Polyphenols from Salix tetrasperma Impair Virulence and Inhibit Quorum Sensing of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results
2.1. Chemical Composition
2.2. Antibacterial Activities
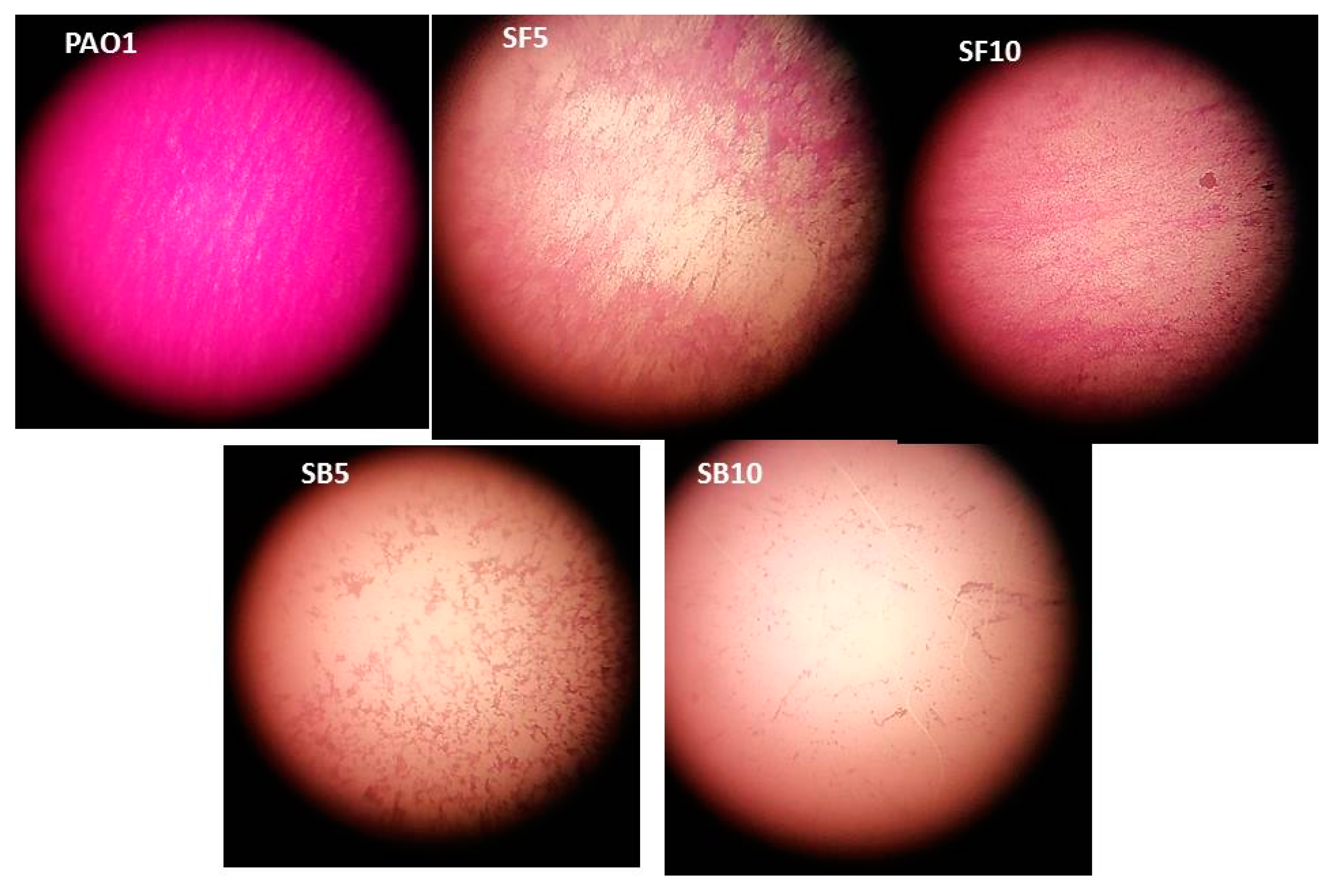
2.3. Stem Bark and Flower Extracts as Biofilm Inhibitors
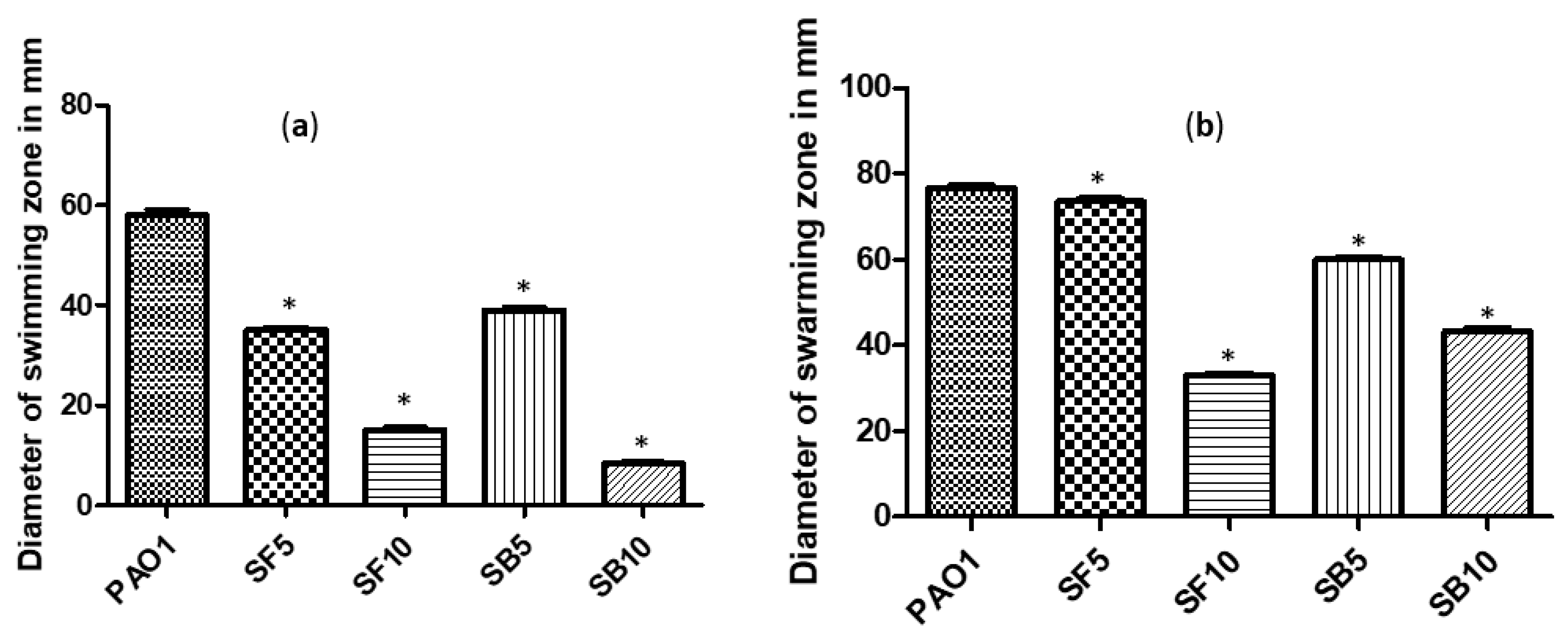
2.4. Effect on Swimming and Swarming Motilities
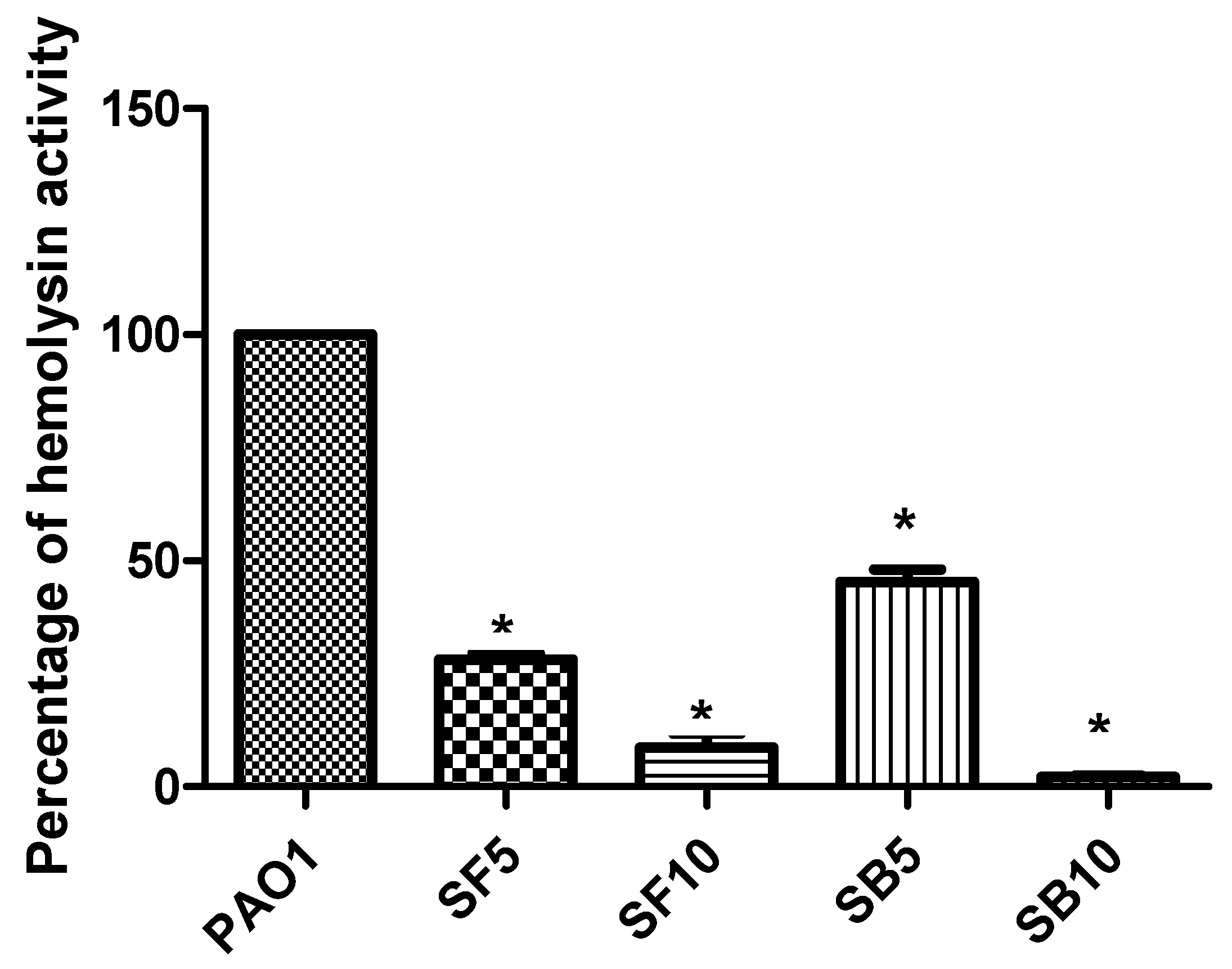
2.5. Inhibition of Protease Production and Hemolytic Activity
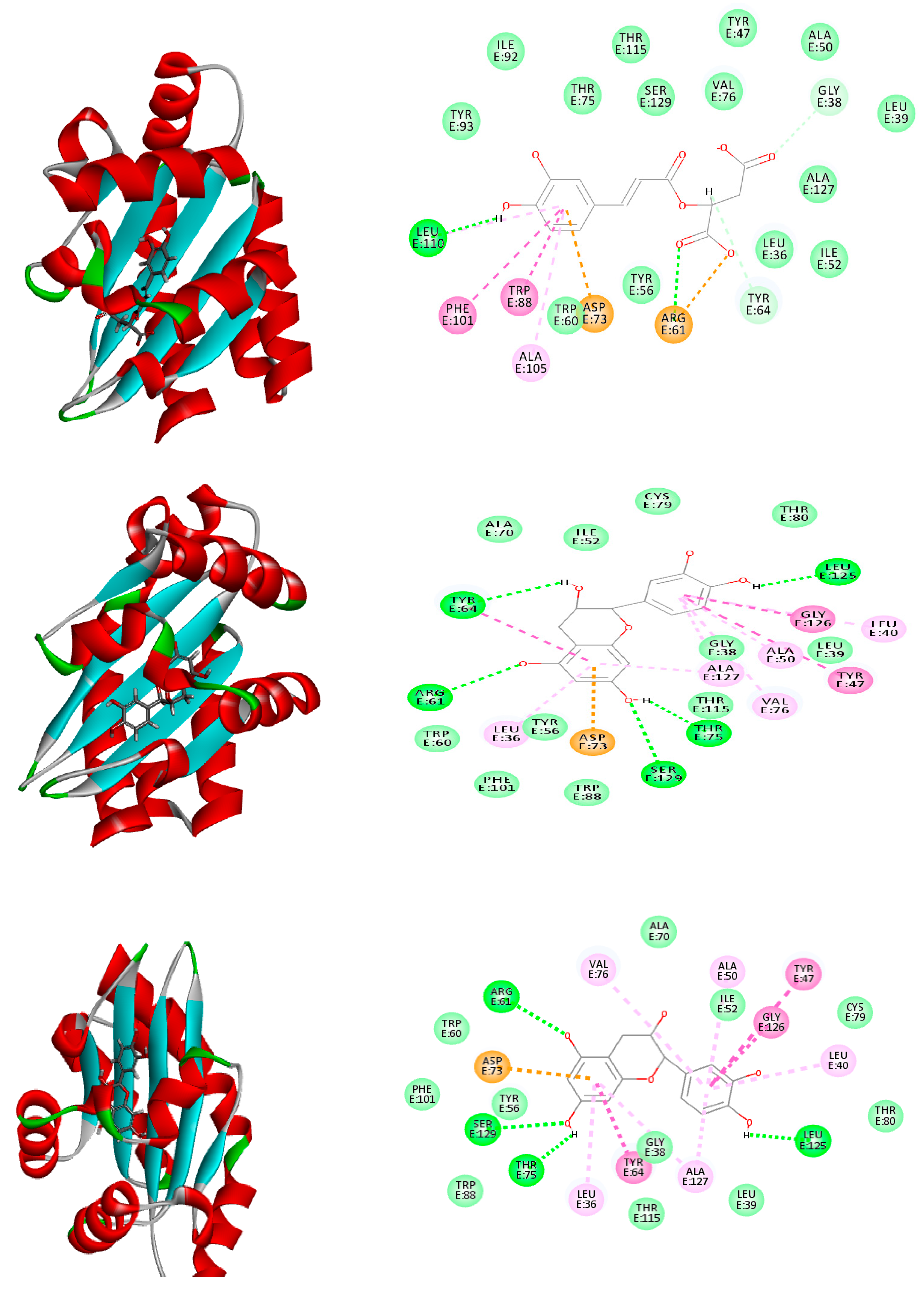
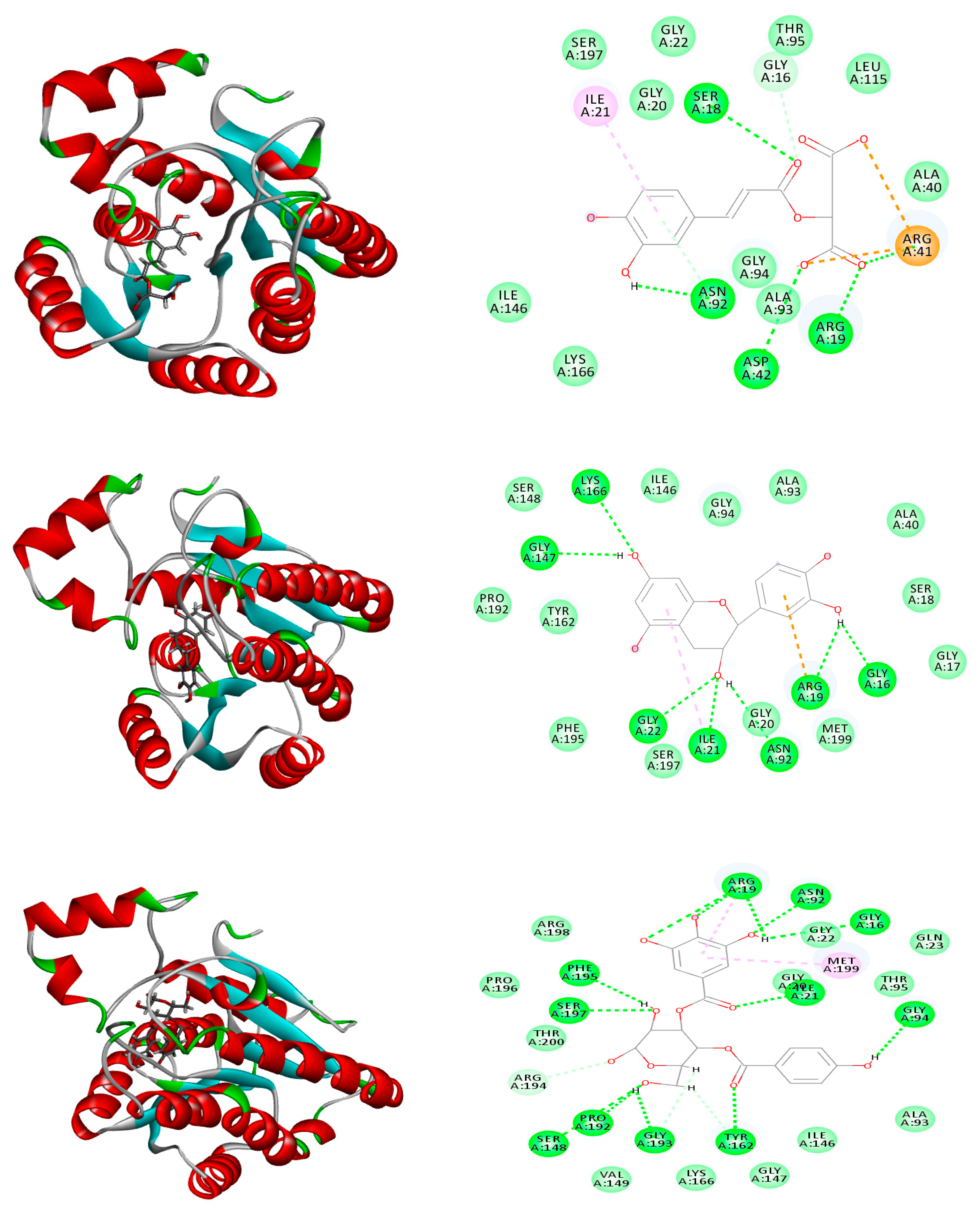
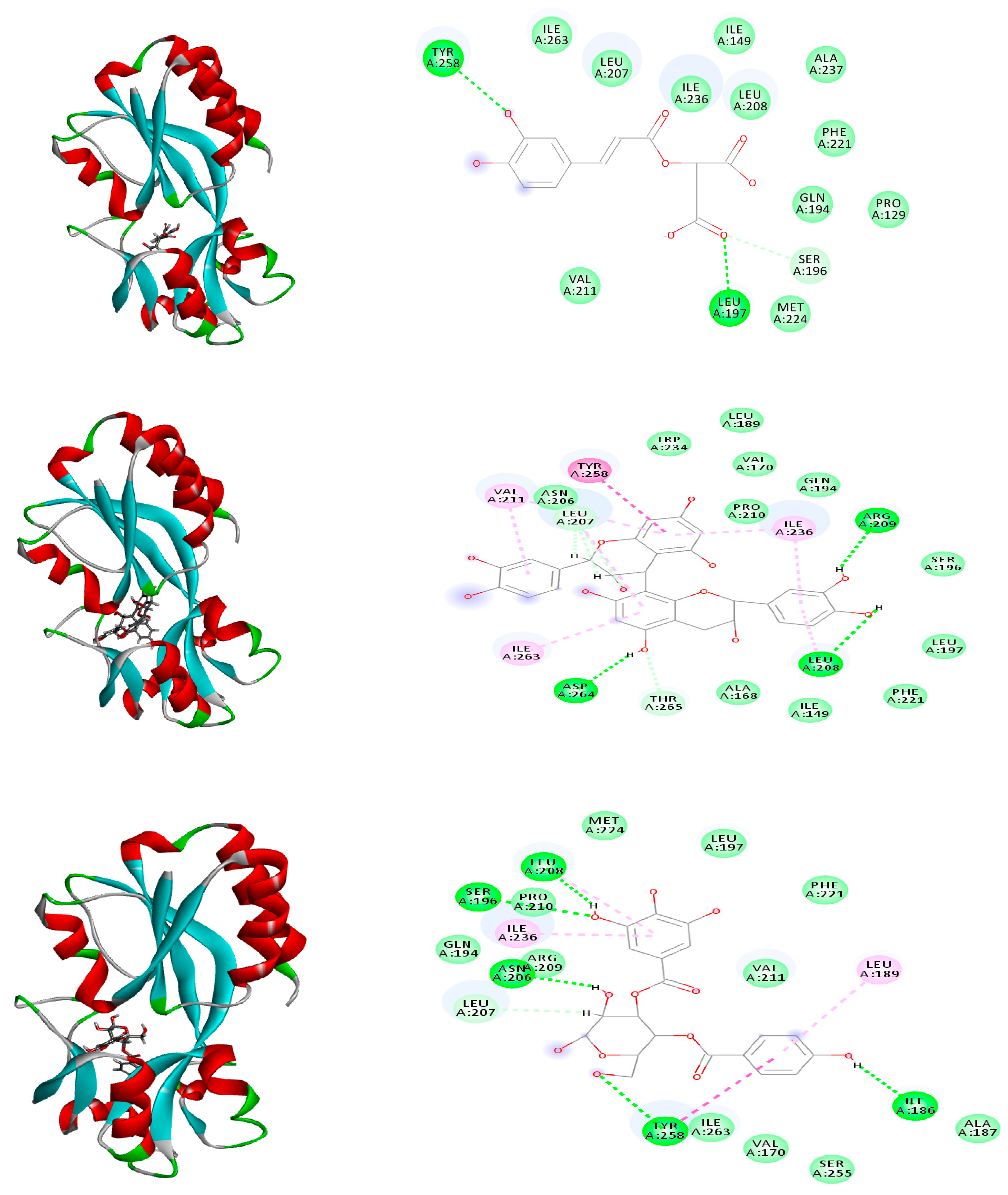
2.6. Molecular Modeling
3. Discussion
4. Materials and Methods
4.1. Plant Material and Extraction
4.2. HPLC-MS Analysis
4.3. Media and Chemicals
4.4. Minimum Inhibitory Concentration (MIC) Determination and Subinhibitory Concentration Effect on Bacterial Growth
4.5. Measurement of Biofilm Inhibition
4.6. Measurement of Swimming and Swarming Motilities
4.7. Measurement of Protease Inhibition
4.8. Measurement of Hemolysis Inhibition
4.9. Molecular Modelling Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mashhady, M.A.; Abkhoo, J.; Jahani, S.; Abyar, S.; Khosravani, F. Inhibitory effects of plant extracts on Pseudomonas aeruginosa biofilm formation. Life 2016, 3, e38199. [Google Scholar] [CrossRef]
- Fletcher, J.; Leach, J.E.; Eversole, K.; Tauxe, R. Human pathogens on plants: Designing a multidisciplinary strategy for research. Phytopathology 2013, 103, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.S.; Bais, H.P.; Déziel, E.; Schweizer, H.P.; Rahme, L.G.; Fall, R.; Vivanco, J.M. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004, 134, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef]
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef]
- Abbas, H.A.; Abdo, I.M.; Moustafa, M.Z. In vitro antibacterial and antibiofilm activities of Hibiscus sabdariffa L. Extract and apple vinegar against bacteria isolated from diabetic foot infections. Res. J. Pharm. Technol. 2014, 7, 131–136. [Google Scholar]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Alavi, M.; Karimi, N.; Valadbeigi, T. Antibacterial, antibiofilm, antiquorum sensing, antimotility, and antioxidant activities of green fabricated Ag, Cu, TiO2, ZnO, and Fe3O4 NPs via Protoparmeliopsis muralis lichen aqueous extract against multi-drug-resistant bacteria. ACS Biomater. Sci. Eng. 2019, 5, 4228–4243. [Google Scholar] [CrossRef]
- Abbas, H.A.; Elsherbini, A.M.; Shaldam, M.A. Repurposing metformin as a quorum sensing inhibitor in Pseudomonas aeruginosa. Afr. Health Sci. 2017, 17, 808–819. [Google Scholar] [CrossRef]
- Pesci, E.C.; Pearson, J.P.; Seed, P.C.; Iglewski, B.H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3127–3132. [Google Scholar] [CrossRef]
- Whiteley, M.; Lee, K.M.; Greenberg, E. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 13904–13909. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.M.; Soliman, W.E.E.; Shaldam, M.A. Perturbation of quorum sensing in Pseudomonas aeruginosa by febuxostat. Adv. Microbiol. 2018, 8, 650. [Google Scholar] [CrossRef]
- Kumar, N.V.; Murthy, P.S.; Manjunatha, J.; Bettadaiah, B. Synthesis and quorum sensing inhibitory activity of key phenolic compounds of ginger and their derivatives. Food Chem. 2014, 159, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rajaonson, S.; Diallo, B.; Mol, A.; El Jaziri, M.; Baucher, M. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa pao1. Appl. Environ. Microbiol. 2010, 76, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Enayat, S.; Banerjee, S. Comparative antioxidant activity of extracts from leaves, bark and catkins of Salix aegyptiaca sp. Food Chem. 2009, 116, 23–28. [Google Scholar] [CrossRef]
- Hussain, H.; Badawy, A.; Elshazly, A.; Elsayed, A.; Krohn, K.; Riaz, M.; Schulz, B. Chemical constituents and antimicrobial activity of Salix subserrata. Rec. Nat. Prod. 2011, 5, 133. [Google Scholar]
- Van Wyk, B.E.; Wink, M. Phytomedicines, Herbal Drugs & Plant Poisons; Briza Publications: Pretoria, South Africa, 2015. [Google Scholar]
- Sobeh, M.; Mahmoud, M.F.; Rezq, S.; Alsemeh, A.E.; Sabry, O.M.; Mostafa, I.; Abdelfattah, M.A.; Ait El-Allem, K.; El-Shazly, A.M.; Yasri, A. Salix tetrasperma roxb. extract alleviates neuropathic pain in rats via modulation of the nf-κb/tnf-α/nox/inos pathway. Antioxidants 2019, 8, 482. [Google Scholar] [CrossRef]
- El-Shazly, A.; El-Sayed, A.; Fikrey, E. Bioactive secondary metabolites from Salix tetrasperma Roxb. Z. Naturforsch. C. 2012, 67, 353–359. [Google Scholar] [CrossRef]
- Tawfeek, N.; Sobeh, M.; Hamdan, D.I.; Farrag, N.; Roxo, M.; El-Shazly, A.M.; Wink, M. Phenolic compounds from Populus alba L. and Salix subserrata Willd. (Salicaceae) counteract oxidative stress in caenorhabditis elegans. Molecules 2019, 24, 1999. [Google Scholar] [CrossRef]
- Zhao, T.; Liu, Y. N-acetylcysteine inhibit biofilms produced by Pseudomonas aeruginosa. BMC Microbiol. 2010, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.; Hodgkinson, J.T.; Bowden, S.; Welch, M.; Spring, D.R. Applications of small molecule activators and inhibitors of quorum sensing in gram-negative bacteria. Trends Microbiol. 2012, 20, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.; Shaldam, M.A. Glyceryl trinitrate is a novel inhibitor of quorum sensing in Pseudomonas aeruginosa. Afr. Health Sci. 2016, 16, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.; Rukayadi, Y.; Hwang, J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef]
- Adonizio, A.; Kong, K.-F.; Mathee, K. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by south florida plant extracts. Antimicrob. Agents Chemother. 2008, 52, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, T.; Yin, W.F.; Chan, K.G. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa pao1 by ayurveda spice clove (Syzygium aromaticum) bud extract. Sensors 2012, 12, 4016–4030. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stevigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa pao1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef]
- Kirisits, M.J.; Parsek, M.R. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 2006, 8, 1841–1849. [Google Scholar] [CrossRef]
- Onsare, J.; Arora, D. Antibiofilm potential of flavonoids extracted from Moringa oleifera seed coat against Staphylococcus aureus, Pseudomonas aeruginosa and Candida albicans. J. Appl. Microbiol. 2015, 118, 313–325. [Google Scholar] [CrossRef]
- Glessner, A.; Smith, R.S.; Iglewski, B.H.; Robinson, J.B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 1999, 181, 1623–1629. [Google Scholar] [CrossRef]
- Heydorn, A.; Ersbøll, B.; Kato, J.; Hentzer, M.; Parsek, M.R.; Tolker-Nielsen, T.; Givskov, M.; Molin, S. Statistical analysis of Pseudomonas aeruginosa biofilm development: Impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 2002, 68, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Setia, S.; Harjai, K. Expression of quorum sensing and virulence factors are interlinked in Pseudomonas aeruginosa: An in vitro approach. Am. J. Biomed. Sci. 2011, 3, 116–125. [Google Scholar] [CrossRef]
- Kazemian, H.; Ghafourian, S.; Heidari, H.; Amiri, P.; Yamchi, J.K.; Shavalipour, A.; Houri, H.; Maleki, A.; Sadeghifard, N. Antibacterial, anti-swarming and anti-biofilm formation activities of Chamaemelum nobile against Pseudomonas aeruginosa. Rev. Soc. Bras. Med. Trop. 2015, 48, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Musthafa, K.S.; Ravi, A.V.; Annapoorani, A.; Packiavathy, I.S.V.; Pandian, S.K. Evaluation of anti-quorum-sensing activity of edible plants and fruits through inhibition of the n-acyl-homoserine lactone system in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 2010, 56, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Singh, H.; Singh, A.; Singh, B.R.; Mishra, A.; Nautiyal, C. Lagerstroemia speciosa fruit extract modulates quorum sensing-controlled virulence factor production and biofilm formation in Pseudomonas aeruginosa. Microbiology 2012, 158, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Maisuria, V.B.; Lopez-de Los Santos, Y.; Tufenkji, N.; Déziel, E. Cranberry-derived proanthocyanidins impair virulence and inhibit quorum sensing of Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 30169. [Google Scholar] [CrossRef] [PubMed]
- Rajasekharan, S.K.; Ramesh, S.; Bakkiyaraj, D.; Elangomathavan, R.; Kamalanathan, C. Burdock root extracts limit quorum-sensing-controlled phenotypes and biofilm architecture in major urinary tract pathogens. Urolithiasis 2015, 43, 29–40. [Google Scholar] [CrossRef]
- Sobeh, M.; Hassan, S.A.; El Raey, M.A.; Khalil, W.A.; Hassan, M.A.; Wink, M. Polyphenolics from Albizia harveyi exhibit antioxidant activities and counteract oxidative damage and ultra-structural changes of cryopreserved bull semen. Molecules 2017, 22, 1993. [Google Scholar] [CrossRef]
- Nalca, Y.; Jänsch, L.; Bredenbruch, F.; Geffers, R.; Buer, J.; Häussler, S. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa pao1: A global approach. Antimicrob. Agents Chemother. 2006, 50, 1680–1688. [Google Scholar] [CrossRef]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef]
- Abbas, H.A. Antibacterial, anti-swarming and antibiofilm activities of local Egyptian clover honey against Proteus mirabilis isolated from diabetic foot infection. Asian, J. Pharm. Res. 2013, 3, 114–117. [Google Scholar]
- Vijayaraghavan, P.; Vincent, S.G.P. A simple method for the detection of protease activity on agar plates using bromocresolgreen dye. J. Biochem. Technol. 2013, 4, 628–630. [Google Scholar]
- Rossignol, G.; Merieau, A.; Guerillon, J.; Veron, W.; Lesouhaitier, O.; Feuilloley, M.G.; Orange, N. Involvement of a phospholipase C in the hemolytic activity of a clinical strain of Pseudomonas fluorescens. BMC Microbiol. 2008, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Muraglia, E.; Bazzo, R.; Carfì, A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. [Google Scholar] [CrossRef]
- Miller, D.J.; Zhang, Y.M.; Rock, C.O.; White, S.W. Structure of RhlG, an Essential beta-Ketoacyl Reductase in the Rhamnolipid Biosynthetic Pathway of Pseudomonas aeruginosa. J. Biol. Chem. 2006, 281, 18025–18032. [Google Scholar] [CrossRef]
- Ilangovan, A.; Fletcher, M.; Rampioni, G.; Pustelny, C.; Rumbaugh, K.; Heeb, S.; Camara, M.; Truman, A.; Chhabra, S.R.; Emsley, J.; et al. Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLoS Pathog. 2013, 9, e1003508. [Google Scholar] [CrossRef]
- Youssef, F.S.; Ashour, M.L.; Sobeh, M.; El-Beshbishy, H.A.; Singab, A.N.; Wink, M. Eremophila maculata—isolation of a rare naturally-occurring lignan glycoside and the hepatoprotective activity of the leaf extract. Phytomedicine 2016, 23, 1484–1493. [Google Scholar] [CrossRef]
- Ashour, M.L.; Youssef, F.S.; Gad, H.A.; Wink, M. Inhibition of cytochrome p450 (cyp3A4) activity by extracts from 57 plants used in Traditional Chinese Medicine (TCM). Pharmacogn. Mag. 2017, 13, 300. [Google Scholar]
Sample Availability: Samples of the plant materials are available from the authors. |
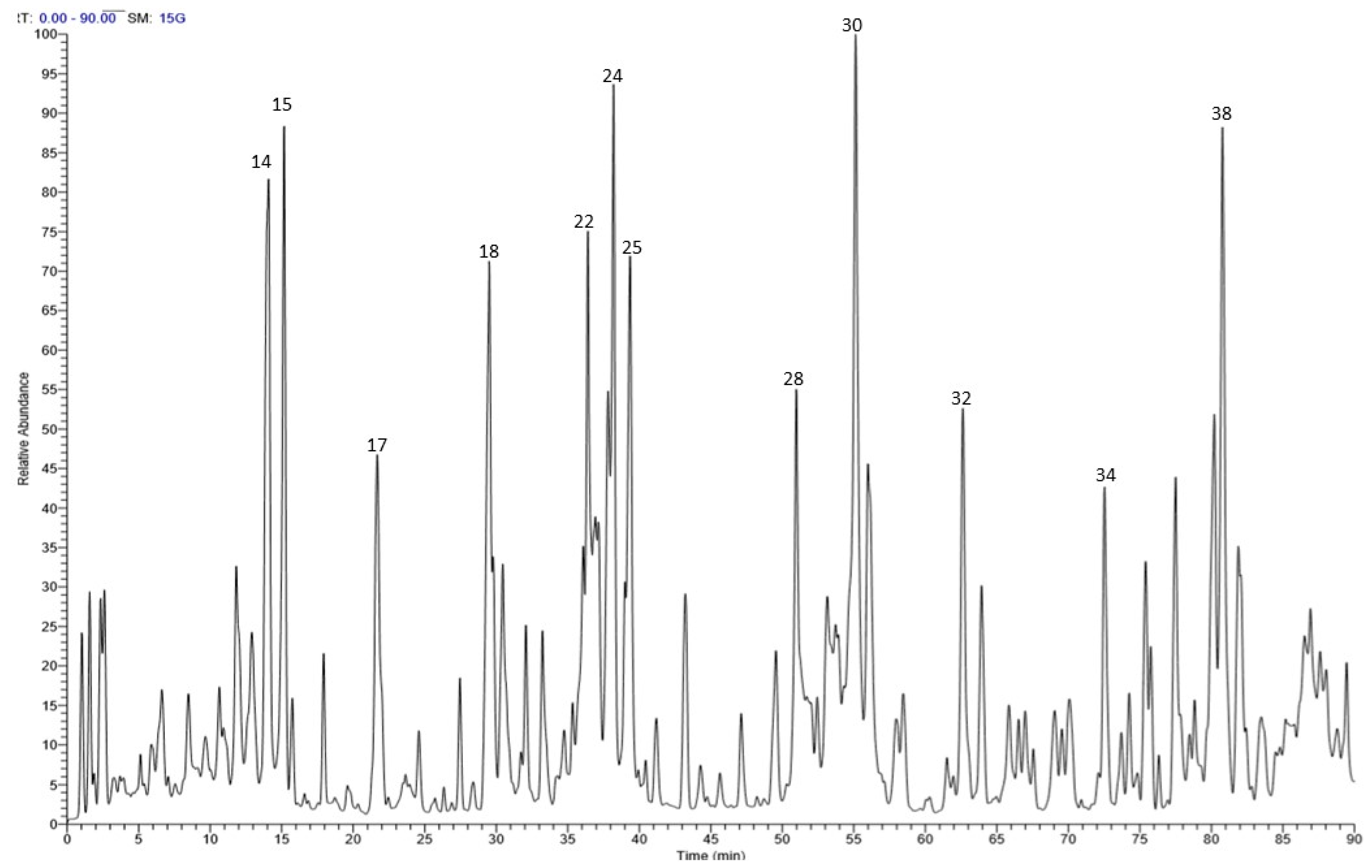

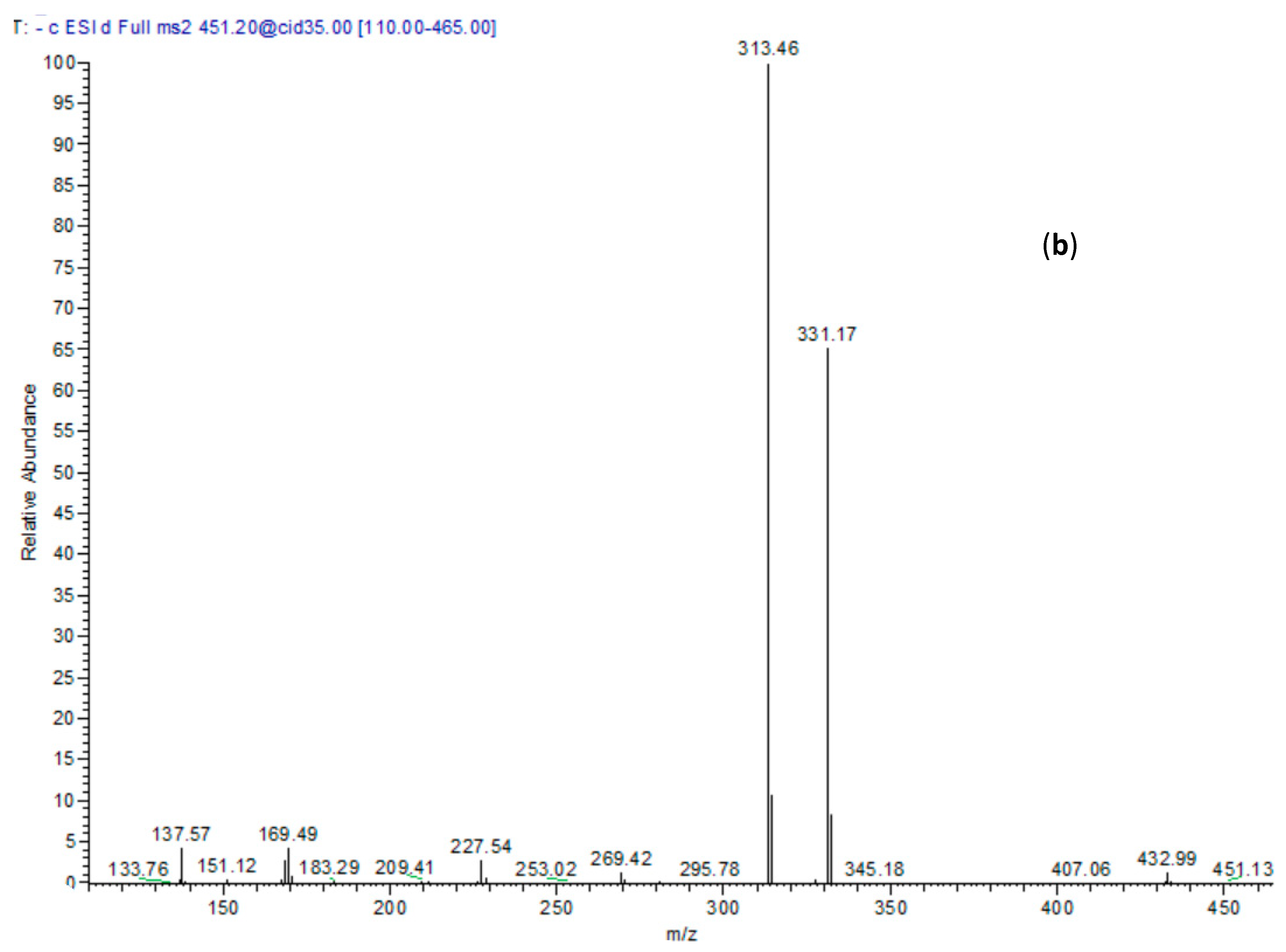
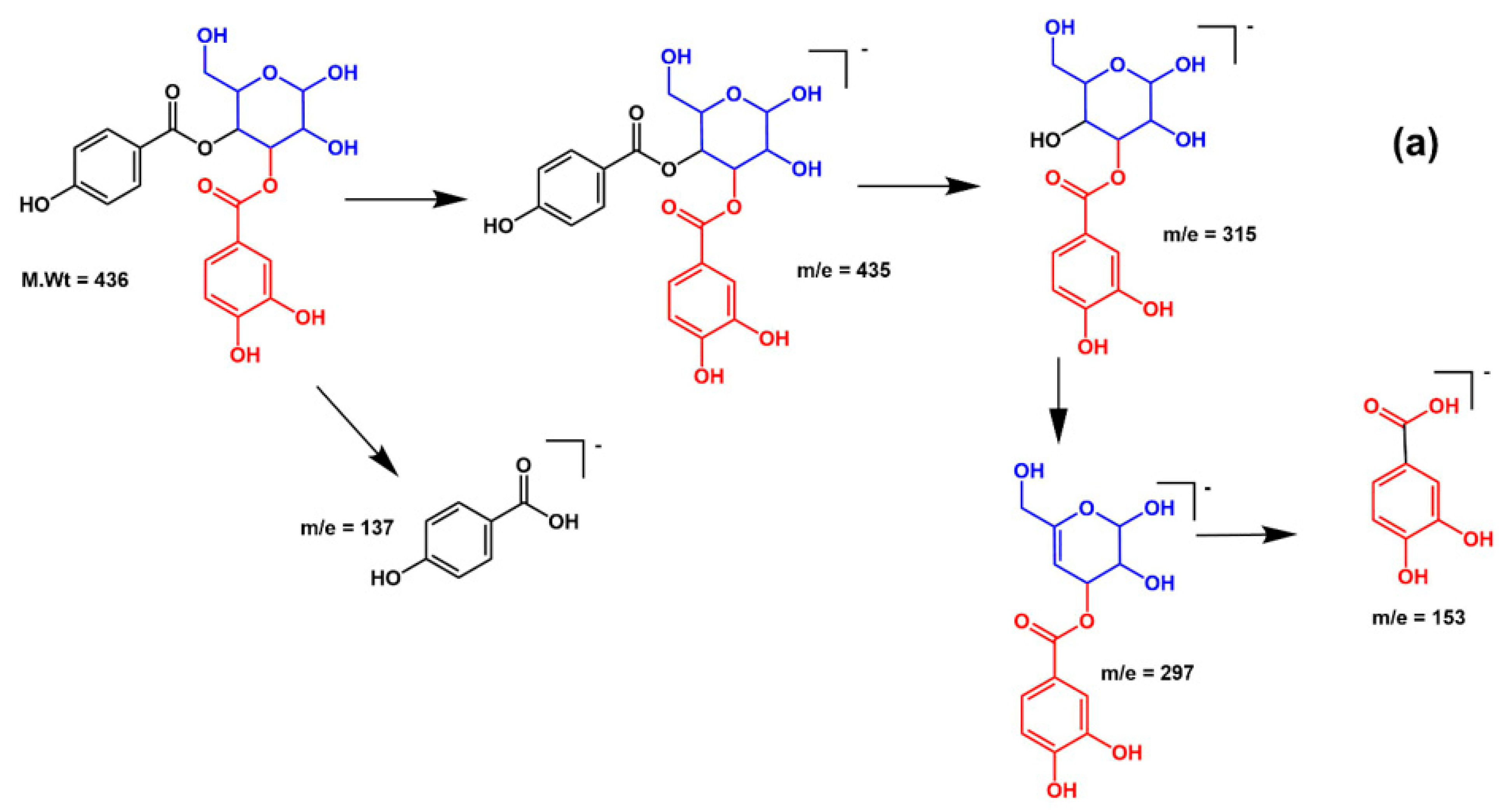
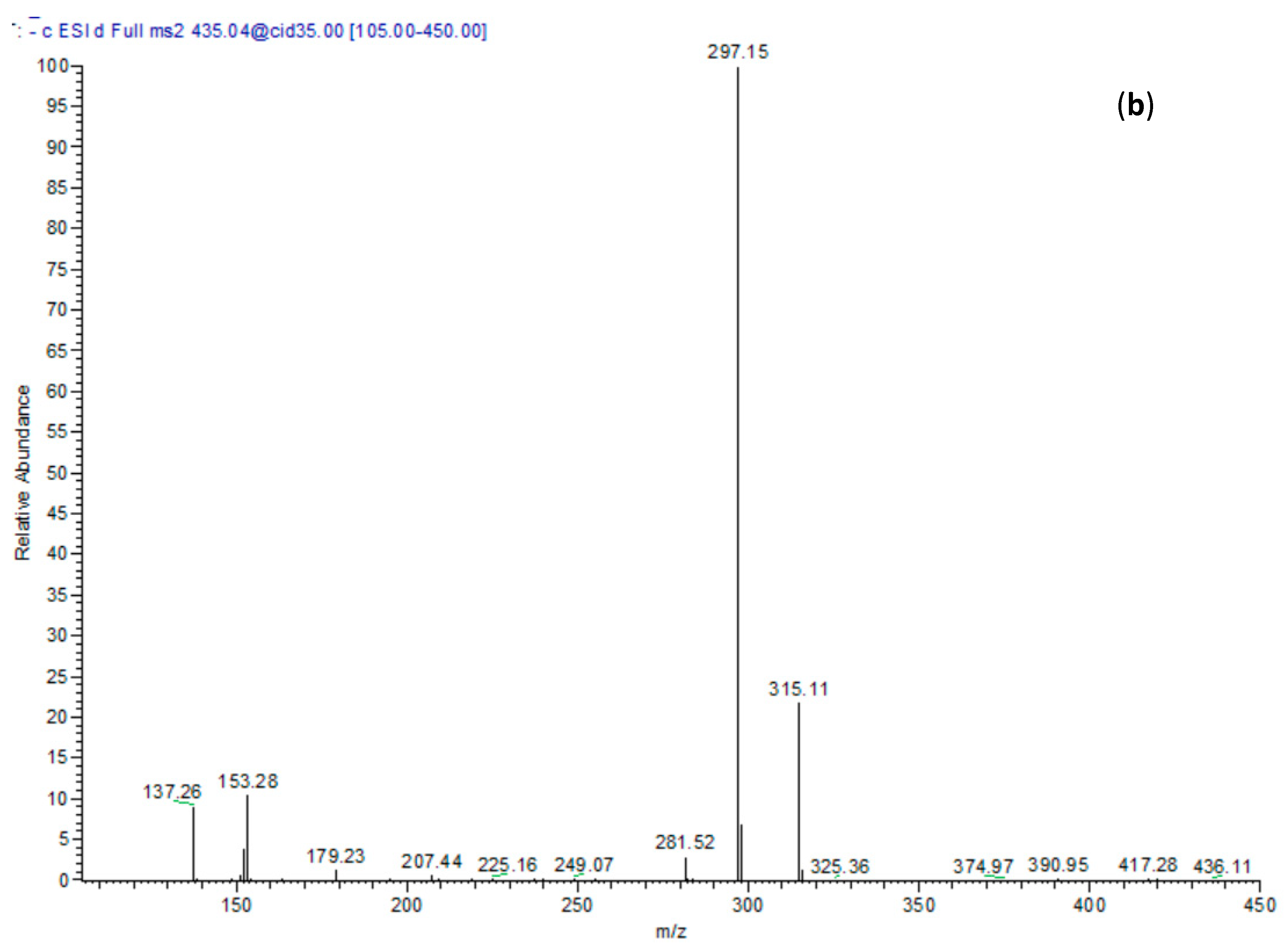

| No. | RT | M-H m/z | MS/MS | Tentatively Identified Compounds |
|---|---|---|---|---|
| 1 | 0.95 | 507 | 169, 345, 417 | Gallic acid glucuronide-glucoside |
| 2 | 1.05 | 133 | - | Malic acid |
| 3 | 1.37 | 191 | 111, 173 | Quinic acid |
| 4 | 2.29 | 315 | 153 | Protocatechuic acid 3-O-hexoside * |
| 5 | 2.60 | 331 | 125, 169, 313 | Gallic acid glucoside |
| 6 | 2.79 | 447 | 163, 315 | Coumaric acid galloyl pentoside |
| 7 | 3.46 | 609 | 305, 441, 457 | (epi)Gallocatechin digallate |
| 8 | 5.24 | 593 | 289, 425, 575 | (epi)Catechin-(epi)-gallocatechin |
| 9 | 5.91 | 865 | 289, 451, 595 | (epi)Catechin-(epi)catechin-(epi)catechin |
| 10 | 6.55 | 609 | 305, 423, 441 | (epi)Gallocatechin-(epi)-gallocatechin |
| 11 | 8.46 | 305 | 179, 221, 287 | (epi)Gallocatechin |
| 12 | 11.82 | 881 | 289, 577, 695 | (epi)Gallocatechin-(epi)catechin-(epi)catechin |
| 13 | 12.81 | 577 | 289, 407, 451 | (epi)Catechin-(epi)catechin # |
| 14 | 13.56 | 577 | 289, 407, 451 | (epi)Catechin-(epi)catechin # |
| 15 | 14.16 | 289 | 179, 285, 245 | Epicatechin |
| 16 | 15.12 | 289 | 179, 285, 245 | Catechin # |
| 17 | 21.73 | 423 | 161, 285 | Salicortin * |
| 18 | 29.58 | 451 | 169, 313, 331 | p-Hydroxy benzoyl galloyl glucose |
| 19 | 30.45 | 435 | 153, 297, 315 | p-Hydroxy benzoyl protocatechuic acid glucose |
| 20 | 32.07 | 423 | 145, 163, 307 | Grandidentatin # |
| 21 | 33.26 | 423 | 145, 163, 307 | Grandidentatin isomer |
| 22 | 36.41 | 477 | 151, 179, 315 | Isorhamnetin-3-O-glucoside |
| 23 | 37.82 | 431 | 145, 163, 307 | Trichocarposide * |
| 24 | 38.19 | 431 | 145, 163, 307 | Trichocarposide isomer |
| 25 | 39.33 | 431 | 145, 163, 307 | Trichocarposide isomer |
| 26 | 43.23 | 435 | 179, 273 | Phlorizin |
| 27 | 49.29 | 569 | 307, 423, 431 | Coumaroyl dihydrobenzoylsalicin * |
| 28 | 5.92 | 631 | 191, 329, 353 | Chlorogenic acid derivative |
| 29 | 53.02 | 569 | 307, 423, 431 | Coumaroyl dihydrobenzoylsalicin isomer * |
| 30 | 55.21 | 527 | 155, 405 | Tremulacin * |
| 31 | 56.32 | 577 | 269 | Terniflorin * |
| 32 | 62.93 | 309 | 171, 251, 291 | Fatty acid derivative |
| 33 | 70.32 | 311 | 293, 311 | Eicosanoic acid |
| 34 | 72.48 | 723 | 269, 453, 559, 577 | Coumaroyl-terniflorin |
| 35 | 75.53 | 271 | 209, 253, 271 | Unidentified |
| 36 | 77.26 | 293 | 171, 235, 275, 293 | Hydroxy-octadecatrienoic acid * |
| 37 | 79.92 | 295 | 171, 277, 295 | Hydroxy-octadecadienoic acid * |
| 38 | 81.01 | 295 | 171, 277, 295 | Hydroxy-octadecadienoic acid isomer |
| 39 | 82.39 | 293 | 113, 249, 293 | Hydroxy-octadecatrienoic acid isomer |
| Compound Name | 2UV0 Protein | 2B4Q Protein | 4JVD Protein | |||
|---|---|---|---|---|---|---|
| Free Binding Energy | Amino Acid Interactions | Free Binding Energy | Amino Acid Interactions | Free Binding Energy | Amino Acid Interactions | |
| Caffeoylmalic acid | −41.66 | ASP E:73, ARG E:61, LEU E:110, TRP E:88, ALA E:105 | −74.39 | ARG A:41, ARG A:19, ASP A:42, ASN A:92, SER A:18, GLY A:16 | −30.09 | TYR:258, LEU: 197, SER:196 |
| Rutin | FD | -- | −1.51 | ARG A:194, ARG A:19, ASP A:42, ASN A: 92, SER A:197, MET A:199, TYR A:162, PHE A:195, GLY A:94, GLY A:20; THR A A:95 | Unfavorable binding | -- |
| Quercetin-3-O-glucoside | Unfavorable binding | -- | −4.86 | GLY A:94, ASN A:92 ARG A:198, GLY A:16, ALA A:93, SER A:197 | −7.04 | ARG A: 209 |
| Kaempferol-3-O-galactoside | Unfavorable binding | -- | −1.82 | GLY A:94, ASN A:92, ARG A:19, MET A:199, ALA A:93, SER A:18 | −4.19 | ARG A: 209, ASP A: 264, GLN A: 194, PRO A: 210 |
| Kaempferide-3-O-glucoside | Unfavorable binding | -- | Unfavorable binding | -- | −3.46 | TYR A: 258, LEU A: 197, LEU A: 208, GLN A: 194 SER A: 196, ARG A: 209 |
| Isorhamnetin-3-O-rhamnoside | Unfavorable binding | -- | Unfavorable binding | -- | −3.41 | TYR A: 258, ASP A: 264, GLU A: 259, ASN A: 206, THR A: 265 |
| (epi)Catechin-(epi)catechin | FD | -- | FD | -- | −36.51 | ARG A: 209, LEU A: 208, ASP A: 264, THR A: 265, ASN A: 206, LEU A: 207 |
| Epicatechin | −39.23 | ASP E:73, SER E:129, THR E:75, LEU E:125 | −37.91 | ARG A:19, GLY A:16, GLY A:22, GLY A:147, ASN A:92, ILE A:21, LYS A:166 | −26.83 | ILE A: 236, ASP A: 264, LEU A: 207 |
| Catechin | −36.66 | ARG E:61, SER E: 129, THR E:75, LEU E:125 | −35.51 | ARG A:19, GLY A:94, ASN A:92, GLY A:147, GLY A:193, SER A:148 | −26.13 | TYR A: 258, LEU A: 208 LEU A: 207 |
| Tremulacin | FD | -- | −12.67 | ARG A:19, ASN A:92, MET A:199, PHE A:195, SER A:197, GLY A:20, TYR A:162 | −6.83 | TYR A: 258, LEU A: 207, LEU A: 208, SER A: 196, LEU A: 207 |
| Salicortin | Unfavorable binding | -- | −6.31 | ARG A:19, ARG A:41, ASN A:92, MET A:199, TYR A:162, GLY A:147, GLY A:16, GLY A:94 | Unfavorable binding | -- |
| Trichocarposide | Unfavorable binding | -- | −15.87 | ARG A:19, ARG A:41, ASN A:92, MET A:199, TYR A:162, GLY A:147, SER A:197 | −12.63 | ILE A:186, LEU A: 207 |
| p-Hydroxy benzoyl galloyl glucose | Unfavorable binding | -- | −33.82 | ARG A:19, GLY A:16, GLY A:22, SER A:197, ILE A:21, TYR A:162, GLY A:94, PHE A:195, ASN A:92 | −33.62 | ILE A: 186, TYR A: 258, ASN A: 206, LEU A: 208, SER A: 196, LEU A: 207 |
| p-Hydroxy benzoyl protocatechuic acid glucose | Unfavorable binding | -- | −30.41 | ARG A:19, GLY A:16, GLY A:22, GLY A:147, ILE A:21, LYS A:164, GLY A:94, | −28.96 | ILE A: 186, TYR A: 258, GLN A: 151, LEU A: 207 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafa, I.; Abbas, H.A.; Ashour, M.L.; Yasri, A.; El-Shazly, A.M.; Wink, M.; Sobeh, M. Polyphenols from Salix tetrasperma Impair Virulence and Inhibit Quorum Sensing of Pseudomonas aeruginosa. Molecules 2020, 25, 1341. https://doi.org/10.3390/molecules25061341
Mostafa I, Abbas HA, Ashour ML, Yasri A, El-Shazly AM, Wink M, Sobeh M. Polyphenols from Salix tetrasperma Impair Virulence and Inhibit Quorum Sensing of Pseudomonas aeruginosa. Molecules. 2020; 25(6):1341. https://doi.org/10.3390/molecules25061341
Chicago/Turabian StyleMostafa, Islam, Hisham A. Abbas, Mohamed L. Ashour, Abdelaziz Yasri, Assem M. El-Shazly, Michael Wink, and Mansour Sobeh. 2020. "Polyphenols from Salix tetrasperma Impair Virulence and Inhibit Quorum Sensing of Pseudomonas aeruginosa" Molecules 25, no. 6: 1341. https://doi.org/10.3390/molecules25061341
APA StyleMostafa, I., Abbas, H. A., Ashour, M. L., Yasri, A., El-Shazly, A. M., Wink, M., & Sobeh, M. (2020). Polyphenols from Salix tetrasperma Impair Virulence and Inhibit Quorum Sensing of Pseudomonas aeruginosa. Molecules, 25(6), 1341. https://doi.org/10.3390/molecules25061341








