UPLC-ESI-MS/MS-Based Widely Targeted Metabolomics Analysis of Wood Metabolites in Teak (Tectona grandis)
Abstract
1. Introduction
2. Materials and Methods
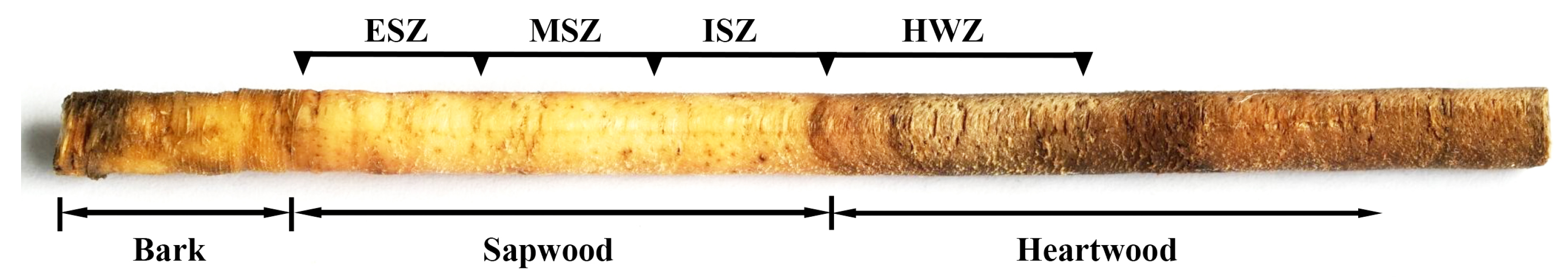
2.1. Plant Materials
2.2. Sample Preparation
2.3. HPLC Conditions
2.4. ESI-Q TRAP-MS/MS
2.5. Qualitative and Quantitative Analysis of Metabolites
2.6. Statistical Analysis
3. Results
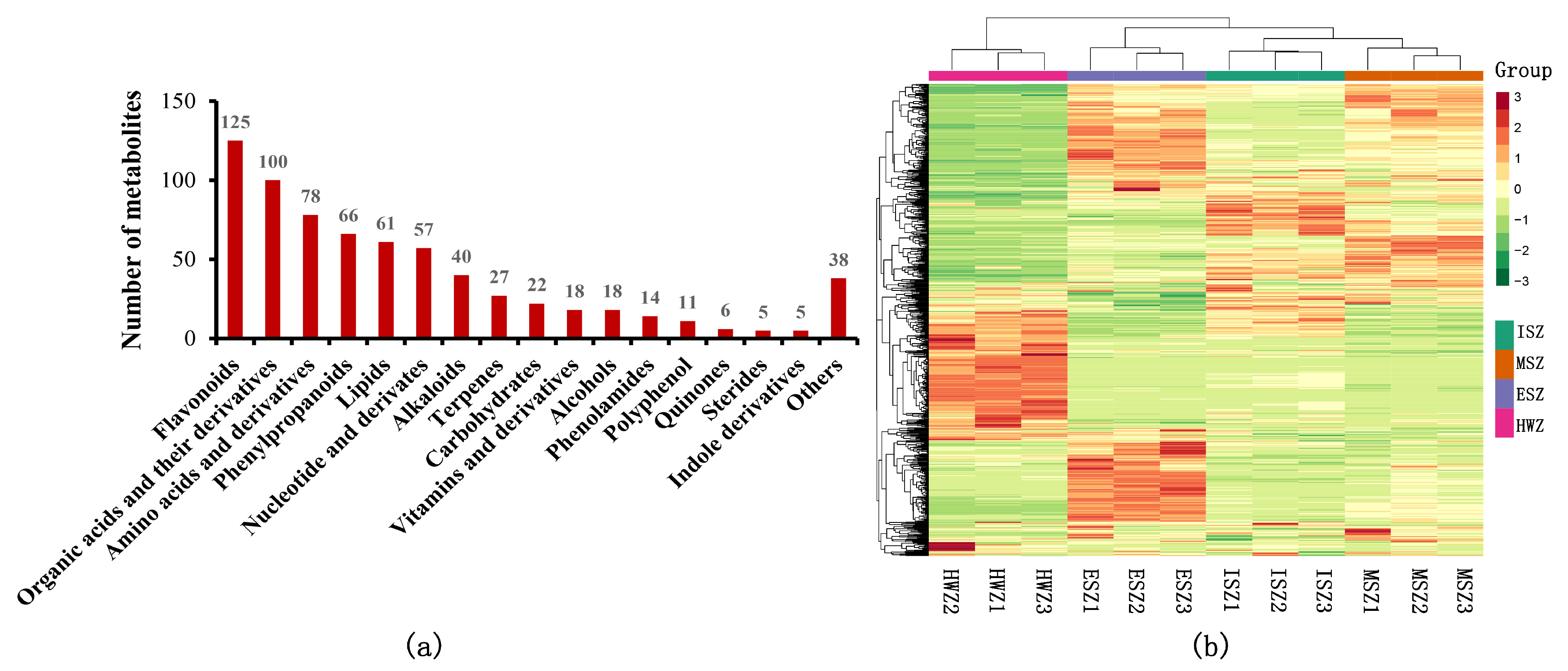
3.1. Widely Targeted Metabolic Profiling of Teak Wood
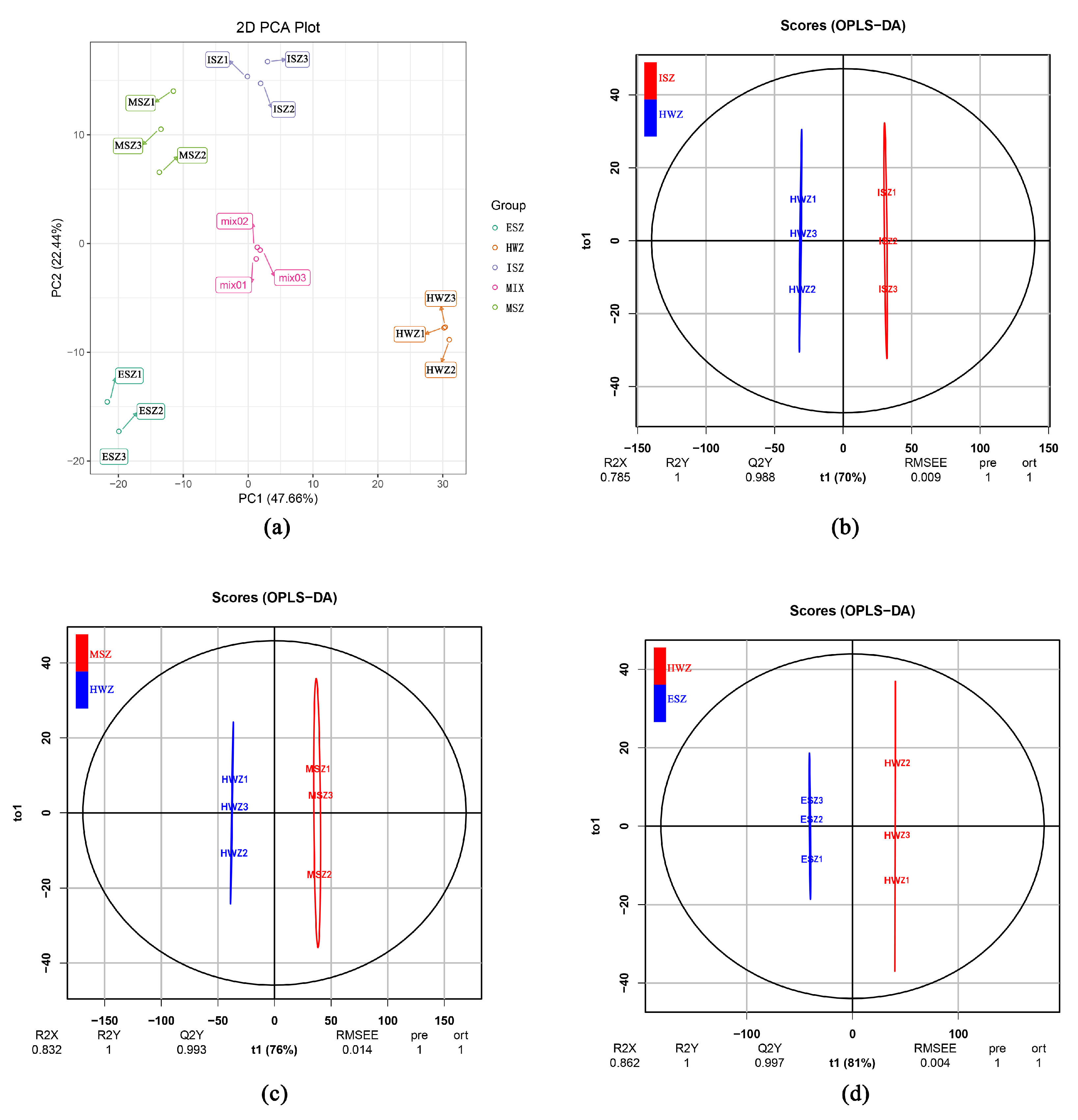
3.2. PCA for All Samples
3.3. OPLS-DA for all Samples
3.4. Differential Metabolite Screening
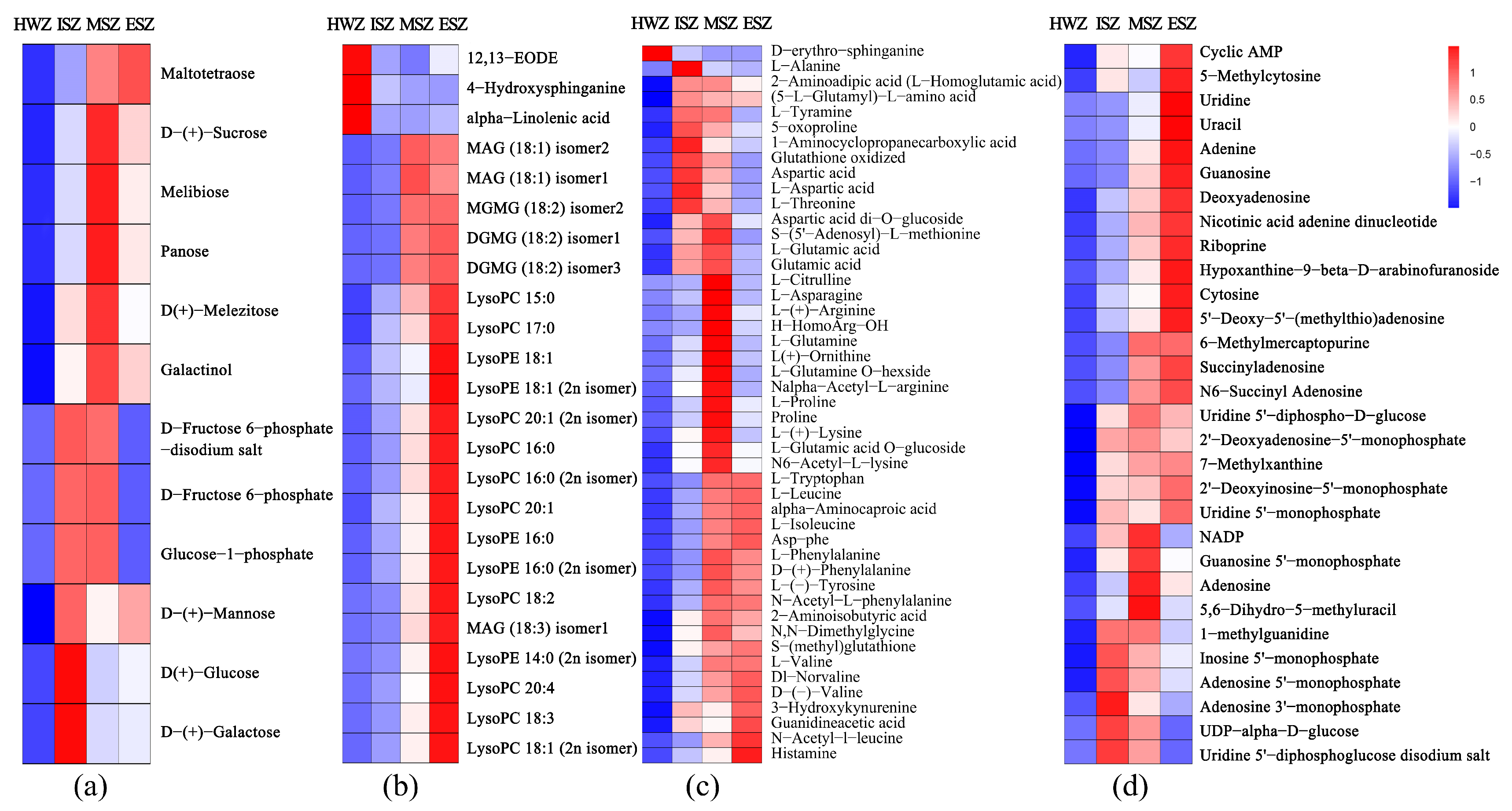
3.5. Differential Primary Metabolites Analysis
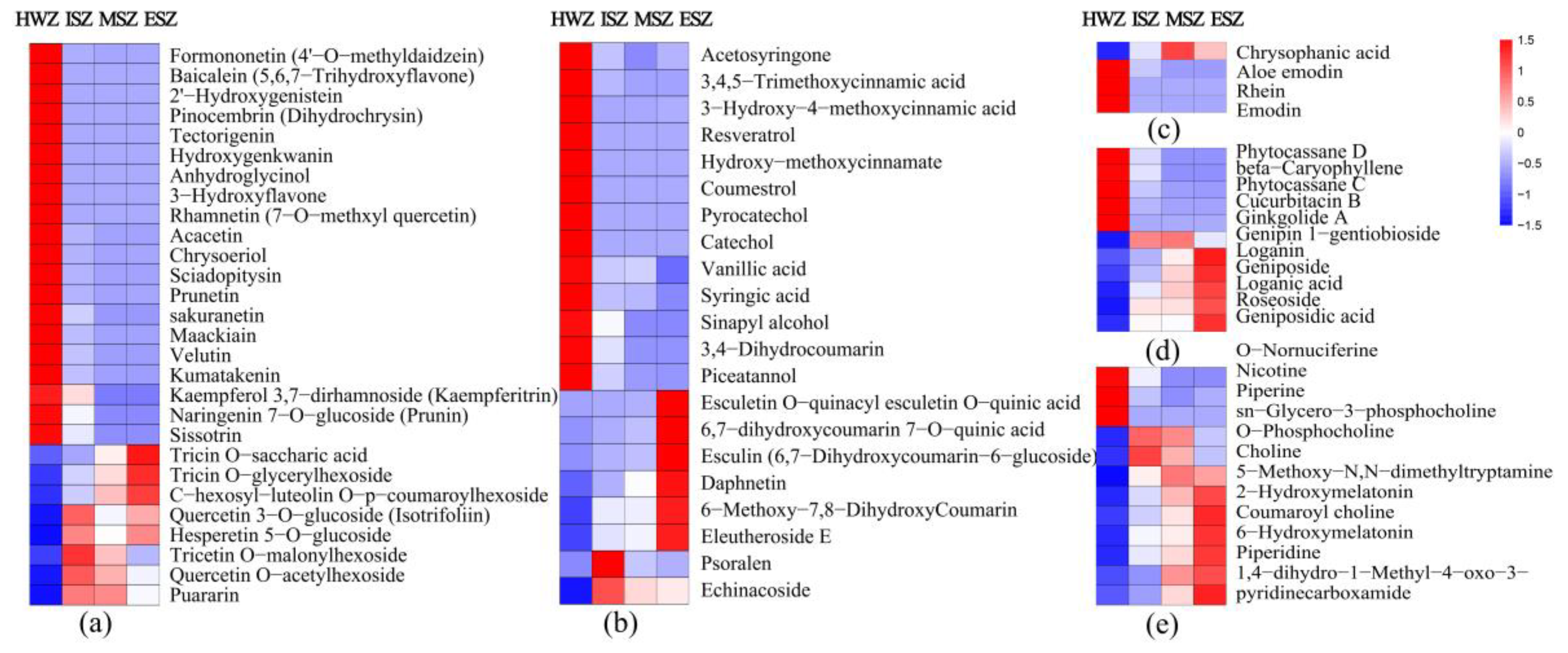
3.6. Differential Secondary Metabolites Analysis
4. Discussion
4.1. Wide Target Metabolome
4.2. Differential Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Palanisamy, K.; Hegde, M.; Yi, J.-S. Teak (Tectona grandis Linn. f.): A renowned commercial timber species. J. For. Environ. Sci. 2009, 25, 1–24. [Google Scholar]
- Kokutse, A.D.; Stokes, A.; Kokutse, N.K.; Kokou, K. Which factors most influence heartwood distribution and radial growth in plantation teak? Ann. For. Sci. 2010, 67, 407. [Google Scholar] [CrossRef]
- Brocco, V.F.; Paes, J.B.; Costa, L.G.d.; Brazolin, S.; Arantes, M.D.C. Potential of teak heartwood extracts as a natural wood preservative. J. Clean. Prod. 2017, 142, 2093–2099. [Google Scholar] [CrossRef]
- Qiu, H.; Liu, R.; Long, L. Analysis of chemical composition of extractives by acetone and the chromatic aberration of Teak (Tectona Grandis L.F.) from China. Molecules 2019, 24, 1989. [Google Scholar] [CrossRef]
- Rodríguez Anda, R.; Koch, G.; Richter, H.-G.; Fuentes Talavera, F.J.; Silva Guzmán, J.A.; Satyanarayana, K.G. Formation of heartwood, chemical composition of extractives and natural durability of plantation-grown teak wood from Mexico. Holzforschung 2019, 73, 547–557. [Google Scholar] [CrossRef]
- Lukmandaru, G.; Takahashi, K. Variation in the natural termite resistance of teak (Tectona grandis Linn. fil.) wood as a function of tree age. Ann. For. Sci. 2008, 65, 708. [Google Scholar] [CrossRef]
- Lukmandaru, G.; Takahashi, K. Radial distribution of quinones in plantation teak (Tectona grandis L.f.). Ann. For. Sci. 2009, 66, 605. [Google Scholar] [CrossRef]
- Haupt, M.; Leithoff, H.; Meier, D.; Puls, J.; Richter, H.G.; Faix, O. Heartwood extractives and natural durability of plantation-grown teakwood (Tectona grandis L.)? A case study. Holz Als Roh- Und Werkst. 2003, 61, 473–474. [Google Scholar] [CrossRef]
- Niamké, F.B.; Amusant, N.; Charpentier, J.-P.; Chaix, G.; Baissac, Y.; Boutahar, N.; Adima, A.A.; Kati-Coulibaly, S.; Jay-Allemand, C. Relationships between biochemical attributes (non-structural carbohydrates and phenolics) and natural durability against fungi in dry teak wood (Tectona grandis L. f.). Ann. For. Sci. 2011, 68, 201–211. [Google Scholar] [CrossRef]
- Sumthong, P.; Damveld, R.A.; Choi, Y.H.; Arentshorst, M.; Ram, A.F.; van den Hondel, C.A.; Verpoorte, R. Activity of quinones from teak (Tectona grandis) on fungal cell wall stress. Planta Med. 2006, 72, 943–944. [Google Scholar] [CrossRef]
- Ismayati, M.; Nakagawa-Izumi, A.; Kamaluddin, N.N.; Ohi, H. Toxicity and feeding deterrent effect of 2-Methylanthraquinone from the wood extractives of Tectona grandis on the subterranean termites coptotermes formosanus and reticulitermes speratus. Insects 2016, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Thulasidas, P.; Bhat, K. Chemical extractive compounds determining the brown-rot decay resistance of teak wood. Holz Als Roh-Und Werkst. 2007, 65, 121–124. [Google Scholar] [CrossRef]
- Burtin, P.; Jay-Allemand, C.; Charpentier, J.-P.; Janin, G. Natural wood colouring process in Juglans sp. (J. nigra, J. regia and hybrid J. nigra 23× J. regia) depends on native phenolic compounds accumulated in the transition zone between sapwood and heartwood. Trees 1998, 12, 258–264. [Google Scholar] [CrossRef]
- Thulasidas, P.; Bhat, K.; Okuyama, T. Heartwood colour variation in home garden teak (Tectona grandis) from wet and dry localities of Kerala, India. J. Trop. For. Sci. 2006, 18, 51–54. [Google Scholar]
- Moya, R.; Marín, J.D. Grouping of Tectona grandis (L.f.) clones using wood color and stiffness. New For. 2011, 42, 329–345. [Google Scholar] [CrossRef]
- Moya, R.; Berrocal, A. Wood colour variation in sapwood and heartwood of young trees of Tectona grandis and its relationship with plantation characteristics, site, and decay resistance. Ann. For. Sci. 2010, 67, 109. [Google Scholar] [CrossRef]
- Miranda, I.; Sousa, V.; Pereira, H. Wood properties of teak (Tectona grandis) from a mature unmanaged stand in East Timor. J. Wood Sci. 2011, 57, 171–178. [Google Scholar] [CrossRef]
- Gierlinger, N.; Jacques, D.; Grabner, M.; Wimmer, R.; Schwanninger, M.; Rozenberg, P.; Pques, L.E. Colour of larch heartwood and relationships to extractives and brown-rot decay resistance. Trees Struct. Funct. 2004, 18, 102–108. [Google Scholar] [CrossRef]
- Windeisen, E.; Klassen, A.; Wegener, G. On the chemical characterisation of plantation teakwood from Panama. Holz Als Roh- Und Werkst. 2003, 61, 416–418. [Google Scholar] [CrossRef]
- Niamké, F.B.; Amusant, N.; Kadio, A.A.; Thevenon, M.-F.; Nourissier, S.; Adima, A.A.; Jay-Allemand, C.; Chaix, G. Rapid prediction of phenolic compounds as chemical markers for the natural durability of teak (Tectona Grandis Linn f.) Heartwood by near infrared spectroscopy. J. Near Infrared Spectrosc. 2014, 22, 35–43. [Google Scholar] [CrossRef]
- Vyas, P.; Yadav, D.K.; Khandelwal, P. Tectona grandis (teak)—A review on its phytochemical and therapeutic potential. Nat. Prod. Res. 2019, 33, 2338–2354. [Google Scholar] [CrossRef] [PubMed]
- Romanis, R. LXXXVIII.—Certain products from teak. Preliminary notice. J. Chem. Soc. Trans. 1887, 51, 868–871. [Google Scholar] [CrossRef][Green Version]
- Sandermann, W.; Dietrichs, H. Chemical studies on tropical woods. IV. Chemical researches on teakwood. Holzforschung 1959, 13, 137–148. [Google Scholar] [CrossRef]
- Macías, F.A.; Lacret, R.; Varela, R.M.; Nogueiras, C.; Molinillo, J.M. Isolation and phytotoxicity of terpenes from Tectona grandis. J. Chem. Ecol. 2010, 36, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Dayal, R.; Seshadri, T. Chemical investigation of Tectona grandis (roots). J. Indian Chem. Soc. 1979. [Google Scholar]
- Rudman, P. Anthraquinones of Teak (Tectona grandis LF). Chem. Ind. 1960, 44, 1356–1357. [Google Scholar]
- Sandermann, W.; Simatupang, M. On the chemistry and biochemistry of teakwood. Holz Als Roh-Und Werkst. 1966, 24, 190–204. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Wang, F.; Chen, L.; Chen, H.; Chen, S.; Liu, Y. Analysis of flavonoid metabolites in citrus peels (Citrus reticulata “Dahongpao”) using UPLC-ESI-MS/MS. Molecules 2019, 24, 2680. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, L.; Huang, X.; Wang, X.; Yang, R.; Mao, J.; Wang, X.; Wang, X.; Zhang, Q.; Li, P. Identification of nutritional components in black sesame determined by widely targeted metabolomics and traditional Chinese medicines. Molecules 2018, 23, 1180. [Google Scholar] [CrossRef]
- Wang, A.; Li, R.; Ren, L.; Gao, X.; Zhang, Y.; Ma, Z.; Ma, D.; Luo, Y. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam). Food Chem 2018, 260, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Thevenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Nobuchi, T.; Okada, N.; Nishida, M.; Siripatanadilok, S.; Veenin, T.; Tobing, T.; Sahri, M. Some characteristics of wood formation in Teak (Tectona grandis) with special reference to water conditions. Qual. Timber Prod. Teak Sustain. For. Management. India 2005, 495–499. [Google Scholar]
- Datta, S.; Kumar, A. Histochemical studies of the transition from sapwood to heartwood in Tectona grandis. Iawa J. 1987, 8, 363–368. [Google Scholar] [CrossRef]
- Niamke, B.F.; Adima, A.A.; Seraphin, K.-C.; Amusant, N.; Jay-Allemand, C. Heartwood formation process in teak (Tectona grandis L. f): Fate of non-structural carbohydrates and characterization of forsythoside B. Int. J. Biol. Chem. Sci. 2018, 12, 1102–1112. [Google Scholar] [CrossRef][Green Version]
- Kampe, A.; Magel, E. New insights into heartwood and heartwood formation. In Cellular Aspects of Wood Formation; Springer: Berlin, Germany, 2013; pp. 71–95. [Google Scholar] [CrossRef]
- Nobuchi, T.; Janmahasatien, S.; Sakai, M. Seasonal changes of wood formation and some characteristics of heartwood formation in teak (Tectona grandis L.) plantation. Kasetsart J. Nat. Sci. 1996, 30, 254–263. [Google Scholar]
- Nair, M.; Shah, J.; Pandalai, R. Wood anatomy and histochemical changes of sapwood during heartwood formation in Bridelia retusa Spreng. Proc.: Plant. Sci. 1981, 90, 425–433. [Google Scholar] [CrossRef]
- Hillis, W.; Hasegawa, M. The formation of polyphenols in trees I. Administration of c glucose and subsequent distribution of radioactivity. Phytochemistry 1963, 2, 195–199. [Google Scholar] [CrossRef]
- Hillis, W.E. Heartwood and Tree Exudates; Springer Science & Business Media: Berlin, Germany, 2012; Volume 4. [Google Scholar]
- La Villa, G.; Marra, F.; Laffi, G.; Belli, B.; Meacci, E.; Fascetti, P.; Gentilini, P. Effects of rhein on renal arachidonic acid metabolism and renal function in patients with congestive heart failure. Eur. J. Clin. Pharmacol. 1989, 37, 1–5. [Google Scholar] [CrossRef]
- Zhang, H.-Q.; Zhou, C.-H.; Wu, Y.-Q. Effect of emodin on small intestinal peristalsis of mice and relevant mechanism. World J. Gastroenterol. Wjg 2005, 11, 3147. [Google Scholar] [CrossRef]
- Pecere, T.; Gazzola, M.V.; Mucignat, C.; Parolin, C.; Dalla Vecchia, F.; Cavaggioni, A.; Basso, G.; Diaspro, A.; Salvato, B.; Carli, M. Aloe-emodin is a new type of anticancer agent with selective activity against neuroectodermal tumors. Cancer Res. 2000, 60, 2800–2804. [Google Scholar] [PubMed]
- Floridi, A.; Castiglione, S.; Bianchi, C.; Mancini, A. Effect of rhein on the glucose metabolism of Ehrlich ascites tumor cells. Biochem. Pharmacol. 1990, 40, 217–222. [Google Scholar] [CrossRef]
- Johansson, C.I.; Saddler, J.N.; Beatson, R.P. Characterization of the polyphenolics related to the colour of western red cedar (Thuja plicata Donn) heartwood. Holzforschung 2000, 54, 246–254. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Id | Q1 (Da) | Q3 (Da) | Rt (min) | Molecular Weight (Da) | Compounds | Class |
|---|---|---|---|---|---|---|
| pmb0423 | 195.1 | 145.3 | 3.22 | 194.1 | Hydroxy-methoxycinnamate | Phenylpropanoids |
| pme0307 | 227.1 | 185 | 4.78 | 228.08 | Resveratrol | Phenylpropanoids |
| pmf0117 | 266.9 | 211.1 | 5.75 | 268.04 | Coumestrol | Phenylpropanoids |
| pme1510 | 269.1 | 251 | 5.94 | 270.05 | Baicalein (5,6,7-Trihydroxyflavone) | Flavone |
| pme3137 | 239 | 165 | 7.73 | 238.06 | 3-Hydroxyflavone | Flavonol |
| pme3369 | 315 | 165 | 6.43 | 316.06 | Rhamnetin (7-O-methxyl quercetin) | Flavonol |
| pme2979 | 255.1 | 151 | 7.04 | 256.07 | Pinocembrin (Dihydrochrysin) | Flavanone |
| pme3279 | 287 | 153 | 4.89 | 286.05 | 2’-Hydroxygenistein | Isoflavone |
| pmf0111 | 253 | 224.8 | 6.08 | 254.06 | Anhydroglycinol | Flavonoid |
| pmf0363 | 301.1 | 258 | 6.28 | 300.06 | Hydroxygenkwanin | Flavonoid |
| pmf0567 | 299.1 | 211 | 5.72 | 300.06 | Tectorigenin | Flavonoid |
| pmf0428 | 407.1 | 363.1 | 5.24 | 408.14 | Ginkgolide A | Terpene |
| pmf0520 | 283 | 239 | 6.58 | 284.03 | Rhein | Quinones |
| pmf0612 | 286.1 | 171.2 | 7.42 | 285.14 | Piperine | Alkaloids |
| pmf0299 | 257.1 | 240 | 7.52 | 256.11 | Pterostilbene | Others |
| pma6625 | 405.1 | 243.1 | 4.63 | 406.13 | E-3,4,5’-Trihydroxy-3’-glucopyranosylstilbene | Others |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Liang, K.; Zhou, Z.; Wang, X.; Huang, G. UPLC-ESI-MS/MS-Based Widely Targeted Metabolomics Analysis of Wood Metabolites in Teak (Tectona grandis). Molecules 2020, 25, 2189. https://doi.org/10.3390/molecules25092189
Yang G, Liang K, Zhou Z, Wang X, Huang G. UPLC-ESI-MS/MS-Based Widely Targeted Metabolomics Analysis of Wood Metabolites in Teak (Tectona grandis). Molecules. 2020; 25(9):2189. https://doi.org/10.3390/molecules25092189
Chicago/Turabian StyleYang, Guang, Kunnan Liang, Zaizhi Zhou, Xiyang Wang, and Guihua Huang. 2020. "UPLC-ESI-MS/MS-Based Widely Targeted Metabolomics Analysis of Wood Metabolites in Teak (Tectona grandis)" Molecules 25, no. 9: 2189. https://doi.org/10.3390/molecules25092189
APA StyleYang, G., Liang, K., Zhou, Z., Wang, X., & Huang, G. (2020). UPLC-ESI-MS/MS-Based Widely Targeted Metabolomics Analysis of Wood Metabolites in Teak (Tectona grandis). Molecules, 25(9), 2189. https://doi.org/10.3390/molecules25092189




