and antitrypanosomal activity of novel 1,4-dissubstituted-triazole compounds based on 2-nitroimidazole scaffold: A structure-activity relationship (SAR) study. ChemMedChem 2020, 15, 2019–2028. [Google Scholar] [CrossRef]
- Tautz, M.; Torras, J.; Grijalvo, S.; Eritja, R.; Saldias, C.; Aleman, C.; Diaz, D.D. Expanding the limits of amide-triazole isosteric substitution in bisamide-based physical gels. RSC Adv. 2019, 9, 20841–20851. [Google Scholar] [CrossRef]
- Sahu, A.; Das, D.; Agrawal, R.K.; Gajbhiye, A. Bio-isosteric of amide group with 1,2,3-triazole in phenacetin iéproves the toxicology and efficacy of phenacetin-triazole conjugates (PhTCs). Life Sci. 2019, 228, 176–188. [Google Scholar] [CrossRef]
- Sahu, A.; Das, D.; Sahu, P.; Mishra, S.; Sakthivel, A.; Gajbhiye, A.; Agrawal, R. Bioisosteric replacement of amide group with 1,2,3-triazoles in acetaminophen addresses reactive oxygen species-mediated hepatotoxic insult in wistar albino rats. Chem. Res. Toxicol. 2020, 33, 522–535. [Google Scholar] [CrossRef]
- Peng, X.; Wang, Q.; Mishra, Y.; Xu, J.; Reichert, D.E.; Malik, M.; Taylor, M.; Luedtke, R.R.; Mach, R.H. Synthesis, pharmacological evaluation and molecular modeling studies of triazole containing dopamine D3 receptor ligands. Bioorg. Med. Chem. Lett. 2015, 25, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.; Tu, Z.; McElveen, E.; Xu, J.; Taylor, M.; Luedtke, R.R.; Mach, R.H. Synthesis and in vitro binding of N-phenyl piperazine analogs as potential dopamine D3 receptor ligands. Bioorg. Med. Chem. 2005, 13, 77–87. [Google Scholar] [CrossRef]
- Altimari, J.M.; Niranjan, B.; Risbridger, G.P.; Schweiker, S.S.; Lohning, A.E.; Henderson, L.C. Synthesis and preliminary investigations into novel 1,2,3-triazole-derived androgen receptor antagonists inspired by bicalutamide. Bioorg. Med. Chem. Lett. 2014, 24, 4948–4953. [Google Scholar] [CrossRef] [PubMed]
- Colombo, F.; Tintori, C.; Furlan, A.; Borrelli, S.; Christodoulou, M.S.; Dono, R.; Maina, F.; Botta, M.; Amat, M.; Bosch, J.; et al. ‘Click’ synthesis of a triazole-based inhibitor of Met functions in cancer cells. Bioorg. Med. Chem. Lett. 2012, 22, 4693–4696. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Tjhin, E.T.; Howieson, V.M.; Kittikool, T.; Spry, C.; Saliba, K.J.; Auclair, K. Structure-activity relationships of antiplasmodial pantothenamide analogues reveal a new way by which triazoles mimic amide bonds. ChemMedChem 2018, 13, 2677–2683. [Google Scholar] [CrossRef] [PubMed]
- Howieson, V.M.; Tran, E.; Hoegl, A.; Fam, H.L.; Fu, J.; Sivonen, K.; Li, X.X.; Auclair, K.; Saliba, K.J. Triazole substitution of a labile amide bond stabilizes pantothenamides and improves their antiplasmodial potency. Antimicrob. Agents Chemother. 2016, 60, 7146–7152. [Google Scholar] [CrossRef] [PubMed]







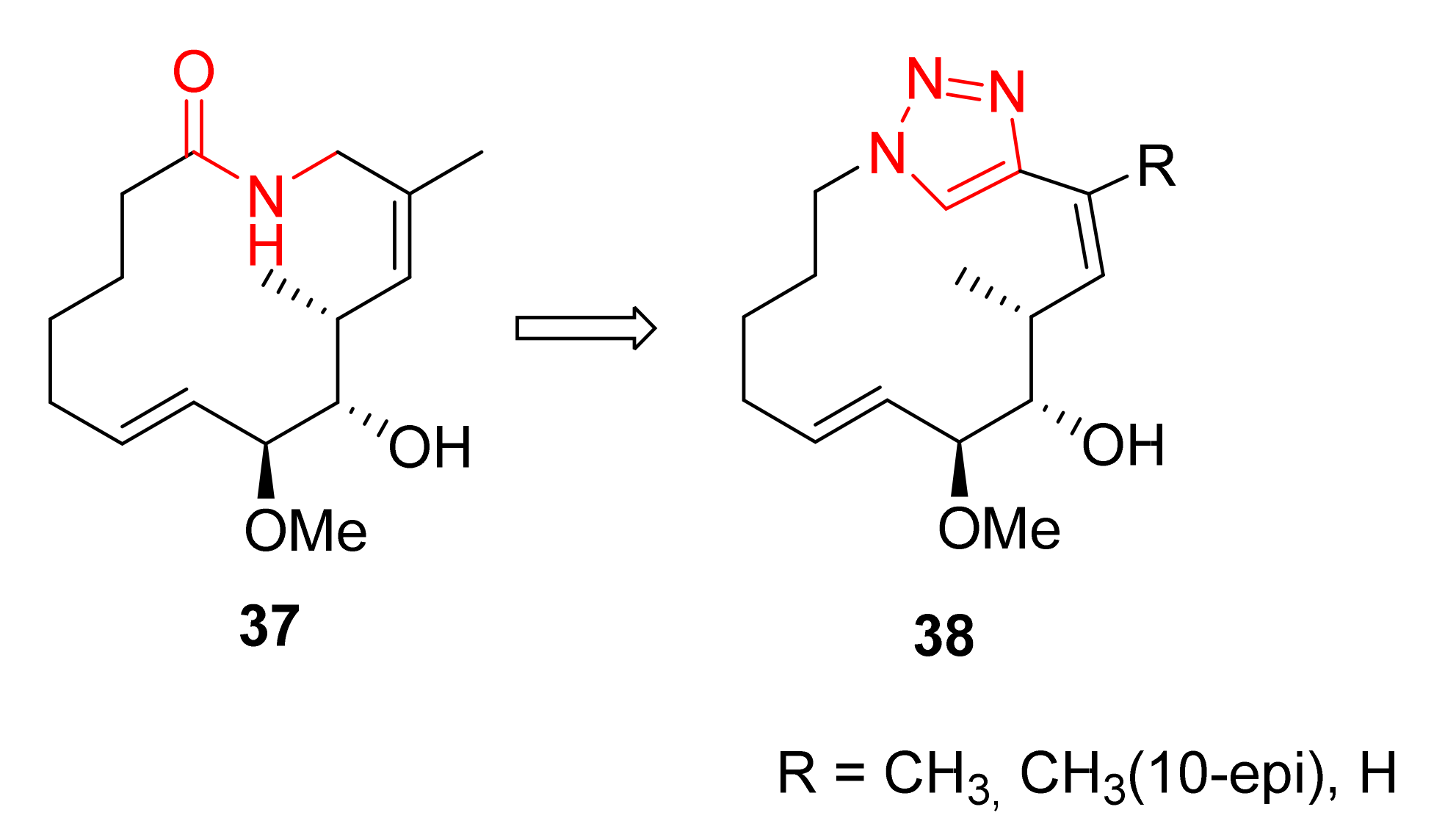
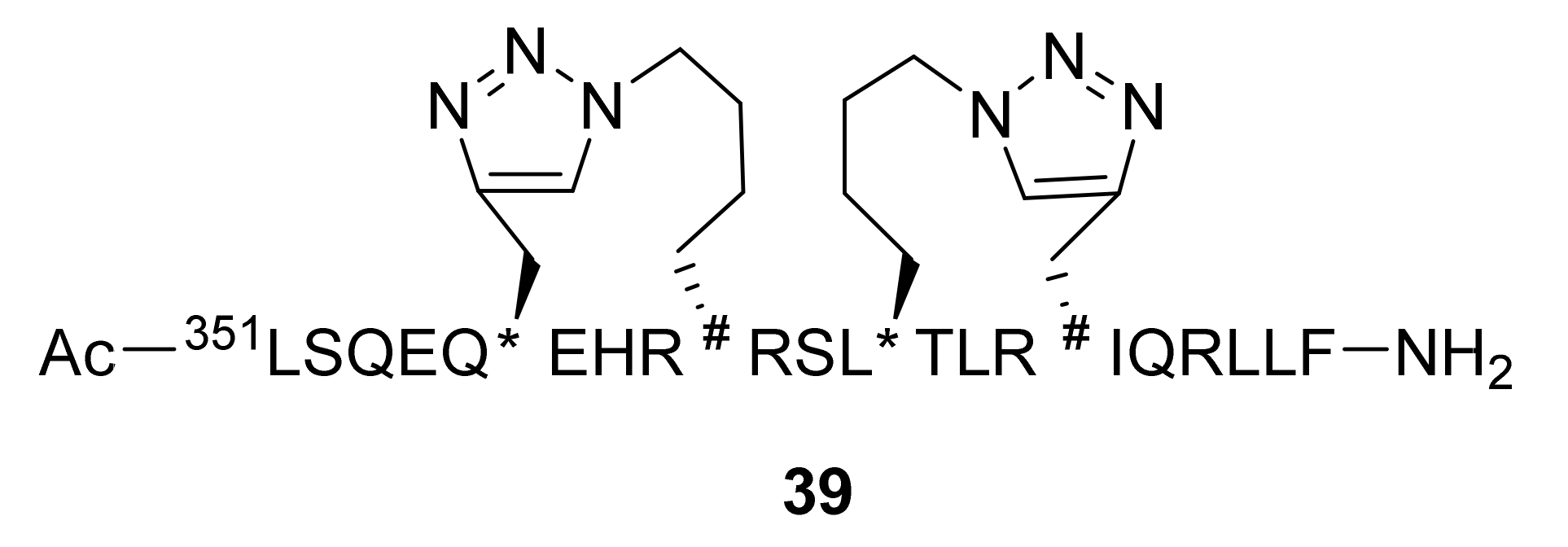
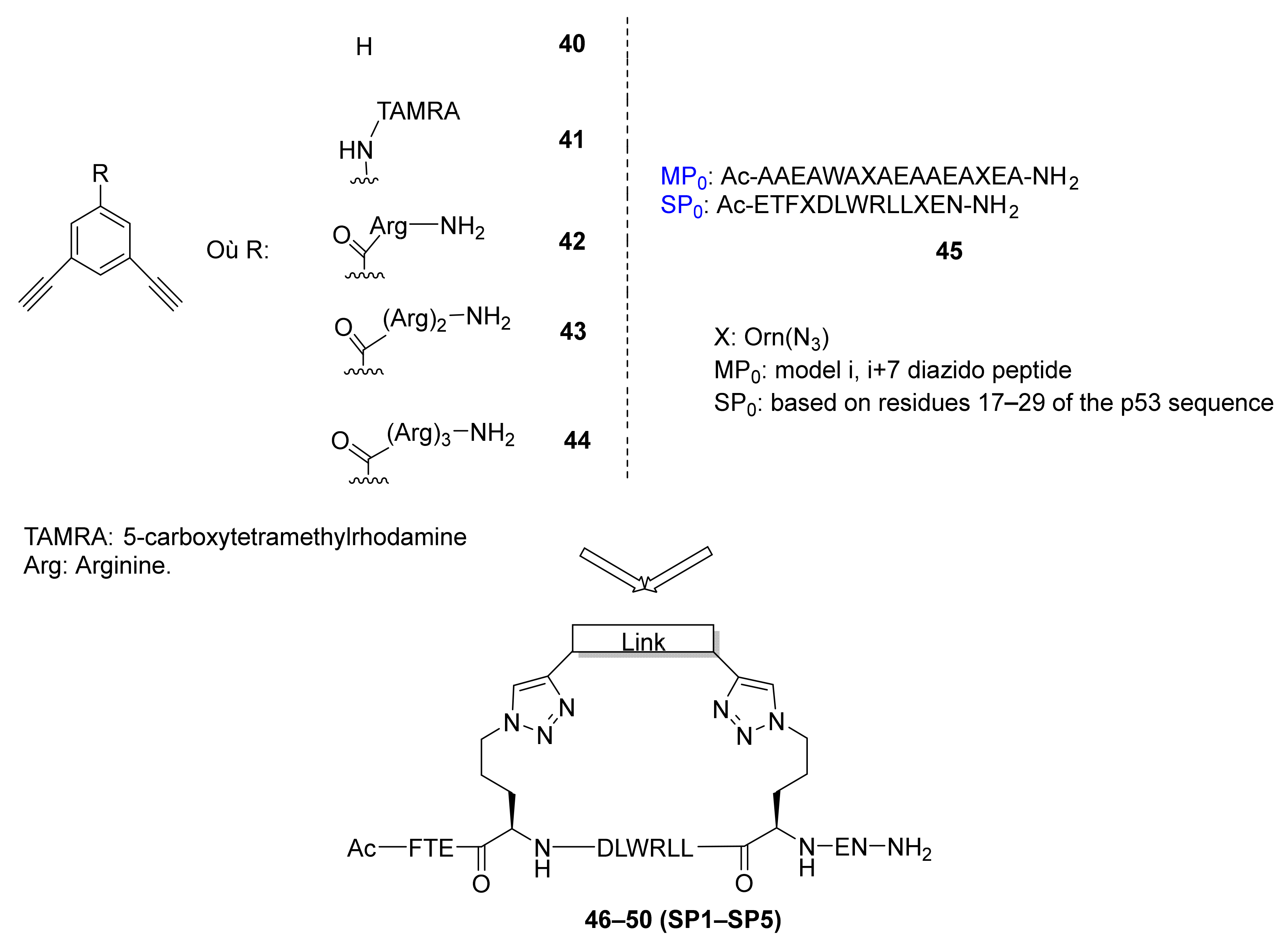

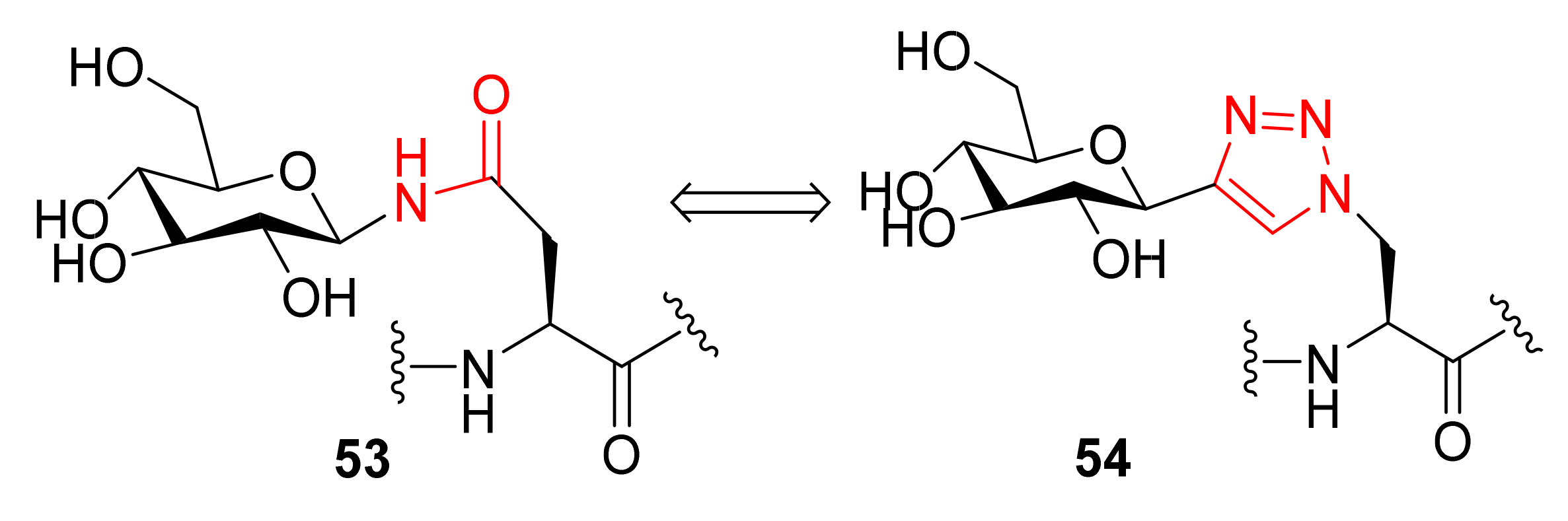



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agouram, N.; El Hadrami, E.M.; Bentama, A. 1,2,3-Triazoles as Biomimetics in Peptide Science. Molecules 2021, 26, 2937. https://doi.org/10.3390/molecules26102937
Agouram N, El Hadrami EM, Bentama A. 1,2,3-Triazoles as Biomimetics in Peptide Science. Molecules. 2021; 26(10):2937. https://doi.org/10.3390/molecules26102937
Chicago/Turabian StyleAgouram, Naima, El Mestafa El Hadrami, and Abdeslem Bentama. 2021. "1,2,3-Triazoles as Biomimetics in Peptide Science" Molecules 26, no. 10: 2937. https://doi.org/10.3390/molecules26102937
APA StyleAgouram, N., El Hadrami, E. M., & Bentama, A. (2021). 1,2,3-Triazoles as Biomimetics in Peptide Science. Molecules, 26(10), 2937. https://doi.org/10.3390/molecules26102937