Repurposing Zileuton as a Depression Drug Using an AI and In Vitro Approach
Abstract
1. Introduction
2. Results
2.1. RNA-seq Data Analysis
2.2. AI Embedding, Similarity, and Relevance Model Performance
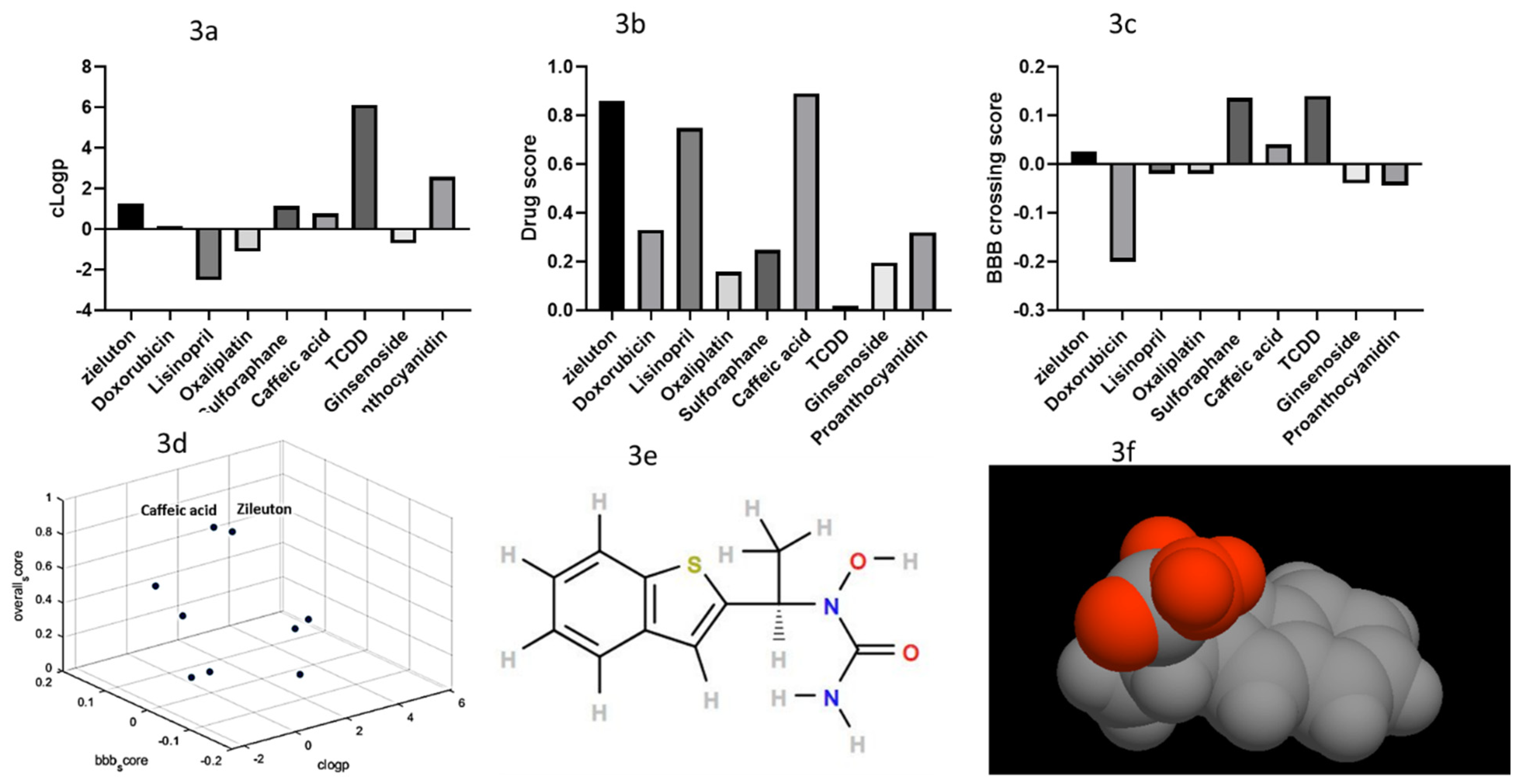
2.3. Filtered Phase Results
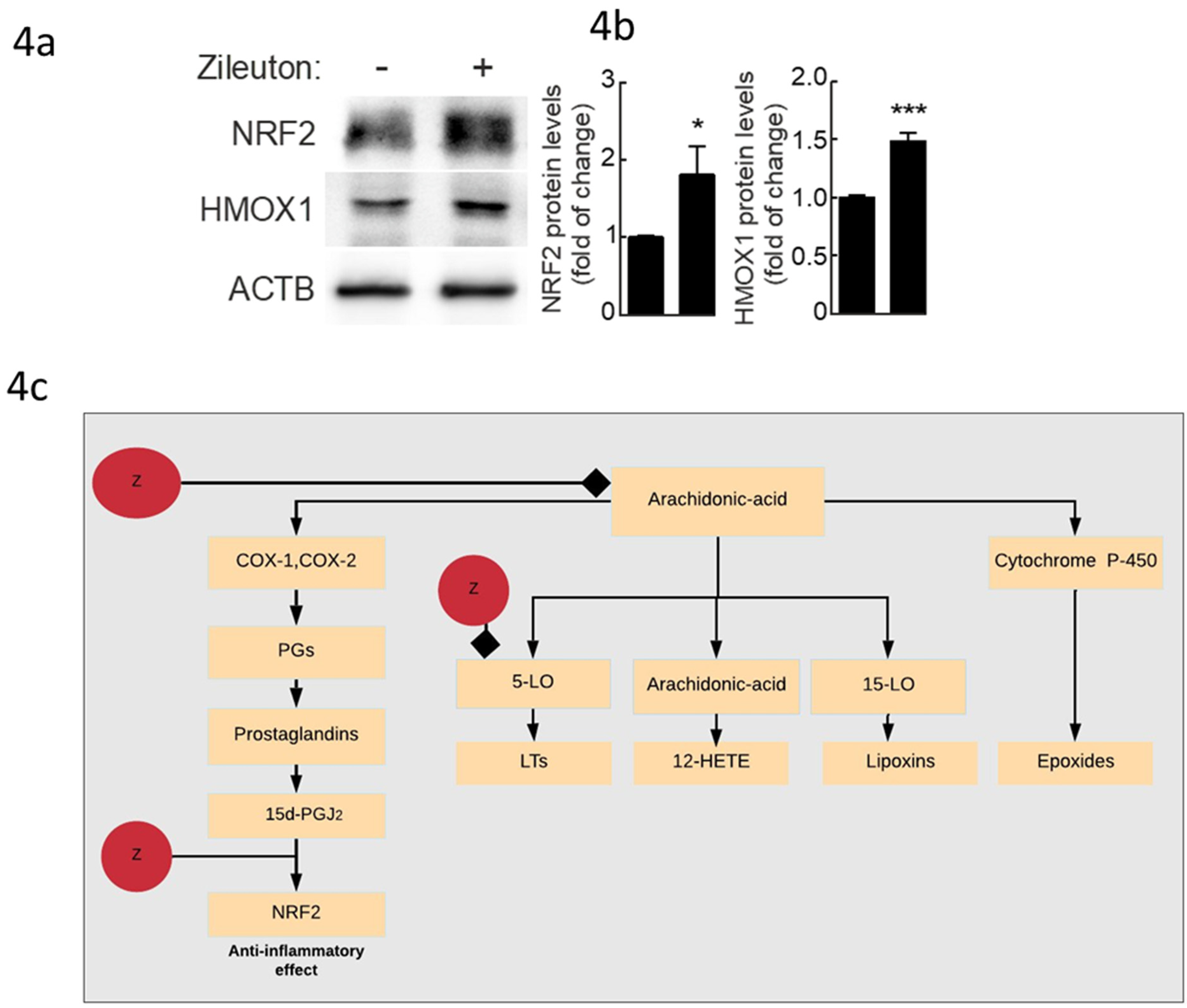
2.4. Experimental Validation Results
3. Discussion
Limitations and Future Development
4. Methods
4.1. Analysis of RNA-seq Data
4.2. Building a Text Mining Deep Learning Neural Network
4.3. The First Phase Used a DAN Neural Network Approach
4.4. The Second Phase Used a Differential Convolution Network (DCN)
4.5. In Silico Prediction of Blood–Brain Barrier Diffusion
4.6. Physicochemical Properties
4.7. Experimental Validation
5. Conclusion
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [PubMed]
- Collier, R. Drug development cost estimates hard to swallow. CMAJ 2009, 180, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Lavecchia, A.; di Giovanni, C. Virtual Screening Strategies in Drug Discovery: A Critical Review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.R.; Sullivan, D.J. New uses for old drugs. Nature 2007, 448, 645–646. [Google Scholar] [CrossRef]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of Drug Repositioning Approaches and Resources. Int. J. Biol. Sci. 2018, 14, 1232–1244. [Google Scholar] [CrossRef]
- Miller, A.H.; Raison, C.L. The role of inflammation in depression: From evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 2016, 16, 22. [Google Scholar] [CrossRef]
- Smith, R.S. The macrophage theory of depression. Med. Hypotheses 1991, 35, 298–306. [Google Scholar] [CrossRef]
- Dey, A.; Giblin, P.A.H. Insights into macrophage heterogeneity and cytokine-induced neuroinflammation in major depressive disorder. Pharmaceuticals 2018, 11, 64. [Google Scholar] [CrossRef]
- Miller, A.H.; Maletic, V.; Raison, C.L. Inflammation and Its Discontents: The Role of Cytokines in the Pathophysiology of Major Depression. Biol. Psychiatry 2009, 65, 732–741. [Google Scholar] [CrossRef]
- Singhal, G.; Baune, B.T. Inflammatory Abnormalities in Major Depressive Disorder. In Major Depressive Disorder; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Maes, M.; Bosmans, E.; de Jongh, R.; Kenis, G.; Vandoolaeghe, E.; Neels, H. Increased serum IL-6 and IL-1 receptor antagonist concentrations in major depression and treatment resistant depression. Cytokine 1997, 9, 853–858. [Google Scholar] [CrossRef]
- Mostafavi, S.; Battle, A.; Zhu, X.; Potash, J.B.; Weissman, M.M.; Shi, J.; Beckman, K.; Haudenschild, C.; McCormick, C.; Mei, R.; et al. Type I interferon signaling genes in recurrent major depression: Increased expression detected by whole-blood RNA sequencing. Mol. Psychiatry 2014, 19, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Carboni, L.; McCarthy, D.J.; Delafont, B.; Filosi, M.; Ivanchenko, E.; Ratti, E.; Learned, S.M.; Alexander, R.; Domenici, E. Biomarkers for response in major depression: Comparing paroxetine and venlafaxine from two randomised placebo-controlled clinical studies. Transl. Psychiatry 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.N.; Rizavi, H.S.; Bhaumik, R.; Ren, X. Innate immunity in the postmortem brain of depressed and suicide subjects: Role of Toll-like receptors. Brain. Behav. Immun. 2019, 75, 101–111. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Tomaz, V.; Chaves Filho, A.J.M.; Cordeiro, R.C.; Jucá, P.M.; Soares, M.V.R.; Barroso, P.N.; Cristino, L.M.F.; Jiang, W.; Teixeira, A.L.; de Lucena, D.F.; et al. Antidepressants of different classes cause distinct behavioral and brain pro- and anti-inflammatory changes in mice submitted to an inflammatory model of depression. J. Affect. Disord. 2020, 268, 188–200. [Google Scholar] [CrossRef]
- Peretti, S.; Judge, R.; Hindmarch, I. Safety and tolerability considerations: Tricyclic antidepressants vs. selective serotonin reuptake inhibitors. Acta Psychiatr. Scand. 2000, 101, 17–25. [Google Scholar] [CrossRef]
- Ferguson, J.M. SSRI antidepressant medications: Adverse effects and tolerability. Prim. Care Companion J. Clin. Psychiatry 2001, 3, 22. [Google Scholar] [CrossRef]
- Kaneko, J.; Tanimukai, H. Monoamine Oxidase Inhibitors (MAO). Sogo Rinsho 1964, 13, 833–846. [Google Scholar]
- Boulenger, J.P.; Loft, H.; Olsen, C.K. Efficacy and safety of vortioxetine (Lu AA21004), 15 and 20 mg/day: A randomized, double-blind, placebo-controlled, duloxetine-referenced study in the acute treatment of adult patients with major depressive disorder. Int. Clin. Psychopharmacol. 2014, 29, 138. [Google Scholar] [CrossRef]
- Dhir, A. Vortioxetine for the treatment of major depression. Drugs Today 2013, 49, 781–790. [Google Scholar] [CrossRef]
- Regan, T.; Gill, A.C.; Clohisey, S.M.; Barnett, M.W.; Pariante, C.M.; Harrison, N.A. MRC Immunopsychiatry Consortium; Hume, D.A.; Bullmore, E.T.; Freeman, T.C. Effects of anti-inflammatory drugs on the expression of tryptophan-metabolism genes by human macrophages. J. Leukoc. Biol. 2018, 103, 681–692. [Google Scholar] [CrossRef]
- Feng, R.; Morine, Y.; Ikemoto, T.; Imura, S.; Iwahashi, S.; Saito, Y.; Shimada, M. Nrf2 activation drive macrophages polarization and cancer cell epithelial-mesenchymal transition during interaction. Cell Commun. Signal. 2018, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.H.; Kuo, C.H.; Kuo, W.W.; Ho, T.J.; Pai, P.; Chen, W.K.; Pan, L.F.; Wang, C.C.; Padma, V.V.; Huang, C.Y. NFIL3 suppresses hypoxia-induced apoptotic cell death by targeting the insulin-like growth factor 2 receptor. J. Cell. Biochem. 2015, 116, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Matsuoka, K.; Sheikh, S.Z.; Elloumi, H.Z.; Kamada, N.; Hisamatsu, T.; Hansen, J.J.; Doty, K.R.; Pope, S.D.; Smale, S.T.; et al. NFIL3 Is a Regulator of IL-12 p40 in Macrophages and Mucosal Immunity. J. Immunol. 2011, 186, 4649–4655. [Google Scholar] [CrossRef] [PubMed]
- Diniz, L.R.L.; Souza, M.T.d.; Barboza, J.N.; de Almeida, R.N.; de Sousa, D.P. Antidepressant potential of cinnamic acids: Mechanisms of action and perspectives in drug development. Molecules 2019, 24, 4469. [Google Scholar] [CrossRef] [PubMed]
- Pawar, A.; Mago, V. IEEE Transactions on knowledge and data engineering Calculating the similarity between words and sentences using a lexical database and corpus statistics. arXiv 2018, arXiv:1802.05667. [Google Scholar]
- Salim, S. Oxidative Stress and Psychological Disorders. Curr. Neuropharmacol. 2014, 12, 140–147. [Google Scholar] [CrossRef]
- Popa-Wagner, A.; Mitran, S.; Sivanesan, S.; Chang, E.; Buga, A.-M. ROS and brain diseases: The good, the bad, and the ugly. Oxid. Med. Cell. Longev. 2013, 2013, 963520. [Google Scholar] [CrossRef]
- Joshi, G.; Johnson, J.A. The Nrf2-ARE pathway: A valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat. CNS Drug Discov. 2012, 7, 218–229. [Google Scholar] [CrossRef]
- Kim, G.H.; Kim, J.E.; Rhie, S.J.; Yoon, S. The Role of Oxidative Stress in Neurodegenerative Diseases. Exp. Neurobiol. 2015, 24, 325–340. [Google Scholar] [CrossRef]
- Costarelli, L.; Malavolta, M.; Giacconi, R.; Provinciali, M. Dysfunctional macrophages in Alzheimer Disease: Another piece of the “macroph-aging” puzzle? Aging (Albany NY) 2017, 9, 1865–1866. [Google Scholar] [CrossRef] [PubMed]
- Bakunina, N.; Pariante, C.M.; Zunszain, P.A. Immune mechanisms linked to depression via oxidative stress and neuroprogression. Immunology 2015, 144, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, P. Oxidative stress and macrophage function: A failure to resolve the inflammatory response. Biochem. Soc. Trans. 2007, 35, 284–287. [Google Scholar] [CrossRef] [PubMed]
- van Horssen, J.; Drexhage, J.A.R.; Flor, T.; Gerritsen, W.; van der Valk, P.; de Vries, H.E. Nrf2 and DJ1 are consistently upregulated in inflammatory multiple sclerosis lesions. Free Radic. Biol. Med. 2010, 49, 1283–1289. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2017, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef]
- Hashimoto, K. Essential role of Keap1-Nrf2 signaling in mood disorders: Overview and future perspective. Front. Pharmacol. 2018, 9, 1182. [Google Scholar] [CrossRef]
- Luo, J.F.; Shen, X.Y.; Lio, C.K.; Dai, Y.; Cheng, C.S.; Liu, J.X.; Yao, Y.D.; Yu, Y.; Xie, Y.; Luo, P.; et al. Activation of Nrf2/HO-1 Pathway by Nardochinoid C Inhibits Inflammation and Oxidative Stress in Lipopolysaccharide-Stimulated Macrophages. Front. Pharmacol. 2018, 9, 911. [Google Scholar] [CrossRef]
- Gold, R.; Kappos, L.; Arnold, D.L.; Bar-Or, A.; Giovannoni, G.; Selmaj, K.; Tornatore, C.; Sweetser, M.T.; Yang, M.; Sheikh, S.I.; et al. Placebo-Controlled Phase 3 Study of Oral BG-12 for Relapsing Multiple Sclerosis. N. Engl. J. Med. 2012, 367, 1098–1107. [Google Scholar] [CrossRef]
- Deeks, E.D. Nivolumab: A Review of Its Use in Patients with Malignant Melanoma. Drugs 2014, 74, 1233–1239. [Google Scholar] [CrossRef]
- Martín-de-Saavedra, M.D.; Budni, J.; Cunha, M.P.; Gómez-Rangel, V.; Lorrio, S.; Del Barrio, L.; Lastres-Becker, I.; Parada, E.; Tordera, R.M.; Rodrigues, A.L.S.; et al. Nrf2 participates in depressive disorders through an anti-inflammatory mechanism. Psychoneuroendocrinology 2013, 38, 2010–2022. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.L.; Fernández-Alonso, J. Acute tubulointerstitial nephritis associated with the selective COX-2 enzyme inhibitor, rofecoxib. Lancet 2001, 357, 1946–1947. [Google Scholar] [CrossRef]
- Swanson, D.R. Medical literature as a potential source of new knowledge. BMLA 1990, 78, 29. [Google Scholar] [PubMed]
- Swanson, D.R. Fish Oil, Raynaud’s Syndrome, and Undiscovered Public Knowledge. Perspect. Biol. Med. 2015, 30, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Tari, L.B.; Patel, J.H. Systematic Drug Repurposing Through Text Mining. Methods Mol. Biol. (Clifton, N.J.) 2014, 1159, 253–267. [Google Scholar]
- Fu, C.; Jin, G.; Gao, J.; Zhu, R.; Ballesteros-Villagrana, E.; Wong, S.T.C. DrugMap Central: An on-line query and visualization tool to facilitate drug repositioning studies. Bioinformatics 2013, 29, 1834–1836. [Google Scholar] [CrossRef]
- Mihalcea, R.; Corley, C.; Strapparava, C. Corpus-based and Knowledge-based Measures of Text Semantic Similarity. In Proceedings of the 21st national conference on Artificial intelligence, Boston, MA, USA, 16–20 July 2006; Volume 6, pp. 775–780. [Google Scholar]
- Kusner, M.J.; Sun, Y.; Kolkin, N.I.; Weinberger, K.Q. From Word Embeddings to Document Distances. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Zelikman, E. Context is Everything: Finding Meaning Statistically in Semantic Spaces. arXiv 2018, arXiv:1803.08493. [Google Scholar]
- Cer, D.; Yang, Y.; Kong, S.Y.; Hua, N.; Limtiaco, N.; John, R.S.; Constant, N.; Guajardo-Cespedes, M.; Yuan, S.; Tar, C.; et al. Universal Sentence Encoder. arXiv 2018, arXiv:1803.11175. [Google Scholar]
- Villar, S.; Bandeira, A.S.; Blumberg, A.J.; Ward, R. A Polynomial-time relaxation of the gromov-hausdorff distance. arXiv 2016, arXiv:1610.05214. [Google Scholar]
- Zhang, J.C.; Yao, W.; Dong, C.; Yang, C.; Ren, Q.; Ma, M.; Han, M.; Wu, J.; Ushida, Y.; Suganuma, H.; et al. Prophylactic effects of sulforaphane on depression-like behavior and dendritic changes in mice after inflammation. J. Nutr. Biochem. 2017, 39, 134–144. [Google Scholar] [CrossRef]
- Tofthagen, C.; Donovan, K.A.; Morgan, M.A.; Shibata, D.; Yeh, Y. Oxaliplatin-induced peripheral neuropathy’s effects on health-related quality of life of colorectal cancer survivors. Support. Care Cancer 2013, 21, 3307–3313. [Google Scholar] [CrossRef] [PubMed]
- Balboa, M.A.; Balsinde, J. Oxidative stress and arachidonic acid mobilization. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2006, 1761, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Hanna, V.S.; Hafez, E.A.A. Synopsis of arachidonic acid metabolism: A review. J. Adv. Res. 2018, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Harizi, H.; Corcuff, J.B.; Gualde, N. Arachidonic-acid-derived eicosanoids: Roles in biology and immunopathology. Trends Mol. Med. 2008, 14, 461–469. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Cyclooxygenases: Structural and functional insights. J. Lipid Res. 2009, 50, S29–S34. [Google Scholar] [CrossRef]
- Chu, J.; Praticò, D. The 5-Lipoxygenase as a Common Pathway for Pathological Brain and Vascular Aging. Cardiovasc. Psychiatry Neurol. 2009. [Google Scholar] [CrossRef]
- Hedi, H.; Norbert, G. 5-Lipoxygenase Pathway, Dendritic Cells, and Adaptive Immunity. J. Biomed. Biotechnol. 2004, 2004, 99. [Google Scholar] [CrossRef]
- Chensue, S.W.; Kunkel, S.L. Arachidonic acid metabolism and macrophage activation. Clin. Lab. Med. 1983, 3, 677–694. [Google Scholar] [CrossRef]
- di Meco, A.; Lauretti, E.; Vagnozzi, A.N.; Praticò, D. Zileuton restores memory impairments and reverses amyloid and tau pathology in aged Alzheimer’s disease mice. Neurobiol. Aging 2014, 35, 2458–2464. [Google Scholar] [CrossRef]
- Wang, L.; Du, C.; Lv, J.; Wei, W.; Cui, Y.; Xie, X. Antiasthmatic drugs targeting the cysteinyl leukotriene receptor 1 alleviate central nervous system inflammatory cell infiltration and pathogenesis of experimental autoimmune encephalomyelitis. J. Immunol. 2011, 187, 2336–2345. [Google Scholar] [CrossRef]
- Rossi, A.; Pergola, C.; Koeberle, A.; Hoffmann, M.; Dehm, F.; Bramanti, P.; Cuzzocrea, S.; Werz, O.; Sautebin, L. The 5-lipoxygenase inhibitor, zileuton, suppresses prostaglandin biosynthesis by inhibition of arachidonic acid release in macrophages. Br. J. Pharmacol. 2010, 161, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Piantadosi, C.A.; Withers, C.M.; Bartz, R.R.; MacGarvey, N.C.; Fu, P.; Sweeney, T.E.; Welty-Wolf, K.E.; Suliman, H.B. Heme Oxygenase-1 Couples Activation of Mitochondrial Biogenesis to Anti-inflammatory Cytokine Expression. J. Biol. Chem. 2011, 286, 16374–16385. [Google Scholar] [CrossRef] [PubMed]
- Roman, A.; Kreiner, G.; Nalepa, I. Macrophages and depression—A misalliance or well-arranged marriage? Pharmacol. Rep. 2013, 65, 1663–1672. [Google Scholar] [CrossRef]
- Conneau, A.; Kiela, D.; Schwenk, H.; Barrault, L.; Bordes, A. Supervised Learning of Universal Sentence Representations from Natural Language Inference Data. arXiv 2017, arXiv:1705.02364. [Google Scholar]
- Mickael, M.E.; Rajput, A.; Steyn, J.; Wiemerslage, L.; Bürglin, T. An optimised phylogenetic method sheds more light on the main branching events of rhodopsin-like superfamily. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 85–94. [Google Scholar] [CrossRef]
- Reichenberg, A.; Yirmiya, R.; Schuld, A.; Kraus, T.; Haack, M.; Morag, A.; Pollmächer, T. Cytokine-associated emotional and cognitive disturbances in humans. Arch. Gen. Psychiatry 2001, 58, 445–452. [Google Scholar] [CrossRef]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage polarization: Different gene signatures in M1(Lps+) vs. Classically and M2(LPS-) vs. Alternatively activated macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Yirmiya, R.; Rimmerman, N.; Reshef, R. Depression as a Microglial Disease. Trends Neurosci. 2015, 38, 637–658. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data BT—Bioinformatics and Computational Biology Solutions Using R and Bioconductor. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005. [Google Scholar]
- Shiel, B.P.; Hall, N.E.; Cooke, I.R.; Robinson, N.A.; Stone, D.A.J.; Strugnell, J.M. The effect of commercial, natural and grape seed extract supplemented diets on gene expression signatures and survival of greenlip abalone (Haliotis laevigata) during heat stress. Aquaculture 2017, 479, 798–807. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Zhang, M.; Li, Q.; Yu, D.; Yao, B.; Guo, W.; Xie, Y.; Xiao, G. GeNeCK: A web server for gene network construction and visualization. BMC Bioinform. 2019, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, F.; Berquand, A.; Kumar, K.; Riccardi, A.; Soares, T.; Gerené, S.; Brauer, N. Knowledge-based information extraction from datasheets of space parts. In Proceedings of the 8th International Systems & Concurrent Engineering for Space Applications Conference, Glasgow, Scotland, 26–28 September 2018. [Google Scholar]
- Kumar, N.; Dangeti, P.; Bhavsar, K. Natural Language Processing with Python (CookBook); Packt: Birmingham, UK, 2017. [Google Scholar]
- Natural Language Toolkit—NLTK 3.3 Documentation. Available online: https://www.nltk.org/ (accessed on 26 October 2018).
- Zakas, N.C. JSON. In Professional Javascript® for Web Developers; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Friesen, J. Introducing JSON. In Java XML and JSON; Apress: New York, NY, USA, 2019. [Google Scholar]
- Iyyer, M.; Manjunatha, V.; Boyd-Graber, J.; Daumé, H., III. Deep Unordered Composition Rivals Syntactic Methods for Text Classification. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers); Association for Computational Linguistics: Beijing, China, 2015; pp. 1681–1691. [Google Scholar]
- Wang, Y.; Yin, G.; Cai, Z.; Dong, Y.; Dong, H. A trust-based probabilistic recommendation model for social networks. J. Netw. Comput. Appl. 2015, 55, 59–67. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Advances in Neural Information Processing Systems; MIT Press: Vancouver, BC Canada, 2007. [Google Scholar]
- Faizi, M.; Jahani, R.; Ebadi, S.A.; Tabatabai, S.A.; Rezaee, E.; Lotfaliei, M.; Amini, M.; Almasirad, A. Novel 4-thiazolidinone derivatives as agonists of benzodiazepine receptors: Design, synthesis and pharmacological evaluation. EXCLI J. 2017, 16, 52. [Google Scholar] [PubMed]
- Mickael, M.-E.; Pajares, M.; Enache, I.; Manda, G.; Cuadrado, A. NRF2 drug repurposing using a question-answer artificial intelligence system. BioRxiv 2009. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compound Zileuton are available from the authors. |




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubick, N.; Pajares, M.; Enache, I.; Manda, G.; Mickael, M.-E. Repurposing Zileuton as a Depression Drug Using an AI and In Vitro Approach. Molecules 2020, 25, 2155. https://doi.org/10.3390/molecules25092155
Kubick N, Pajares M, Enache I, Manda G, Mickael M-E. Repurposing Zileuton as a Depression Drug Using an AI and In Vitro Approach. Molecules. 2020; 25(9):2155. https://doi.org/10.3390/molecules25092155
Chicago/Turabian StyleKubick, Norwin, Marta Pajares, Ioana Enache, Gina Manda, and Michel-Edwar Mickael. 2020. "Repurposing Zileuton as a Depression Drug Using an AI and In Vitro Approach" Molecules 25, no. 9: 2155. https://doi.org/10.3390/molecules25092155
APA StyleKubick, N., Pajares, M., Enache, I., Manda, G., & Mickael, M.-E. (2020). Repurposing Zileuton as a Depression Drug Using an AI and In Vitro Approach. Molecules, 25(9), 2155. https://doi.org/10.3390/molecules25092155






