Discrimination of Gentiana and Its Related Species Using IR Spectroscopy Combined with Feature Selection and Stacked Generalization
Abstract
1. Introduction
2. Results and Discussion
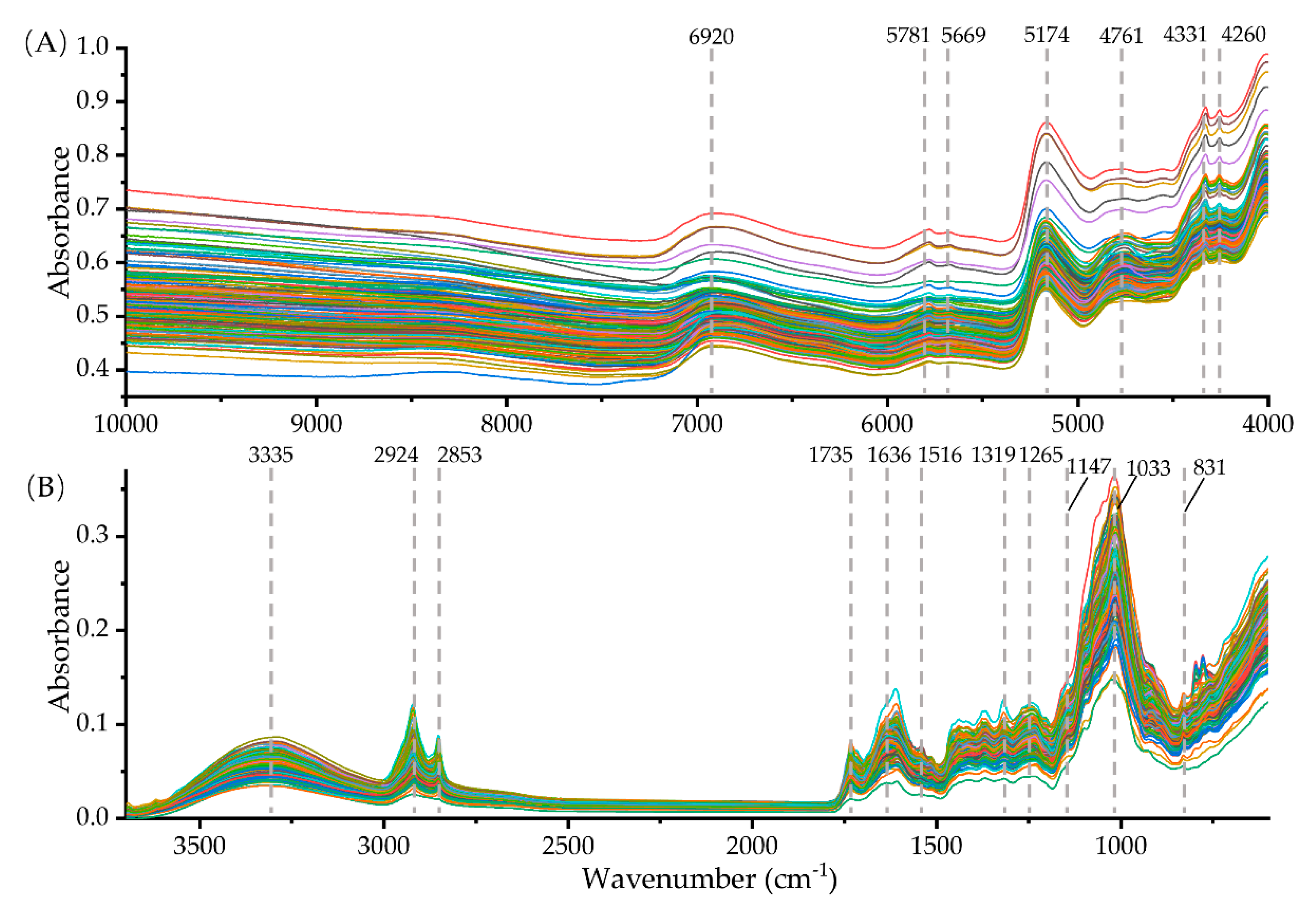
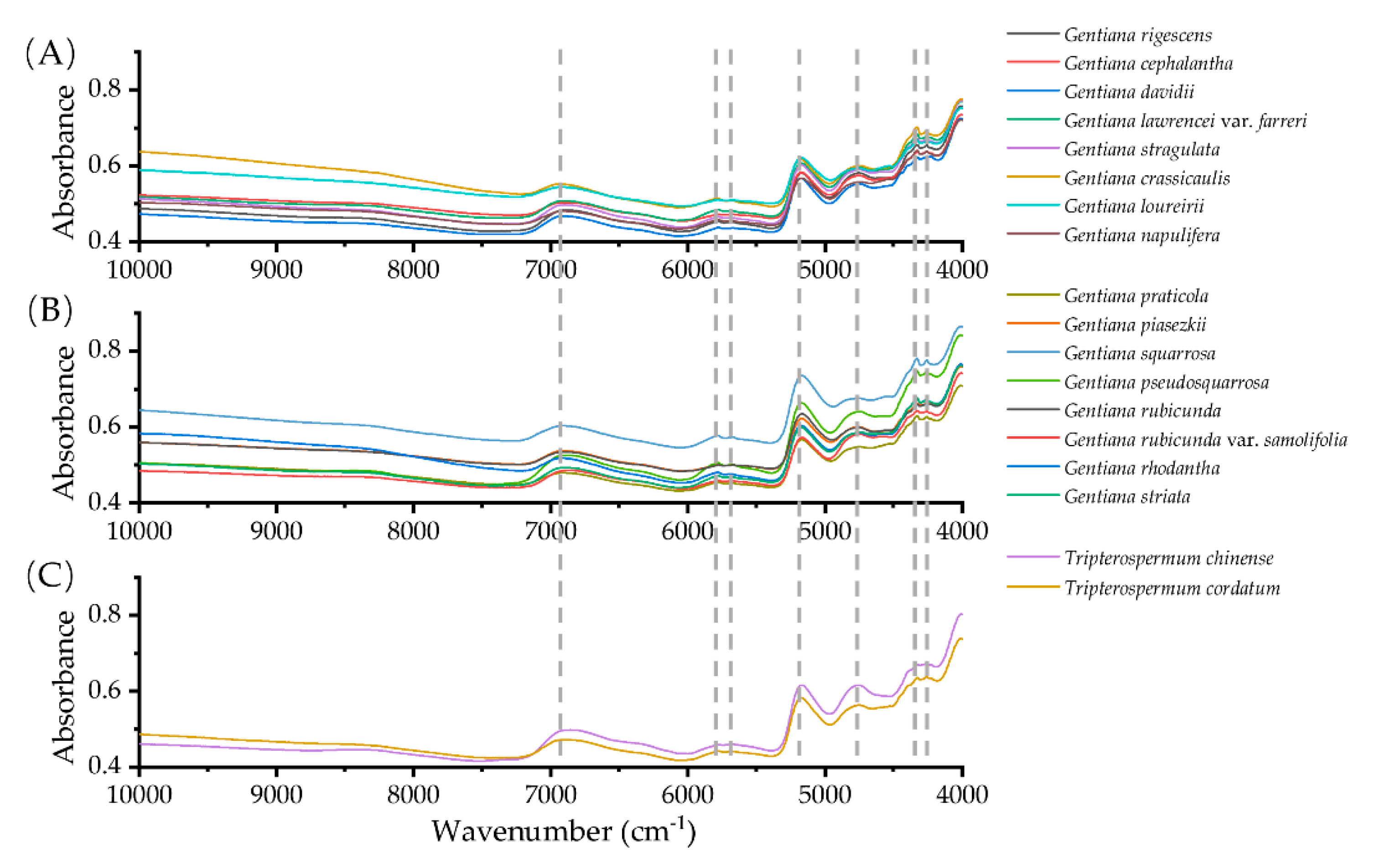
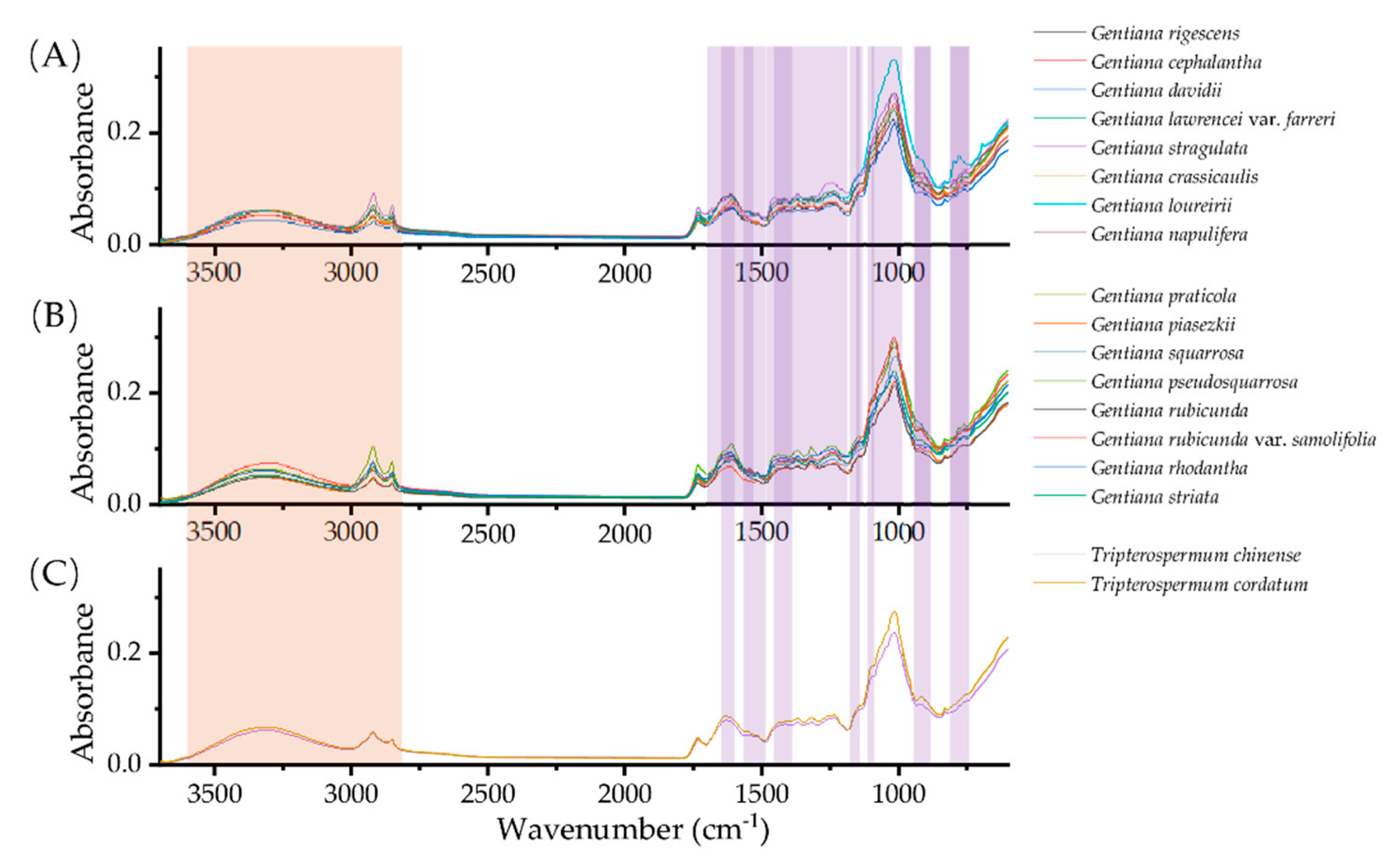
2.1. Spectral Fingerprint of NIR and FT-MIR
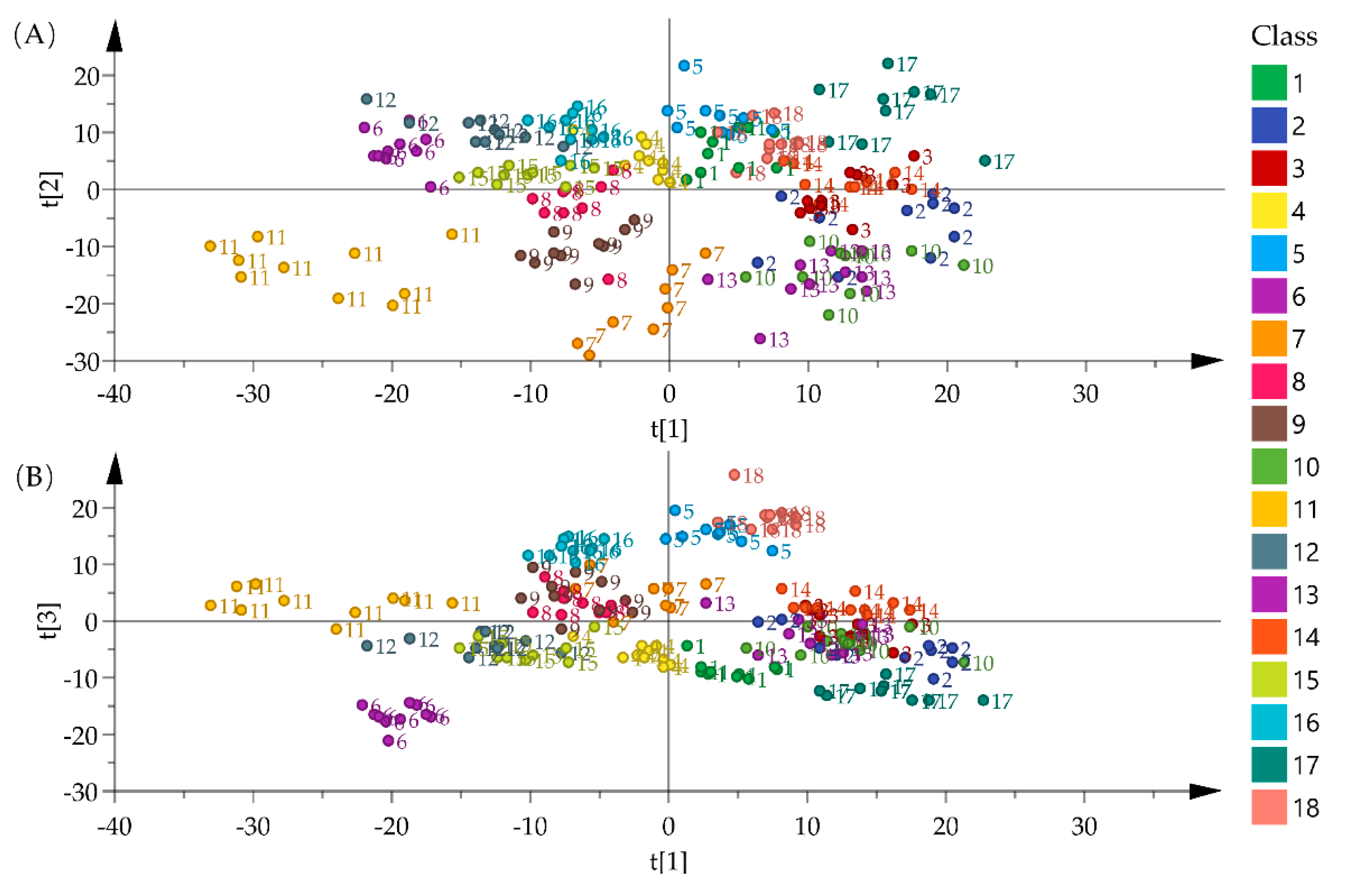
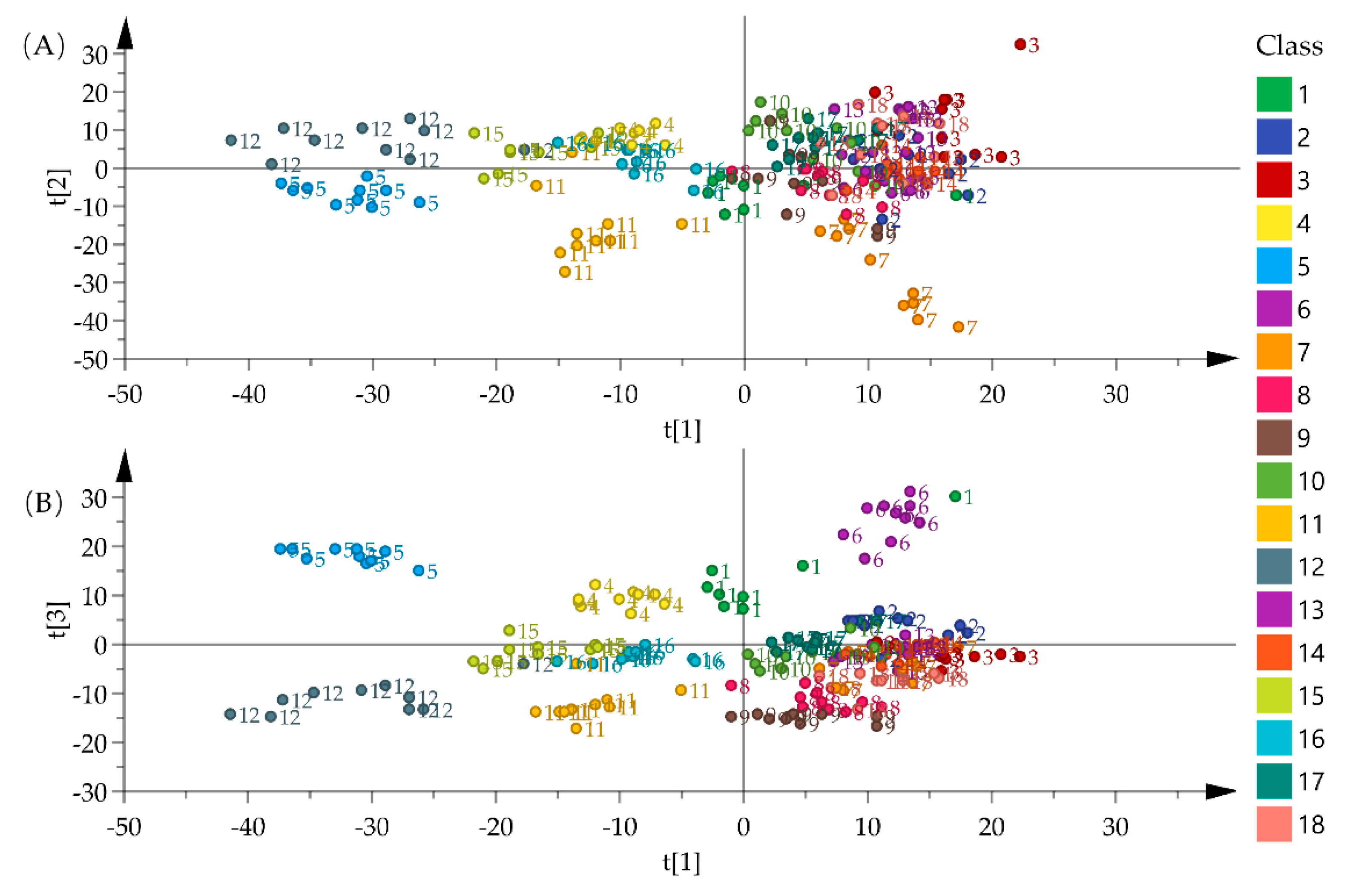
2.2. Exploratory Statistical Analysis
2.3. Single Block Models for Sample Classification
2.3.1. Classification Based on Full Spectra
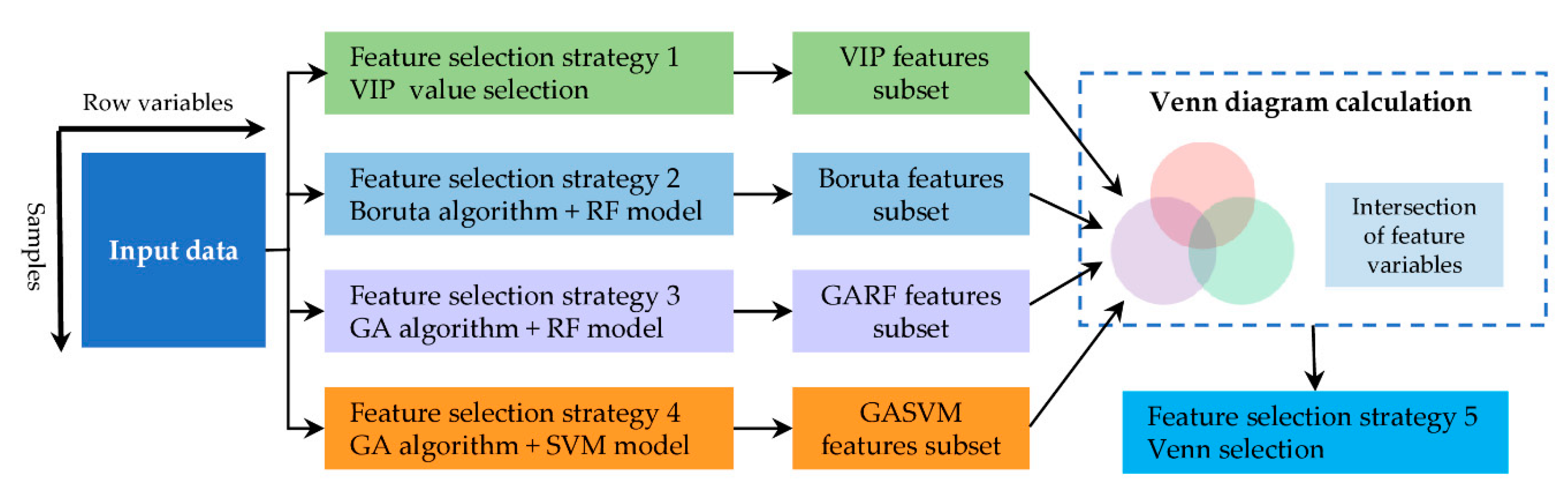
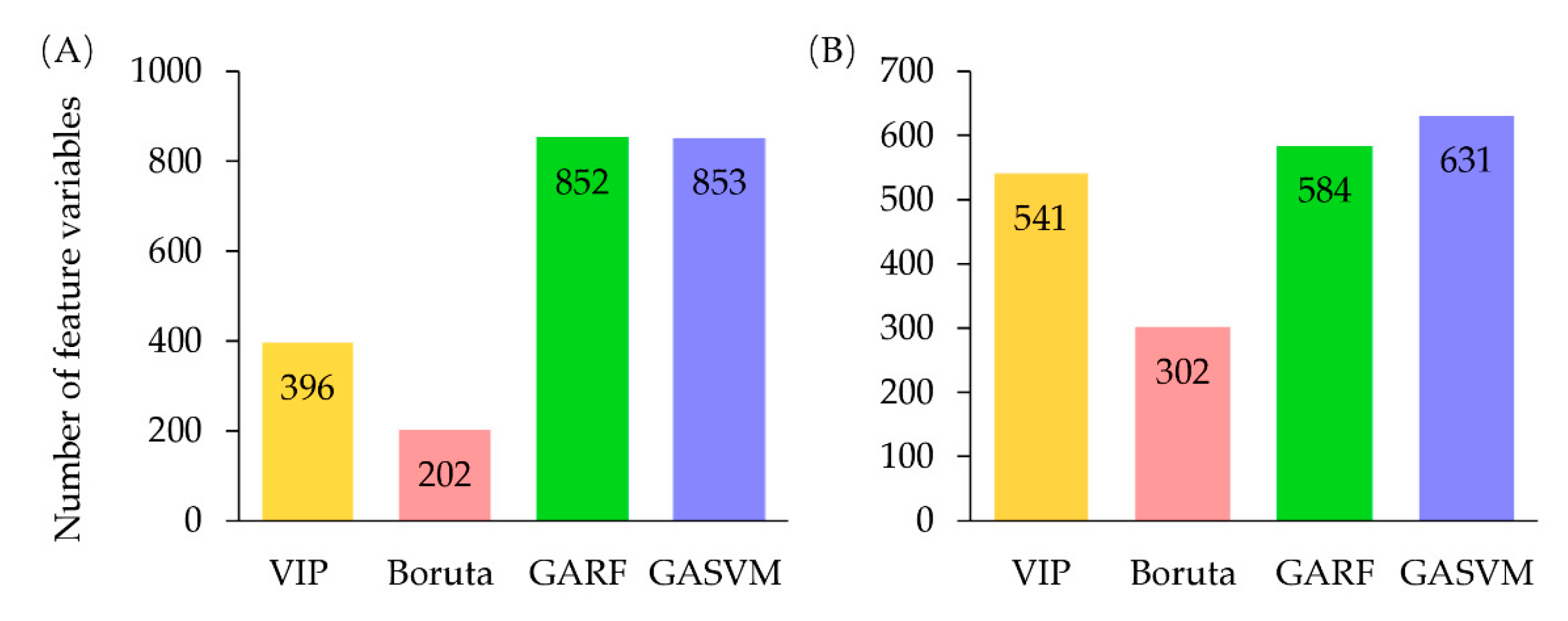
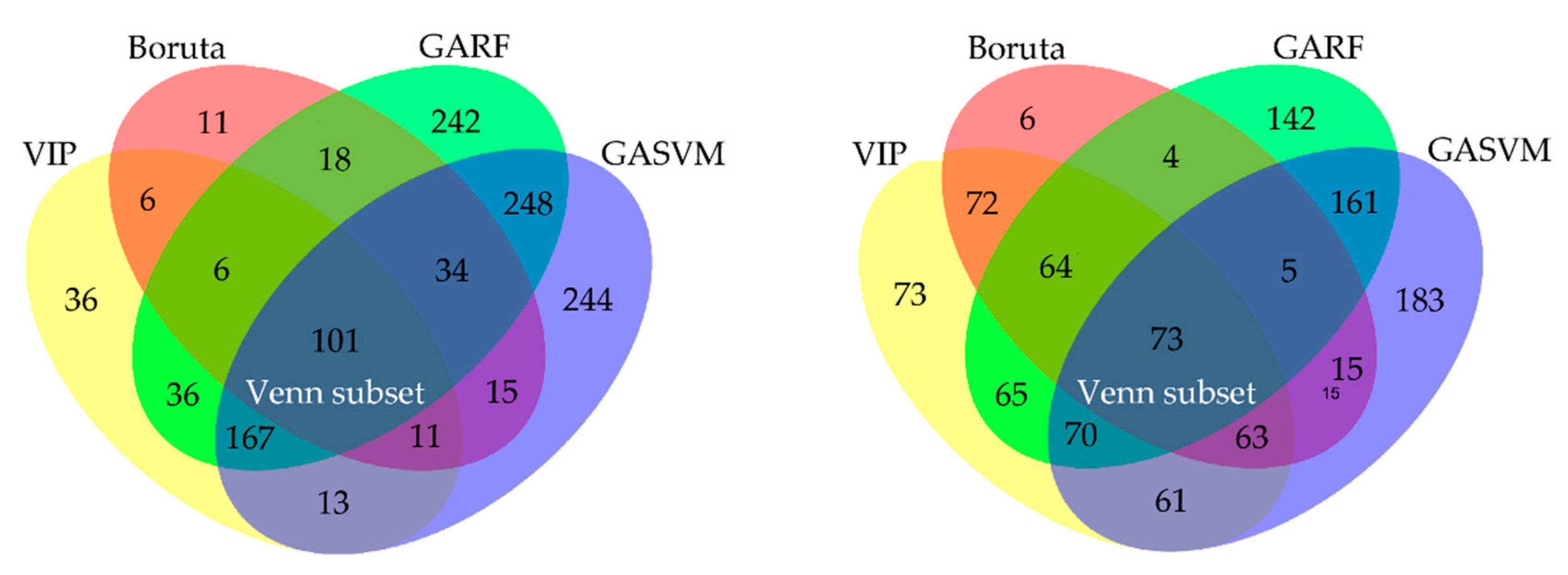
2.3.2. Feature Selection
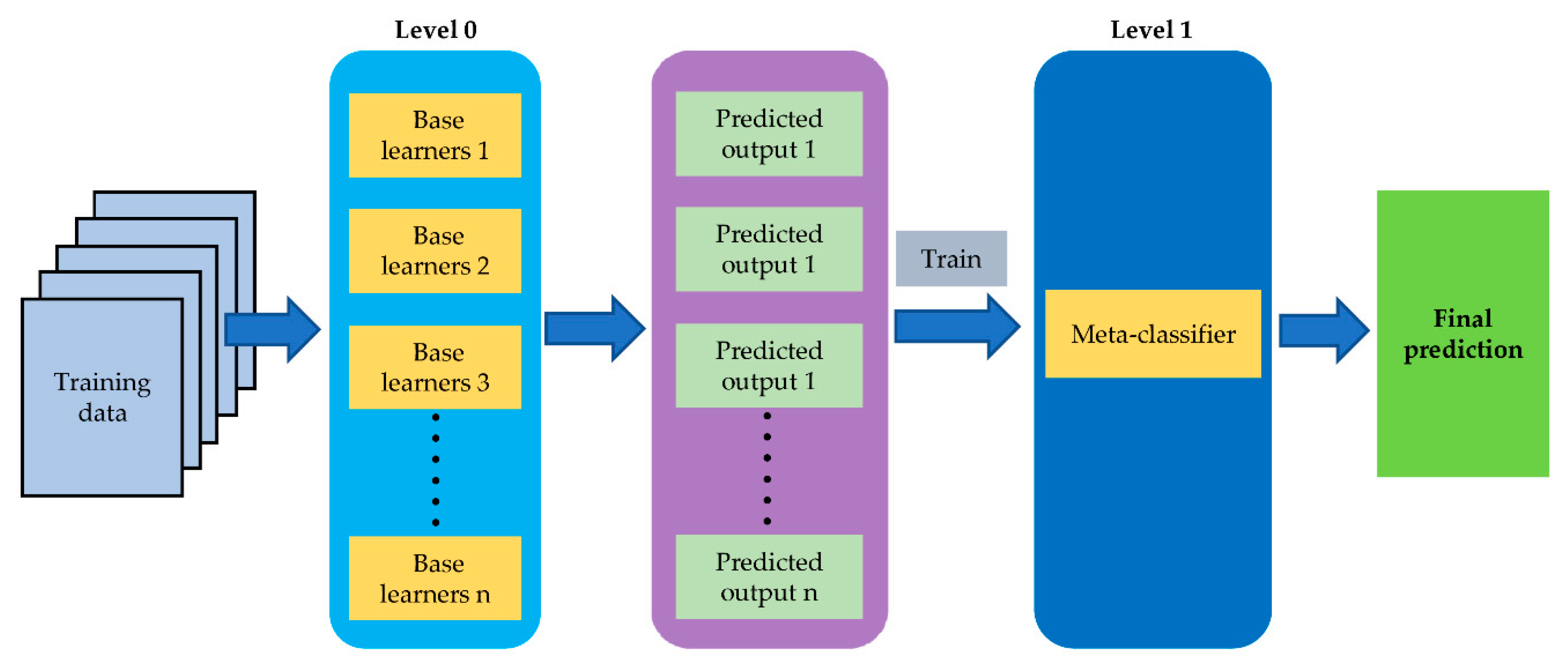
2.4. Model Stacking for Sample Classification
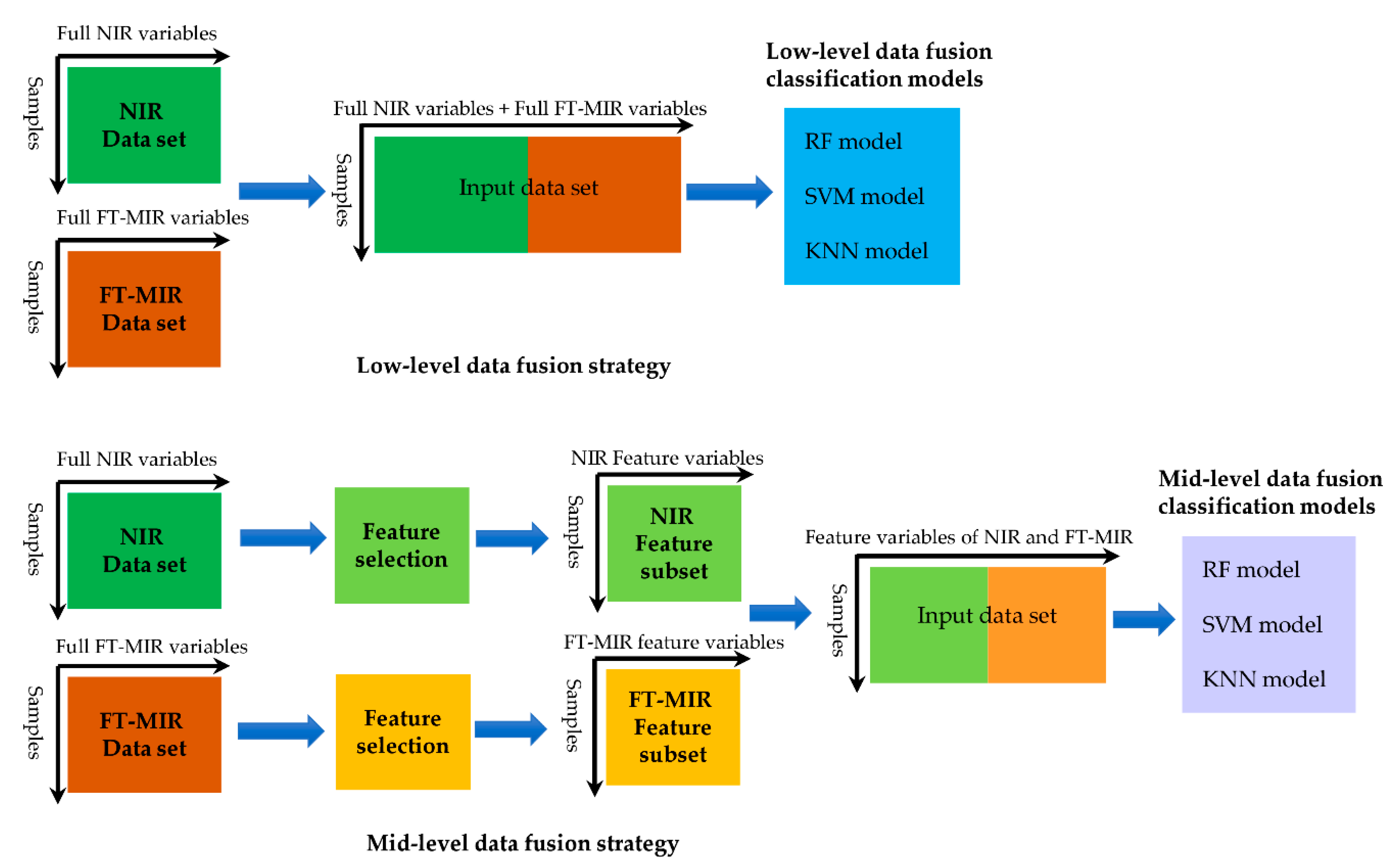
2.5. Are Model Stacking Better than Data Fusion for Gentiana Species Discrimination?
3. Materials and Methods
3.1. Plant Material Collection
3.2. Near Infrared (FT-NIR)
3.3. Fourier Transform Mid Infrared (FT-MIR)
3.4. Statistical Analysis
3.5. Model Stacking and Data Fusion
3.6. Model Evaluation
3.7. Software
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ho, T.N.; James, S.P. Flora of China (Gentianaceae through Boraginaceae); Science Press, Beijing and Missouri Botanical Garden Press: St. Louis, MO, USA, 1995; Volume 16. [Google Scholar]
- Pan, Y.; Zhao, Y.L.; Zhang, J.; Li, W.Y.; Wang, Y.Z. Phytochemistry and pharmacological activities of the genus Gentiana (Gentianaceae). Chem. Biodivers. 2016, 13, 107–150. [Google Scholar] [CrossRef]
- Mirzaee, F.; Hosseini, A.; Jouybari, H.B.; Davoodi, A.; Azadbakht, M. Medicinal, biological and phytochemical properties of Gentiana species. J. Tradit. Complement. Med. 2017, 7, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.M.; Caprioli, G.; Dikmen, M.; Kaya, E.; Maggi, F.; Sagratini, G.; Vittori, S.; Öztürk, Y. Evaluation of neuritogenic activity of cultivated, wild and commercial roots of Gentiana lutea L. J. Funct. Foods 2015, 19, 164–173. [Google Scholar] [CrossRef]
- Mustafa, A.M.; Caprioli, G.; Ricciutelli, M.; Maggi, F.; Marín, R.; Vittori, S.; Sagratini, G. Comparative HPLC/ESI-MS and HPLC/DAD study of different populations of cultivated, wild and commercial Gentiana lutea L. Food Chem. 2015, 174, 426–433. [Google Scholar] [CrossRef]
- Wang, Y.M.; Xu, M.; Wang, D.; Zhu, H.T.; Yang, C.R.; Zhang, Y.J. Review on “Long-Dan”, one of the traditional Chinese medicinal herbs recorded in Chinese pharmacopoeia. Nat. Prod. Bioprospect. 2012, 2, 1–10. [Google Scholar] [CrossRef]
- Kletter, C.; Glasl, S.; Thalhammer, T.; Narantuya, S. Traditional Mongolian medicine—A potential for drug discovery. Sci. Pharm. 2008, 76, 49–64. [Google Scholar] [CrossRef]
- Yang, B.; Kim, S.; Kim, J.; Lim, C.; Kim, H.; Cho, S. Gentiana scabra Bunge roots alleviates skin lesions of contact dermatitis in mice. J. Ethnopharmacol. 2019, 233, 141–147. [Google Scholar] [CrossRef]
- China Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China; China Medicinal Science Press: Beijing, China, 2015. [Google Scholar]
- Xu, Y.; Li, Y.; Maffucci, K.; Huang, L.F.; Zeng, R. Analytical methods of phytochemicals from the Genus Gentiana. Molecules 2017, 22, 2080. [Google Scholar] [CrossRef]
- Huang, J.; Pei, S.Q.; Long, C.L. An ethnobotanical study of medicinal plants used by the Lisu people in Nujiang, Northwest Yunnan, China. Econ. Bot. 2004, 58, S253–S264. [Google Scholar]
- Pei, S.J.; Hamilton, A.C.; Yang, L.X.; Hua, H.Y.; Yang, Z.W.; Gao, F.; Zhang, Q.X. Conservation and development through medicinal plants: A case study from Ludian (Northwest Yunnan, China) and presentation of a general model. Biodivers. Conserv. 2010, 19, 2619–2636. [Google Scholar]
- Yunnan Pharmaceutical Co., Ltd. List of Traditional Chinese Medicine Resources in Yunnan, China; China Science Press: Beijing, China, 1993. [Google Scholar]
- Zhang, X.X.; Zhan, G.Q.; Jin, M.; Zhang, H.; Dang, J.; Zhang, Y.; Guo, Z.J.; Ito, Y. Botany, traditional use, phytochemistry, pharmacology, quality control, and authentication of Radix Gentianae Macrophyllae-A traditional medicine: A review. Phytomedicine 2018, 46, 142–163. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.B.; Wang, X.; Huang, R.; Liu, H.M.; Hu, H.; Hu, W.Y.; Lv, S.T.; Zhao, H.; Chen, G. Seven new chemical constituents from the roots of Gentiana macrophylla pall. Fitoterapia 2020, 141, 104476. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.J.; Li, J.Y.; Qi, J.H. Gentisides A and B, two new neuritogenic compounds from the traditional Chinese medicine Gentiana rigescens Franch. Bioorgan. Med. Chem. 2010, 18, 2131–2134. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.J.; Xiang, L.; Luo, Y.; Wang, G.F.; Li, J.Y.; Qi, J.H. Gentisides C-K: Nine new neuritogenic compounds from the traditional Chinese medicine Gentiana rigescens Franch. Bioorgan. Med. Chem. 2010, 18, 6995–7000. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.W.; Liao, B.S.; Luo, L.; Ren, Y.Y. Species identification of poisonous medicinal plant using DNA barcoding. Chin. J. Nat. Medicines 2019, 17, 585–590. [Google Scholar] [CrossRef]
- Liu, J.; Yang, H.F.; Ge, X.J. The use of DNA barcoding on recently diverged species in the genus Gentiana (Gentianaceae) in China. PLoS ONE 2016, 11, e0153008. [Google Scholar] [CrossRef]
- Tao, Z.; Jian, W.; Jia, Y.; Li, W.L.; Xu, F.S.; Wang, X.M. Comparative chloroplast genome analyses of species in Gentiana section Cruciata (Gentianaceae) and the development of authentication markers. Int. J. Mol. Sci. 2018, 19, 1962. [Google Scholar]
- Zheng, P.; Zhang, K.J.; Wang, Z.Z. Genetic diversity and gentiopicroside content of four Gentiana species in China revealed by ISSR and HPLC methods. Biochem. Syst. Ecol. 2011, 39, 704–710. [Google Scholar] [CrossRef]
- Liu, F.F.; Wang, Y.M.; Zhu, H.T.; Wang, D.; Yang, C.R.; Xu, M.; Zhang, Y.J. Comparative study on “Long-Dan”, “Qin-Jiao” and their adulterants by HPLC Analysis. Nat. Prod. Bioprospect. 2014, 4, 297–308. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, J.; Zhao, Y.L.; Wang, Y.Z.; Jin, H. Chemotaxonomic studies of nine Gentianaceae species from western China based on liquid chromatography tandem mass spectrometry and Fourier transform infrared spectroscopy. Phytochem. Analysis 2016, 27, 158–167. [Google Scholar] [CrossRef]
- Ercioglu, E.; Velioglu, H.M.; Boyaci, I.H. Chemometric evaluation of discrimination of Aromatic plants by Using NIRS, LIBS. Food Anal Method 2018, 11, 1656–1667. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Z.Y.; Li, T.H.; Chen, N.; Xu, W.J.; Liu, S.P. Surface-enhanced Raman scattering spectra revealing the inter-cultivar differences for Chinese ornamental Flos Chrysanthemum: A new promising method for plant taxonomy. Plant Methods 2017, 13, 92. [Google Scholar] [CrossRef]
- Luna, A.S.; Da Silva, A.P.; Da Silva, C.S.; Lima, I.C.A.; de Gois, J.S. Chemometric methods for classification of clonal varieties of green coffee using Raman spectroscopy and direct sample analysis. J. Food Compos. Anal. 2019, 76, 44–50. [Google Scholar] [CrossRef]
- Lang, C.; Almeida, D.R.A.; Costa, F.R.C. Discrimination of taxonomic identity at species, genus and family levels using Fourier transformed near-infrared Spectroscopy (FT-NIR). Forest. Ecol. Manag. 2017, 406, 219–227. [Google Scholar] [CrossRef]
- Guzmán, Q.J.A.; Rivard, B.; Sánchez-Azofeifa, G.A. Discrimination of liana and tree leaves from a neotropical dry forest using visible-near infrared and longwave infrared reflectance spectra. Remote Sens. Environ. 2018, 219, 135–144. [Google Scholar] [CrossRef]
- Borraz-Martínez, S.; Boqué, R.; Simó, J.; Mestre, M.; Gras, A. Development of a methodology to analyze leaves from Prunus dulcis varieties using near infrared spectroscopy. Talanta 2019, 204, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Meenu, M.; Xu, B.J. Application of vibrational spectroscopy for classification, authentication and quality analysis of mushroom: A concise review. Food Chem. 2019, 289, 545–557. [Google Scholar] [CrossRef]
- Chen, Y.F.; Chen, Y.; Feng, X.P.; Yang, X.F.; Zhang, J.N.; Qiu, Z.J.; He, Y. Variety identification of Orchids using Fourier transform infrared spectroscopy combined with stacked sparse auto-encoder. Molecules 2019, 13, 2506. [Google Scholar] [CrossRef]
- Liu, R.H.; Sun, Q.F.; Hu, T.; Li, L.; Nie, L.; Wang, J.Y.; Zhou, W.H.; Zang, H.C. Multi-parameters monitoring during traditional Chinese medicine concentration process with near infrared spectroscopy and chemometrics. Spectrochim. Acta A 2018, 192, 75–81. [Google Scholar] [CrossRef]
- Liu, P.; Wang, J.; Li, Q.; Gao, J.; Tan, X.Y.; Bian, X.Y. Rapid identification and quantification of Panax notoginseng with its adulterants by near infrared spectroscopy combined with chemometrics. Spectrochim. Acta A 2019, 206, 23–30. [Google Scholar] [CrossRef]
- Sousa, C.; Quintelas, C.; Augusto, C.; Ferreira, E.C.; Páscoa, R.N.M.J. Discrimination of Camellia japonica cultivars and chemometric models: An interlaboratory study. Comput. Electron. Agr. 2019, 159, 28–33. [Google Scholar] [CrossRef]
- Wang, Y.; Zuo, Z.T.; Huang, H.Y.; Wang, Y.Z. Original plant traceability of Dendrobium species using multi-spectroscopy fusion and mathematical models. Roy. Soc. Open. Sci. 2019, 6, 190399. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Zuo, Z.T.; Zhang, Q.Z.; Wang, Y.Z. Classification of Paris species according to botanical and geographical origins based on spectroscopic, chromatographic, conventional chemometric analysis and data fusion strategy. Microchem. J. 2018, 143, 367–378. [Google Scholar] [CrossRef]
- Li, T.; Su, C. Authenticity identification and classification of Rhodiola species in traditional Tibetan medicine based on Fourier transform near-infrared spectroscopy and chemometrics analysis. Spectrochim. Acta A 2018, 204, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Li, J.Q.; Liu, H.G.; Wang, Y.Z. Attenuated total reflection-Fourier transform infrared spectroscopy (ATR-FTIR) combined with chemometrics methods for the classification of Lingzhi species. Molecules 2019, 24, 2210. [Google Scholar] [CrossRef] [PubMed]
- Pasquini, C. Near infrared spectroscopy: A mature analytical technique with new perspectives—A review. Anal. Chim. Acta 2018, 1026, 8–36. [Google Scholar] [CrossRef]
- Yun, Y.H.; Li, H.D.; Deng, B.C.; Cao, D.S. An overview of variable selection methods in multivariate analysis of near-infrared spectra. TrAC Trend. Anal. Chem. 2019, 113, 102–115. [Google Scholar] [CrossRef]
- Yang, X.D.; Li, G.L.; Song, J.; Gao, M.J.; Zhou, S.L. Rapid discrimination of Notoginseng powder adulteration of different grades using FT-MIR spectroscopy combined with chemometrics. Spectrochim. Acta A 2018, 205, 457–464. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.Y.; Wang, Y.Z. FT-MIR and NIR spectral data fusion: A synergetic strategy for the geographical traceability of Panax notoginseng. Anal. Bioanal. Chem. 2018, 410, 91–103. [Google Scholar] [CrossRef]
- Wolpert, D.H. Stacked generalization. Neural Networks 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Naimi, A.I.; Balzer, L.B. Stacked generalization: An introduction to super learning. Eur. J. Epidemiol. 2018, 33, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Alexandropoulos, S.A.N.; Aridas, C.K.; Kotsiantis, S.B.; Vrahatis, M.N. Stacking strong ensembles of classifiers. In Nonlinear Model Predictive Control; Springer Science and Business Media LLC: Berlin, Germany, 2019; pp. 545–556. [Google Scholar]
- Shan, P.; Zhao, Y.; Wang, Q.; Sha, X.; Lv, X.; Peng, S.; Ying, Y. Stacked ensemble extreme learning machine coupled with Partial Least Squares-based weighting strategy for nonlinear multivariate calibration. Spectrochim. Acta A 2019, 215, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Sfakianakis, S.; Bei, E.S.; Zervakis, M. Stacking of network based vlassifiers with application in breast cancer classification. In XIV Mediterranean Conference on Medical and Biological Engineering and Computing; Kyriacou, E., Christofides, S., Pattichis, C., Eds.; Springer: Berlin, Germany, 2016. [Google Scholar]
- Wang, Q.Q.; Huang, H.Y.; Wang, Y.Z. Geographical authentication of Macrohyporia cocos by a data fusion method combining ultra-fast liquid chromatography and Fourier transform infrared spectroscopy. Molecules 2019, 24, 1320. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Zuo, Z.T.; Zhang, Q.Z.; Wang, Y.Z. Data fusion of fourier transform mid-infrared (MIR) and near-infrared (NIR) spectroscopies to identify geographical origin of wild Paris polyphylla var. yunnanensis. Molecules 2019, 24, 2559. [Google Scholar] [CrossRef]
- Bischl, B.; Lang, M.; Kotthoff, L.; Schiffne, J.; Richter, J.; Studerus, E.; Casalicchio, G.; Jones, Z.M. mlr: Machine Learning in R. J. Mach. Learn. Res. 2016, 17, 5938–5942. [Google Scholar]
- Chen, W.; Pourghasemi, H.R.; Naghibi, S.A. A comparative study of landslide susceptibility maps produced using support vector machine with different kernel functions and entropy data mining models in China. B. Eng. Geol. Environ. 2018, 77, 647–664. [Google Scholar] [CrossRef]
- Qian, Y.G.; Zhou, W.Q.; Yan, J.L.; Li, W.F.; Han, L.J. Comparing machine learning classifiers for object-based land cover classification using very high resolution imagery. Remote Sens. 2015, 7, 153–168. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Li, T.; Liu, H.G.; Li, J.Q.; Wang, Y.Z. Geographical traceability of wild Boletus edulis based on data fusion of FT-MIR and ICP-AES coupled with data mining methods (SVM). Spectrochim. Acta A 2017, 177, 20–27. [Google Scholar] [CrossRef]
- Li, L.Q.; Xie, S.M.; Ning, J.M.; Chen, Q.S.; Zhang, Z.Z. Evaluating green tea quality based on multisensor data fusion combining hyperspectral imaging and olfactory visualization systems. J. Sci. Food Agr. 2019, 99, 1787–1794. [Google Scholar] [CrossRef]
- Schwolow, S.; Gerhardt, N.; Rohn, S.; Weller, P. Data fusion of GC-IMS data and FT-MIR spectra for the authentication of olive oils and honeys—is it worth to go the extra mile? Anal. Bioanal. Chem. 2019, 411, 6005–6019. [Google Scholar] [CrossRef]
- Ríos-Reina, R.; Callejón, R.M.; Savorani, F.; Amigo, J.M.; Cocchi, M. data fusion approaches in spectroscopic characterization and classification of PDO wine vinegars. Talanta 2019, 198, 560–572. [Google Scholar] [CrossRef]
- Tsakiridis, N.L.; Tziolas, N.V.; Theocharis, J.B.; Zalidis, G.C. A genetic algorithm-based stacking algorithm for predicting soil organic matter from vis–NIR spectral data. Eur. J. Soil. Sci. 2019, 70, 578–590. [Google Scholar] [CrossRef]
- Verma, A.K.; Pal, S. Prediction of skin disease with three different feature selection techniques using stacking ensemble method. Appl. Biochem. Biotech. 2019, 1, 1–20. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, R.R.; Ma, Q.F.; Wang, Y.H.; Wang, Q.Q.; Huang, Z.H.; Huang, L.Y. A feature selection and multi-model fusion-based approach of predicting air quality. ISA T. 2019. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, H.Y.; Zuo, Z.T.; Wang, Y.Z. Comprehensive quality assessment of Dendrubium officinale using ATR-FTIR spectroscopy combined with random forest and support vector machine regression. Spectrochim. Acta A 2018, 205, 637–648. [Google Scholar] [CrossRef]
- Rodríguez, S.D.; Rolandelli, G.; Buera, M.P. Detection of quinoa flour adulteration by means of FT-MIR spectroscopy combined with chemometric methods. Food Chem. 2019, 274, 392–401. [Google Scholar] [CrossRef]
- Horn, B.; Esslinger, S.; Pfister, M.; Fauhl-Hassek, C.; Riedl, J. Non-targeted detection of paprika adulteration using mid-infrared spectroscopy and one-class classification–Is it data preprocessing that makes the performance? Food Chem. 2018, 257, 112–119. [Google Scholar] [CrossRef]
- Mees, C.; Souard, F.; Delporte, C.; Deconinck, E.; Stoffelen, P.; Stévigny, C.; Kauffmann, J.; De Braekeleer, K. Identification of coffee leaves using FT-NIR spectroscopy and SIMCA. Talanta 2018, 177, 4–11. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- De Santana, F.B.; Mazivila, S.J.; Gontijo, L.C.; Neto, W.B.; Poppi, R.J. Rapid Discrimination between authentic and adulterated andiroba oilusing FTIR-HATR spectroscopy and Random Forest. Food Anal. Method 2018, 11, 1927–1935. [Google Scholar] [CrossRef]
- Chapelle, O.; Patrick, H.; Vladimir, N.V. Support vector machines for histogram-based image classification. IEEE Trans. Neural Netw. 1999, 10, 1055–1064. [Google Scholar] [CrossRef]
- Belousov, A.I.; Verzakov, S.A.; von Frese, J. A flexible classification approach with optimal generalisation performance: Support vector machines. Chemometr. Intell. Lab. 2002, 64, 15–25. [Google Scholar] [CrossRef]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens-Basel. 2016, 6, 445. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Y.J.; Li, W.L.; Liu, X.S.; Zheng, J.Y.; Zhang, W.T.; Chen, Y. Determination of geographical origin and icariin content of Herba Epimedii using near infrared spectroscopy and chemometrics. Spectrochim. Acta A 2018, 191, 233–240. [Google Scholar] [CrossRef]
- Altman, N.S. An Introduction to Kernel and Nearest-Neighbor Nonparametric Regression. Am. Stat. 1992, 46, 175–185. [Google Scholar]
- Sharma, P.; Aggarwal, A.; Gupta, A.; Garg, A. Leaf identification using HOG, KNN, and neural networks. In International Conference on Innovative Computing and Communications; Bhattacharyya, S., Hassanien, A., Gupta, D., Khanna, A., Pan, I., Eds.; Springer: Berlin, Germany, 2019. [Google Scholar]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A review of variable selection methods in partial least squares regression. Chemometr. Intell. Lab. 2012, 118, 62–69. [Google Scholar] [CrossRef]
- Casale, M.; Casolino, C.; Oliveri, P.; Forina, M. The potential of coupling information using three analytical techniques for identifying the geographical origin of Liguria extra virgin olive oil. Food Chem. 2010, 118, 163–170. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compound are not available from the authors. |


| Class | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SE | SP | MCC | EFF | ACC (%) | SE | SP | MCC | EFF | |
| 1 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 3 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 6 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 8 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 0.75 | 1.00 | 0.86 | 0.87 |
| 9 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 10 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 11 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 12 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 13 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 14 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 15 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 16 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 17 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 18 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| Class | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SE | SP | MCC | EFF | ACC (%) | SE | SP | MCC | EFF | |
| 1 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 3 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 8 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 0.75 | 1.00 | 0.86 | 0.87 |
| 9 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 10 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 11 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 12 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 0.75 | 1.00 | 0.86 | 0.87 |
| 13 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 14 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 15 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 16 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.50 | 1.00 | 0.70 | 0.71 |
| 17 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 18 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Class | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SE | SP | MCC | EFF | ACC (%) | SE | SP | MCC | EFF | |
| 1 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 0.75 | 1.00 | 0.86 | 0.87 |
| 2 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 3 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 6 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 8 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 0.75 | 1.00 | 0.86 | 0.87 |
| 9 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 10 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 11 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 12 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 13 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 14 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 15 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 16 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 17 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 18 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| Class | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SE | SP | MCC | EFF | ACC (%) | SE | SP | MCC | EFF | |
| 1 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 3 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 8 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 9 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 10 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 11 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 12 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 13 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 14 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 15 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 16 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 17 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 18 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Class | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SE | SP | MCC | EFF | ACC (%) | SE | SP | MCC | EFF | |
| 1 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 2 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.50 | 1.00 | 0.70 | 0.71 |
| 3 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 95.83 | 0.75 | 0.97 | 0.65 | 0.85 |
| 6 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 0.75 | 1.00 | 0.86 | 0.87 |
| 8 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 9 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 10 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 11 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 12 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 95.83 | 0.75 | 0.97 | 0.65 | 0.85 |
| 13 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 14 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 15 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 16 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| 17 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 18 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.75 | 0.99 | 0.74 | 0.86 |
| Class | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | SE | SP | MCC | EFF | ACC (%) | SE | SP | MCC | EFF | |
| 1 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 3 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 5 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 6 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 8 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 1.00 | 0.97 | 0.80 | 0.99 |
| 9 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.50 | 1.00 | 0.70 | 0.71 |
| 10 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 11 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 98.61 | 1.00 | 0.99 | 0.89 | 0.99 |
| 12 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 13 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 14 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 15 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 16 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 97.22 | 0.50 | 1.00 | 0.70 | 0.71 |
| 17 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 18 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 | 100.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Model | Hyperparameters | Calibration Set | Validation Set | |
|---|---|---|---|---|
| Total ACC (%) | Total ACC (%) | K | ||
| VIP-NIR-RF | ntree = 1774, mtry = 14 | 100 | 97.22 | 0.97 |
| Bor-NIR-RF | ntree = 452, mtry = 11 | 100 | 91.67 | 0.91 |
| GARF-NIR-RF | ntree = 678, mtry = 22 | 100 | 91.67 | 0.91 |
| GASVM-NIR-RF | ntree = 1763, mtry = 34 | 100 | 91.67 | 0.91 |
| Ven-NIR-RF | ntree = 1511, mtry = 2 | 100 | 94.44 | 0.94 |
| VIP-NIR-SVM | kernel = linear, cost = 0.01 | 100 | 97.22 | 0.97 |
| Bor-NIR-SVM | kernel = linear, cost = 0.05 | 100 | 98.61 | 0.99 |
| GARF-NIR-SVM | kernel = linear, cost = 0.1 | 100 | 93.06 | 0.93 |
| GASVM-NIR-SVM | kernel = linear, cost = 0.05 | 100 | 91.67 | 0.91 |
| Ven-NIR-SVM | kernel = linear, cost = 0.05 | 100 | 98.61 | 0.99 |
| VIP-NIR-KNN | k = 1 | 100 | 95.83 | 0.96 |
| Bor-NIR-KNN | k = 1 | 100 | 94.44 | 0.94 |
| GARF-NIR-KNN | k = 1 | 100 | 87.50 | 0.87 |
| GASVM-NIR-KNN | k = 1 | 100 | 88.89 | 0.88 |
| Ven-NIR-KNN | k = 1 | 100 | 94.44 | 0.94 |
| Model | Hyperparameter | Calibration Set | Validation Set | |
|---|---|---|---|---|
| Total ACC (%) | Total ACC (%) | K | ||
| VIP-MIR-RF | ntree = 1334, mtry = 23 | 100 | 97.22 | 0.97 |
| Bor-MIR-RF | ntree = 1673, mtry = 13 | 100 | 95.83 | 0.96 |
| GARF-MIR-RF | ntree = 958, mtry = 20 | 100 | 95.83 | 0.96 |
| GASVM-MIR-RF | ntree = 297 mtry = 31 | 100 | 94.44 | 0.94 |
| Ven-MIR-RF | ntree = 190, mtry = 10 | 100 | 98.61 | 0.99 |
| VIP-MIR-SVM | kernel = linear, cost = 0.05 | 100 | 100 | 1.00 |
| Bor-MIR-SVM | kernel = linear, cost = 0.5 | 100 | 100 | 1.00 |
| GARF-MIR-SVM | kernel = linear, cost = 0.10 | 100 | 100 | 1.00 |
| GASVM-MIR-SVM | kernel = linear, cost = 1.00 | 100 | 100 | 1.00 |
| Ven-MIR-SVM | kernel = linear, cost = 1.00 | 100 | 98.61 | 0.99 |
| VIP-MIR-KNN | k = 1 | 100 | 98.61 | 0.99 |
| Bor-MIR-KNN | k = 1 | 100 | 97.22 | 0.97 |
| GARF-MIR-KNN | k = 1 | 100 | 95.83 | 0.96 |
| GASVM-MIR-KNN | k = 1 | 100 | 94.44 | 0.94 |
| Ven-MIR-KNN | k = 1 | 100 | 97.22 | 0.97 |
| Scenario | Data Set | Model | Level 1 | Calibration Set | Validation Set | |
|---|---|---|---|---|---|---|
| Total ACC (%) | Total ACC (%) | K | ||||
| A | Ven-NIR | SG-Ven-NIR- RF | RF | 100.00 | 98.61 | 0.99 |
| B | Ven-NIR | SG-Ven-NIR- SVM | SVM | 100.00 | 97.22 | 0.97 |
| C | Ven-NIR | SG-Ven-NIR- KNN | KNN | 100.00 | 95.83 | 0.96 |
| D | Ven-MIR | SG-Ven-MIR- RF | RF | 100.00 | 94.44 | 0.94 |
| E | Ven-MIR | SG-Ven-MIR- SVM | SVM | 100.00 | 100.00 | 1.00 |
| F | Ven-MIR | SG-Ven-MIR- KNN | KNN | 100.00 | 90.28 | 0.90 |
| Data Fusion Strategy | Number of Variables | Models | Calibration Set | Validation Set | |
|---|---|---|---|---|---|
| Total ACC (%) | Total ACC (%) | K | |||
| Low-level fusion | 2701 | Low-RF | 100.00 | 97.22 | 0.97 |
| Low-level fusion | 2701 | Low-SVM | 100.00 | 100.00 | 1.00 |
| Low-level fusion | 2701 | Low-KNN | 100.00 | 97.22 | 0.97 |
| Mid-level fusion | 174 | Mid-RF | 100.00 | 100.00 | 1.00 |
| Mid-level fusion | 174 | Mid-SVM | 100.00 | 100.00 | 1.00 |
| Mid-level fusion | 174 | Mid-KNN | 100.00 | 100.00 | 1.00 |
| Class | Genus | Species | Geographical Location |
|---|---|---|---|
| 1 | Gentiana | G. rigescens | Yongde, Lincang, Yunnan, China |
| 2 | Gentiana | G. cephalantha | Xuyong, Luzhou, Sichuan, China |
| 3 | Gentiana | G. davidii | Jianghua, Yongzhou, Hunan, China |
| 4 | Gentiana | G. lawrencei var. farreri | Songpan, Aba, Sichuan, China |
| 5 | Gentiana | G. stragulata | Songpan, Aba, Sichuan, China |
| 6 | Gentiana | G. crassicaulis | Lanping, Nujiang, Yunnan, China |
| 7 | Gentiana | G. loureirii | Jianghua, Yongzhou, Hunan, China |
| 8 | Gentiana | G. napulifera | Liping, QianDong-nan, Guizhou, China |
| 9 | Gentiana | G. praticola | Liping, QianDong-nan, Guizhou, China |
| 10 | Gentiana | G. piasezkii | Ningqiang, Hanzhong, Shaanxi, China |
| 11 | Gentiana | G. squarrosa | Songpan, Aba, Sichuan, China |
| 12 | Gentiana | G. pseudosquarrosa | Songpan, Aba, Sichuan, China |
| 13 | Gentiana | G. rubicunda | Xianfeng, Enshi, Hubei, China |
| 14 | Gentiana | G. rubicunda var. samolifolia | Wufeng, Yichang, Hubei, China |
| 15 | Gentiana | G. rhodantha | Nayong, Bijie, Guizhou, China |
| 16 | Gentiana | G. striata | Songpan, Aba, Sichuan, China |
| 17 | Tripterospermum | T. chinense | Tonggu, Yichun, Jiangxi, China |
| 18 | Tripterospermum | T. cordatum | Tonggu, Yichun, Jiangxi, China |
| Species | Chinese Name | Disease | Ch.P. |
| G. rigescens | Dian Longdan | heat-clearing, liver protection, icterohepatitis, Japanese encephalitis, cephalalgia, swelling and pain of eye [9,13] | listed (2015 edition) [9] |
| G. cephalantha | Tou hua Longdan | heat-clearing, icterohepatitis | unlisted |
| G. davidii | Wu ling Longdan | heat-clearing, urinary tract infection, conjunctivitis [13] | unlisted |
| G. lawrencei var. farreri | Xian ye Longdan | trachitis, cough, smallpox [13] | unlisted |
| G. stragulata | Shi e Longdan | none reported | unlisted |
| G. crassicaulis | Cu jing qin jiao | heat-clearing, icterohepatitis, hematochezia, rheumatism [9] | listed (2015 edition) [9] |
| G. loureirii | Hua nan Longdan | heat-clearing, icterohepatitis, diarrhea, swelling and pain of eye [13] | unlisted |
| G. napulifera | Fu gen Longdan | none reported | unlisted |
| G. praticola | Cao dian Longdan | heat-clearing, detumescence analgesic [13] | unlisted |
| G. piasezkii | Shan nan Longdan | none reported | unlisted |
| G. squarrosa | Lin ye Longdan | heat-clearing, acute appendicitis, swelling and pain of eye [13] | unlisted |
| G. pseudosquarrosa | Jia lin ye Longdan | none reported | unlisted |
| G. rubicunda | Shen hong Longdan | dyspepsia, bone fracture, snakebite, diminish inflammation [13] | unlisted |
| G. rubicunda var. samolifolia | Xiao fan lu ye Longdan | none reported | unlisted |
| G. rhodantha | Hong hua Longdan | heat-clearing, diminish inflammation, urinary tract infection, cold, icterohepatitis, diarrhea, scald [9,13] | listed (2015 edition) [9] |
| G. striata | Tiao wen Longdan | none reported | unlisted |
| T. chinense | Shuang hudie | heat-clearing, phthisis, pulmonary abscess, irregular menstruation [13] | unlisted |
| T. cordatum | E mei Shuang hudie | bone fracture [13] | unlisted |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, T.; Yu, H.; Wang, Y.-Z. Discrimination of Gentiana and Its Related Species Using IR Spectroscopy Combined with Feature Selection and Stacked Generalization. Molecules 2020, 25, 1442. https://doi.org/10.3390/molecules25061442
Shen T, Yu H, Wang Y-Z. Discrimination of Gentiana and Its Related Species Using IR Spectroscopy Combined with Feature Selection and Stacked Generalization. Molecules. 2020; 25(6):1442. https://doi.org/10.3390/molecules25061442
Chicago/Turabian StyleShen, Tao, Hong Yu, and Yuan-Zhong Wang. 2020. "Discrimination of Gentiana and Its Related Species Using IR Spectroscopy Combined with Feature Selection and Stacked Generalization" Molecules 25, no. 6: 1442. https://doi.org/10.3390/molecules25061442
APA StyleShen, T., Yu, H., & Wang, Y.-Z. (2020). Discrimination of Gentiana and Its Related Species Using IR Spectroscopy Combined with Feature Selection and Stacked Generalization. Molecules, 25(6), 1442. https://doi.org/10.3390/molecules25061442






