Nanoparticle-Mediated Gene Silencing for Sensitization of Lung Cancer to Cisplatin Therapy
Abstract
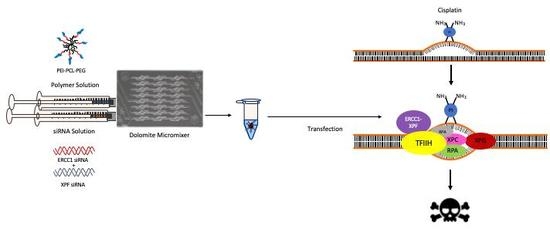
1. Introduction
2. Results
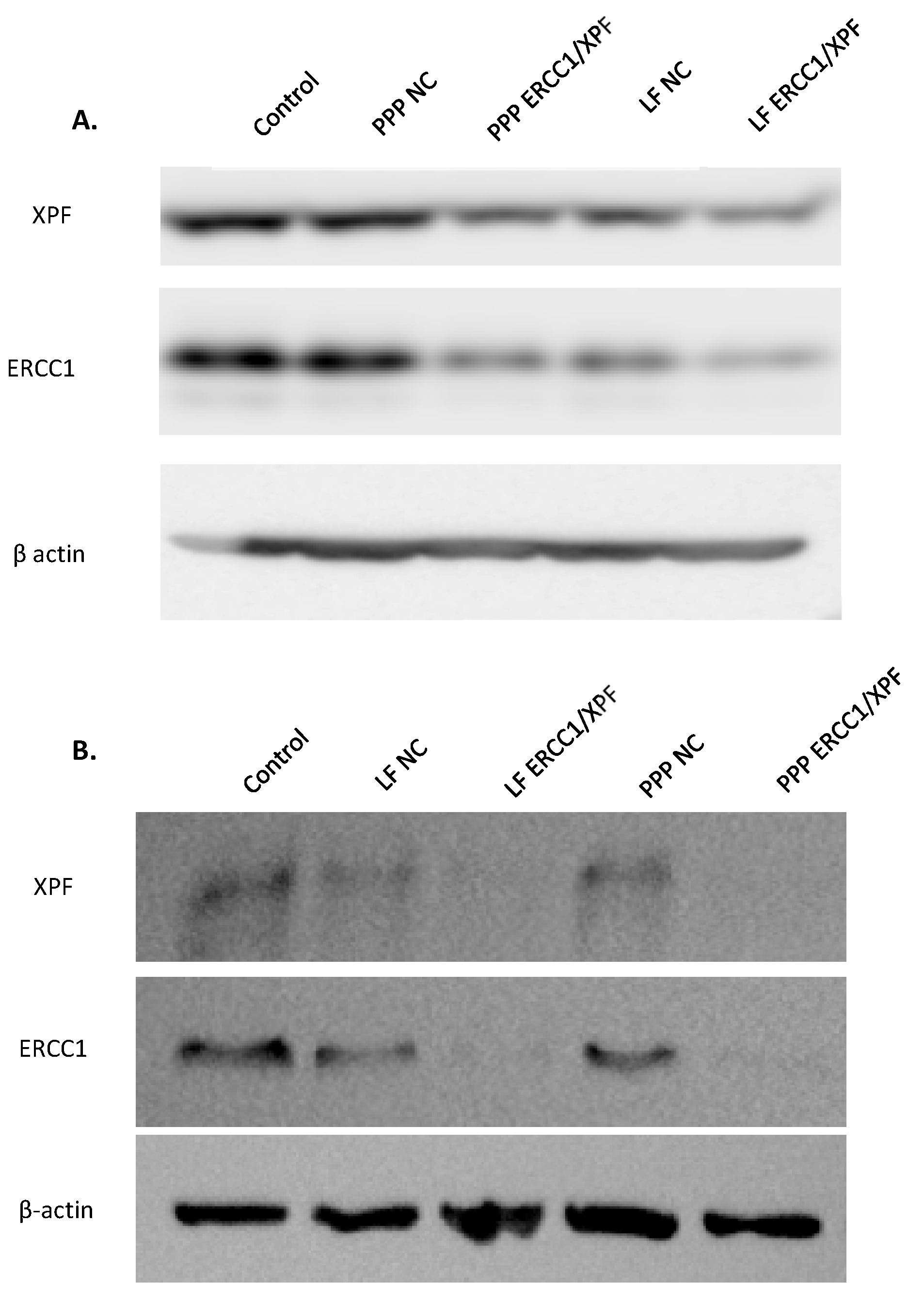
2.1. In Vitro ERCC1-XPF Protein Knockdown
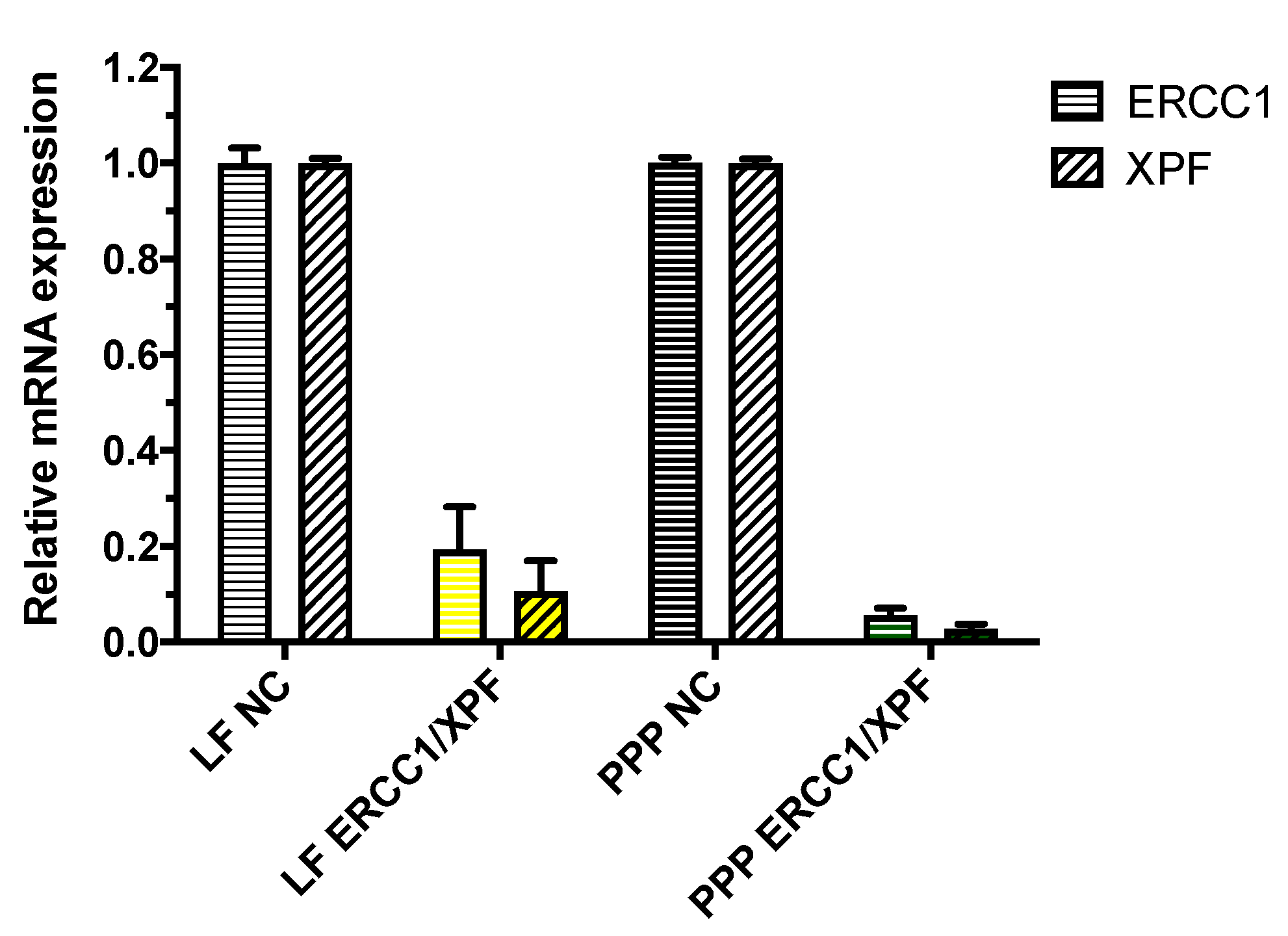
2.2. In Vitro ERCC1-XPF Gene Knockdown
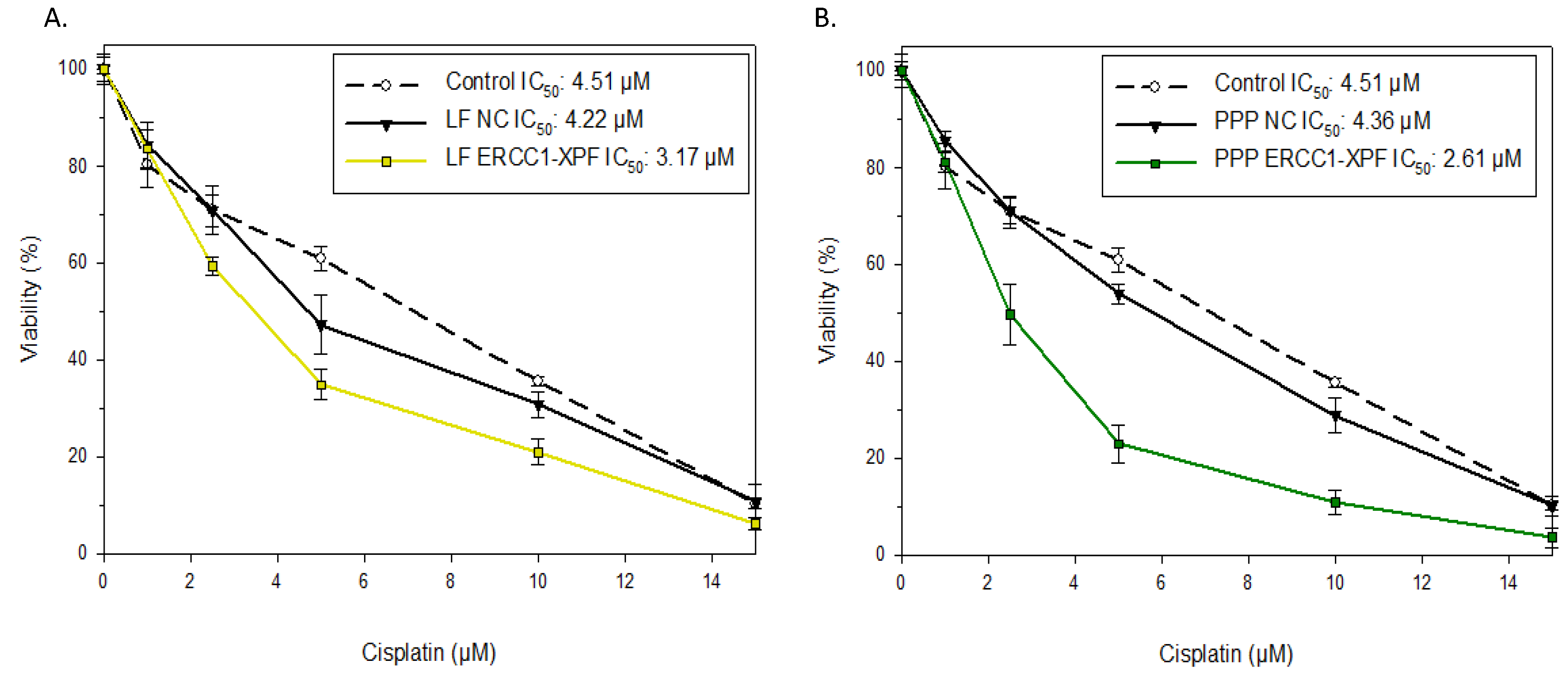
2.3. Colony Survival Assay
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of Triblock Copolymer and Characterization
4.3. Microfluidic Preparation of PEI-PCL-PEG Micelleplexes
4.4. Cell Culture
4.5. In Vitro ERCC1-XPF Protein Knockdown and Western Blot Analysis
4.6. In Vitro ERCC1-XPF Gene Knockdown
4.7. Colony Survival Assay
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cancer Stat Facts: Lung and Bronchus Cancer. Available online: https://seer.cancer.gov/statfacts/html/lungb.html (accessed on 10 February 2020).
- Kumar, V.; Abbas, A.K.; Fausto, N.; Mitchell, R.N. Robbins Basic Pathology; Elsevier Health Sciences: Philadelphia, PA, USA, 2012. [Google Scholar]
- Gandhi, L.; Rodriguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef] [PubMed]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Kothandapani, A.; Tillison, K.; Kalman-Maltese, V.; Patrick, S.M. Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair 2010, 9, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Altaha, R.; Liang, X.; Yu, J.J.; Reed, E. Excision repair cross complementing-group 1: Gene expression and platinum resistance. Int. J. Mol. Med. 2004, 14, 959–970. [Google Scholar]
- Lord, R.V.; Brabender, J.; Gandara, D.; Alberola, V.; Camps, C.; Domine, M.; Cardenal, F.; Sanchez, J.M.; Gumerlock, P.H.; Taron, M.; et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2002, 8, 2286–2291. [Google Scholar]
- Olaussen, K.A.; Dunant, A.; Fouret, P.; Brambilla, E.; Andre, F.; Haddad, V.; Taranchon, E.; Filipits, M.; Pirker, R.; Popper, H.H.; et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006, 355, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Friboulet, L.; Barrios-Gonzales, D.; Commo, F.; Olaussen, K.A.; Vagner, S.; Adam, J.; Goubar, A.; Dorvault, N.; Lazar, V.; Job, B.; et al. Molecular Characteristics of ERCC1-Negative versus ERCC1-Positive Tumors in Resected NSCLC. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 5562–5572. [Google Scholar] [CrossRef] [PubMed]
- Dabholkar, M.; Bostick-Bruton, F.; Weber, C.; Bohr, V.A.; Egwuagu, C.; Reed, E. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J. Natl. Cancer Inst. 1992, 84, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Dabholkar, M.; Vionnet, J.; Bostick-Bruton, F.; Yu, J.J.; Reed, E. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J. Clin. Investig. 1994, 94, 703. [Google Scholar] [CrossRef]
- Joshi, M.B.; Shirota, Y.; Danenberg, K.D.; Conlon, D.H.; Salonga, D.S.; Herndon, J.E., 2nd; Danenberg, P.V.; Harpole, D.H., Jr. High gene expression of TS1, GSTP1, and ERCC1 are risk factors for survival in patients treated with trimodality therapy for esophageal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 2215–2221. [Google Scholar] [CrossRef]
- Langer, R.; Specht, K.; Becker, K.; Ewald, P.; Bekesch, M.; Sarbia, M.; Busch, R.; Feith, M.; Stein, H.J.; Siewert, J.R.; et al. Association of pretherapeutic expression of chemotherapy-related genes with response to neoadjuvant chemotherapy in Barrett carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 7462–7469. [Google Scholar] [CrossRef] [PubMed]
- Reed, E.; Dabholkar, M.; Thornton, K.; Thompson, C.; Yu, J.J.; Bostick-Bruton, F. Evidence for in the appearance of mRNAs of nucleotide excision repair genes, in human ovarian cancer tissues. Oncol. Rep. 2000, 7, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Shirota, Y.; Stoehlmacher, J.; Brabender, J.; Xiong, Y.P.; Uetake, H.; Danenberg, K.D.; Groshen, S.; Tsao-Wei, D.D.; Danenberg, P.V.; Lenz, H.J. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J. Clin. Oncol. 2001, 19, 4298–4304. [Google Scholar] [CrossRef] [PubMed]
- Heyza, J.R.; Lei, W.; Watza, D.; Zhang, H.; Chen, W.; Back, J.B.; Schwartz, A.G.; Bepler, G.; Patrick, S.M. Identification and Characterization of Synthetic Viability with ERCC1 Deficiency in Response to Interstrand Crosslinks in Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 2523–2536. [Google Scholar] [CrossRef]
- Heyza, J.R.; Arora, S.; Zhang, H.; Conner, K.L.; Lei, W.; Floyd, A.M.; Deshmukh, R.R.; Sarver, J.; Trabbic, C.J.; Erhardt, P.; et al. Targeting the DNA Repair Endonuclease ERCC1-XPF with Green Tea Polyphenol Epigallocatechin-3-Gallate (EGCG) and Its Prodrug to Enhance Cisplatin Efficacy in Human Cancer Cells. Nutrients 2018, 10, 1–17. [Google Scholar] [CrossRef]
- Arora, S.; Heyza, J.; Zhang, H.; Kalman-Maltese, V.; Tillison, K.; Floyd, A.M.; Chalfin, E.M.; Bepler, G.; Patrick, S.M. Identification of small molecule inhibitors of ERCC1-XPF that inhibit DNA repair and potentiate cisplatin efficacy in cancer cells. Oncotarget 2016, 7, 75104–75117. [Google Scholar] [CrossRef]
- Chapman, T.M.; Wallace, C.; Gillen, K.J.; Bakrania, P.; Khurana, P.; Coombs, P.J.; Fox, S.; Bureau, E.A.; Brownlees, J.; Melton, D.W.; et al. N-Hydroxyimides and hydroxypyrimidinones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg. Med. Chem. Lett. 2015, 25, 4104–4108. [Google Scholar] [CrossRef]
- Chapman, T.M.; Gillen, K.J.; Wallace, C.; Lee, M.T.; Bakrania, P.; Khurana, P.; Coombs, P.J.; Stennett, L.; Fox, S.; Bureau, E.A.; et al. Catechols and 3-hydroxypyridones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg. Med. Chem. Lett. 2015, 25, 4097–4103. [Google Scholar] [CrossRef]
- McNeil, E.M.; Astell, K.R.; Ritchie, A.M.; Shave, S.; Houston, D.R.; Bakrania, P.; Jones, H.M.; Khurana, P.; Wallace, C.; Chapman, T.; et al. Inhibition of the ERCC1-XPF structure-specific endonuclease to overcome cancer chemoresistance. DNA Repair 2015, 31, 19–28. [Google Scholar] [CrossRef]
- Gentile, F.; Elmenoufy, A.H.; Ciniero, G.; Jay, D.; Karimi-Busheri, F.; Barakat, K.H.; Weinfeld, M.; West, F.G.; Tuszynski, J.A. Computer-Aided Drug Design of Small Molecule Inhibitors of the ERCC1-XPF Protein-Protein Interaction. Chem. Biol. Drug Des. 2019, 95, 460–471. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Rettig, G.R.; Behlke, M.A. Progress toward in vivo use of siRNAs-II. Mol. Ther. J. Am. Soc. Gene Ther. 2012, 20, 483–512. [Google Scholar] [CrossRef] [PubMed]
- Titze-de-Almeida, S.S.; Brandao, P.R.P.; Faber, I.; Titze-de-Almeida, R. Leading RNA Interference Therapeutics Part 1: Silencing Hereditary Transthyretin Amyloidosis, with a Focus on Patisiran. Mol. Diagn. Ther. 2019, 24, 49–59. [Google Scholar] [CrossRef] [PubMed]
- De Paula Brandão, P.; Titze-de-Almeida, S.; Titze-de-Almeida, R. Leading RNA Interference Therapeutics Part 2: Silencing Delta-Aminolevulinic Acid Synthase 1, with a Focus on Givosiran. Mol. Diagn. Ther. 2020, 24, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef]
- Bono, N.; Ponti, F.; Mantovani, D.; Candiani, G. Non-Viral in Vitro Gene Delivery: It is Now Time to Set the Bar! Pharmaceutics 2020, 12. [Google Scholar] [CrossRef]
- Caillaud, M.; El Madani, M.; Massaad-Massade, L. Small interfering RNA from the lab discovery to patients’ recovery. J. Control. Release Off. J. Control. Release Soc. 2020, 321, 616–628. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release Off. J. Control. Release Soc. 2006, 114, 100–109. [Google Scholar] [CrossRef]
- Park, T.G.; Jeong, J.H.; Kim, S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006, 58, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Samsonova, O.; Sproat, B.; Merkel, O.; Kissel, T. Biophysical characterization of hyper-branched polyethylenimine-graft-polycaprolactone-block-mono-methoxyl-poly(ethylene glycol) copolymers (hy-PEI-PCL-mPEG) for siRNA delivery. J. Control. Release Off. J. Control. Release Soc. 2011, 153, 262–268. [Google Scholar] [CrossRef]
- Essex, S.; Navarro, G.; Sabhachandani, P.; Chordia, A.; Trivedi, M.; Movassaghian, S.; Torchilin, V.P. Phospholipid-modified PEI-based nanocarriers for in vivo siRNA therapeutics against multidrug-resistant tumors. Gene 2015, 22, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Merkel, O.M.; Beyerle, A.; Librizzi, D.; Pfestroff, A.; Behr, T.M.; Sproat, B.; Barth, P.J.; Kissel, T. Nonviral siRNA delivery to the lung: Investigation of PEG-PEI polyplexes and their in vivo performance. Mol. Pharm. 2009, 6, 1246–1260. [Google Scholar] [CrossRef]
- Trinh, P.; Atzet, S.; Curtain, S.; Ratner, B.D. Characterization of poly (2-hydroxyethyl methacrylate) scaffolds for tissue engineering. J. Undergrad. Res. Bioeng. 2008, 2010, 98–104. [Google Scholar]
- Feldmann, D.P.; Xie, Y.; Jones, S.K.; Yu, D.; Moszczynska, A.; Merkel, O.M. The impact of microfluidic mixing of triblock micelleplexes on in vitro/in vivo gene silencing and intracellular trafficking. Nanotechnology 2017, 28, 224001. [Google Scholar] [CrossRef]
- Jones, S.K.; Lizzio, V.; Merkel, O.M. Folate Receptor Targeted Delivery of siRNA and Paclitaxel to Ovarian Cancer Cells via Folate Conjugated Triblock Copolymer to Overcome TLR4 Driven Chemotherapy Resistance. Biomacromolecules 2016, 17, 76–87. [Google Scholar] [CrossRef]
- Zheng, M.; Librizzi, D.; Kilic, A.; Liu, Y.; Renz, H.; Merkel, O.M.; Kissel, T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials 2012, 33, 6551–6558. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, M.; Librizzi, D.; Renette, T.; Merkel, O.M.; Kissel, T. Efficient and Tumor Targeted siRNA Delivery by Polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol)-folate (PEI-PCL-PEG-Fol). Mol. Pharm. 2016, 13, 134–143. [Google Scholar] [CrossRef]
- Abstiens, K.; Goepferich, A.M. Microfluidic manufacturing improves polydispersity of multicomponent polymeric nanoparticles. J. Drug Deliv. Sci. Tec. 2019, 49, 433–439. [Google Scholar] [CrossRef]
- Loy, D.M.; Klein, P.M.; Krzysztoń, R.; Lächelt, U.; Rädler, J.O.; Wagner, E. A microfluidic approach for sequential assembly of siRNA polyplexes with a defined structure-activity relationship. PeerJ Mater. Sci. 2019, 1, 1–37. [Google Scholar] [CrossRef]
- Abraham, D.; Stroock, S.K.W.D.; Armand, A.; Igor, M.; . Stone, H.A.; Whitesides, G.M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. [Google Scholar]
- Li, Y.; Huang, X.; Lee, R.J.; Qi, Y.; Wang, K.; Hao, F.; Zhang, Y.; Lu, J.; Meng, Q.; Li, S.; et al. Synthesis of Polymer-Lipid Nanoparticles by Microfluidic Focusing for siRNA Delivery. Molecules 2016, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feldmann, D.P.; Heyza, J.; Zimmermann, C.M.; Patrick, S.M.; Merkel, O.M. Nanoparticle-Mediated Gene Silencing for Sensitization of Lung Cancer to Cisplatin Therapy. Molecules 2020, 25, 1994. https://doi.org/10.3390/molecules25081994
Feldmann DP, Heyza J, Zimmermann CM, Patrick SM, Merkel OM. Nanoparticle-Mediated Gene Silencing for Sensitization of Lung Cancer to Cisplatin Therapy. Molecules. 2020; 25(8):1994. https://doi.org/10.3390/molecules25081994
Chicago/Turabian StyleFeldmann, Daniel P., Joshua Heyza, Christoph M. Zimmermann, Steve M. Patrick, and Olivia M. Merkel. 2020. "Nanoparticle-Mediated Gene Silencing for Sensitization of Lung Cancer to Cisplatin Therapy" Molecules 25, no. 8: 1994. https://doi.org/10.3390/molecules25081994
APA StyleFeldmann, D. P., Heyza, J., Zimmermann, C. M., Patrick, S. M., & Merkel, O. M. (2020). Nanoparticle-Mediated Gene Silencing for Sensitization of Lung Cancer to Cisplatin Therapy. Molecules, 25(8), 1994. https://doi.org/10.3390/molecules25081994