PNA-Based MicroRNA Detection Methodologies
Abstract
1. Introduction
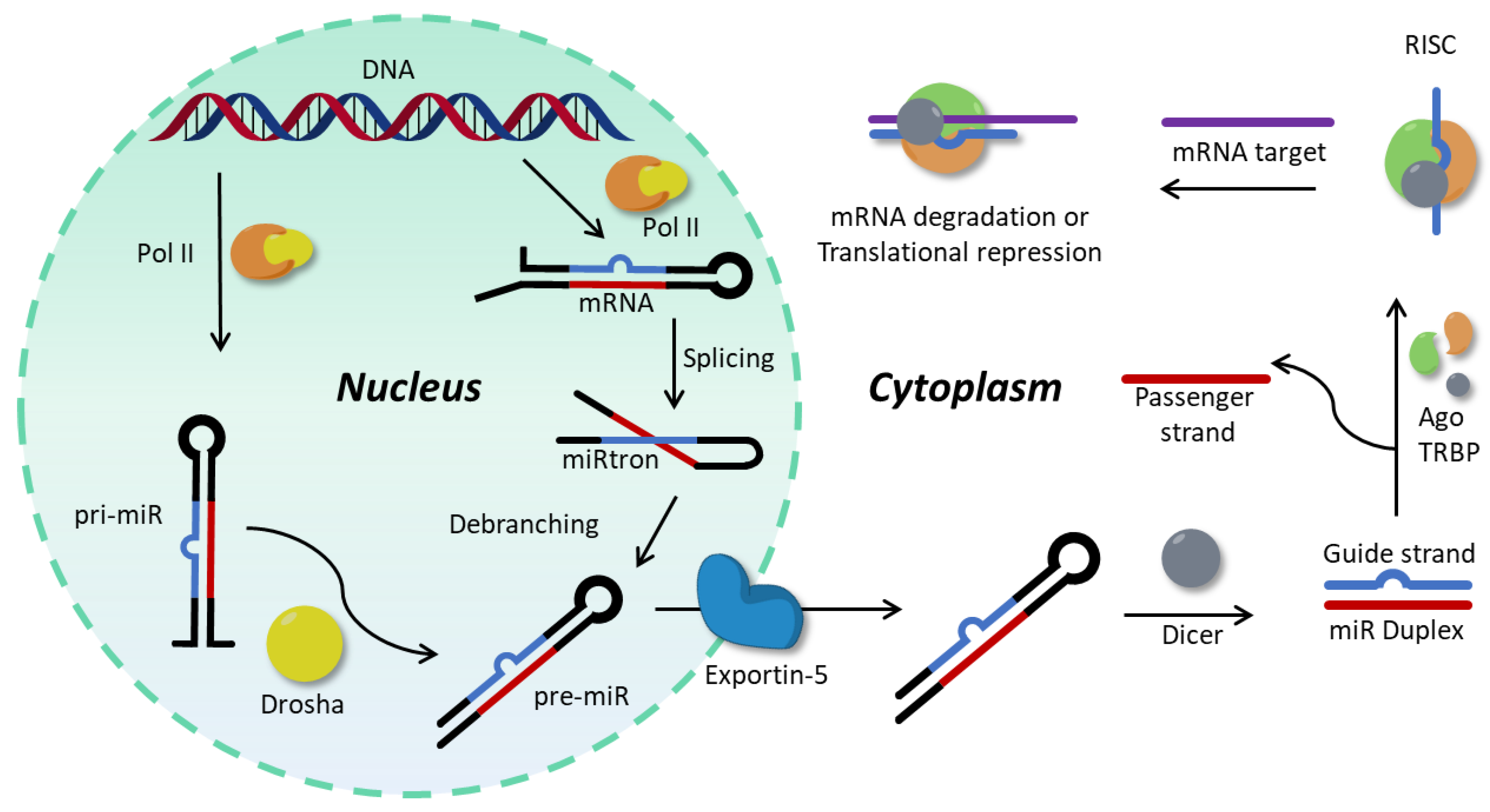
1.1. MicroRNA Background and Importance as Biomarkers
1.2. Peptide Nucleic Acids
2. Electrochemical Detection Methodologies
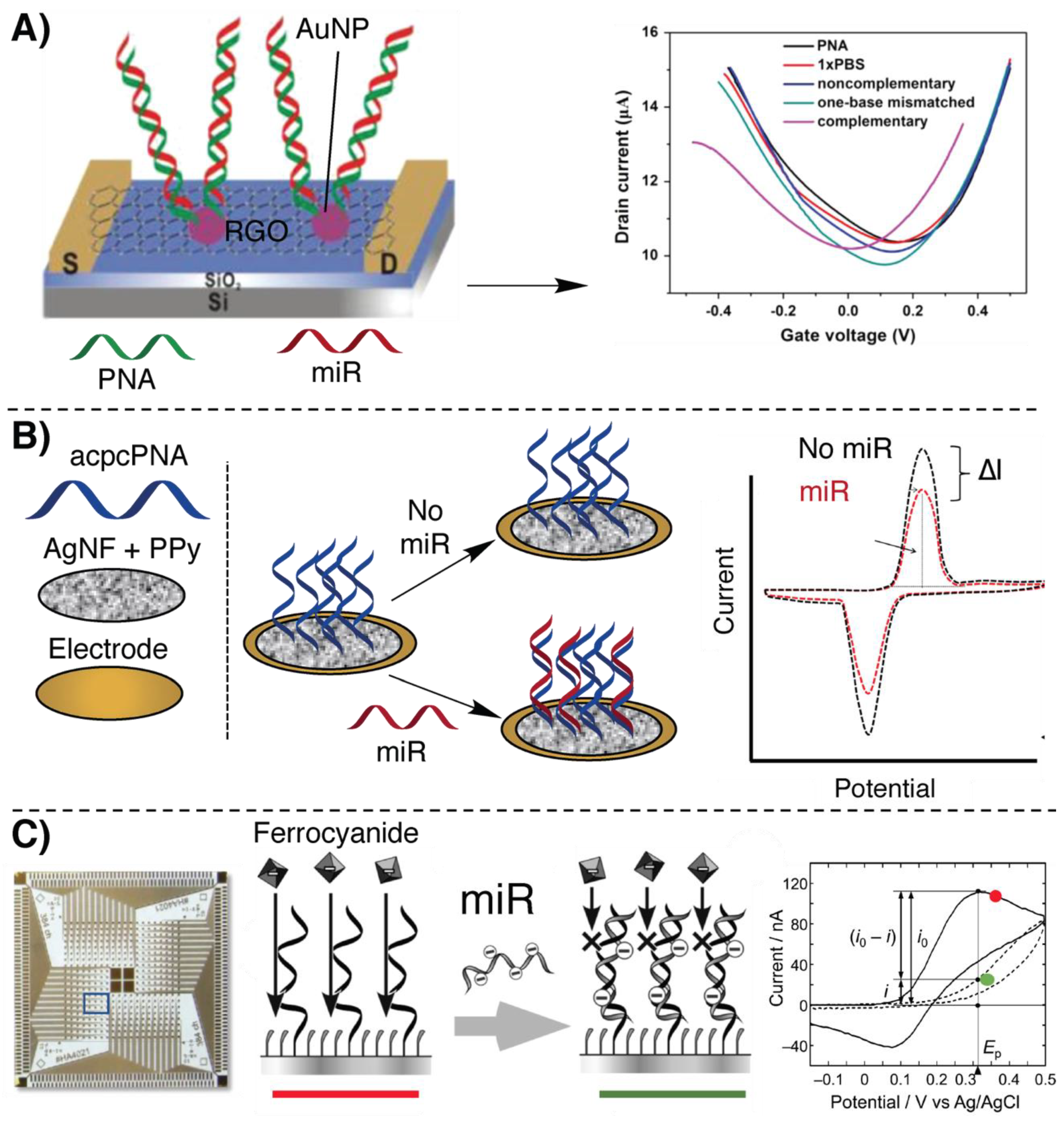
2.1. Electrochemical Detection Methodologies Based on Hybridization
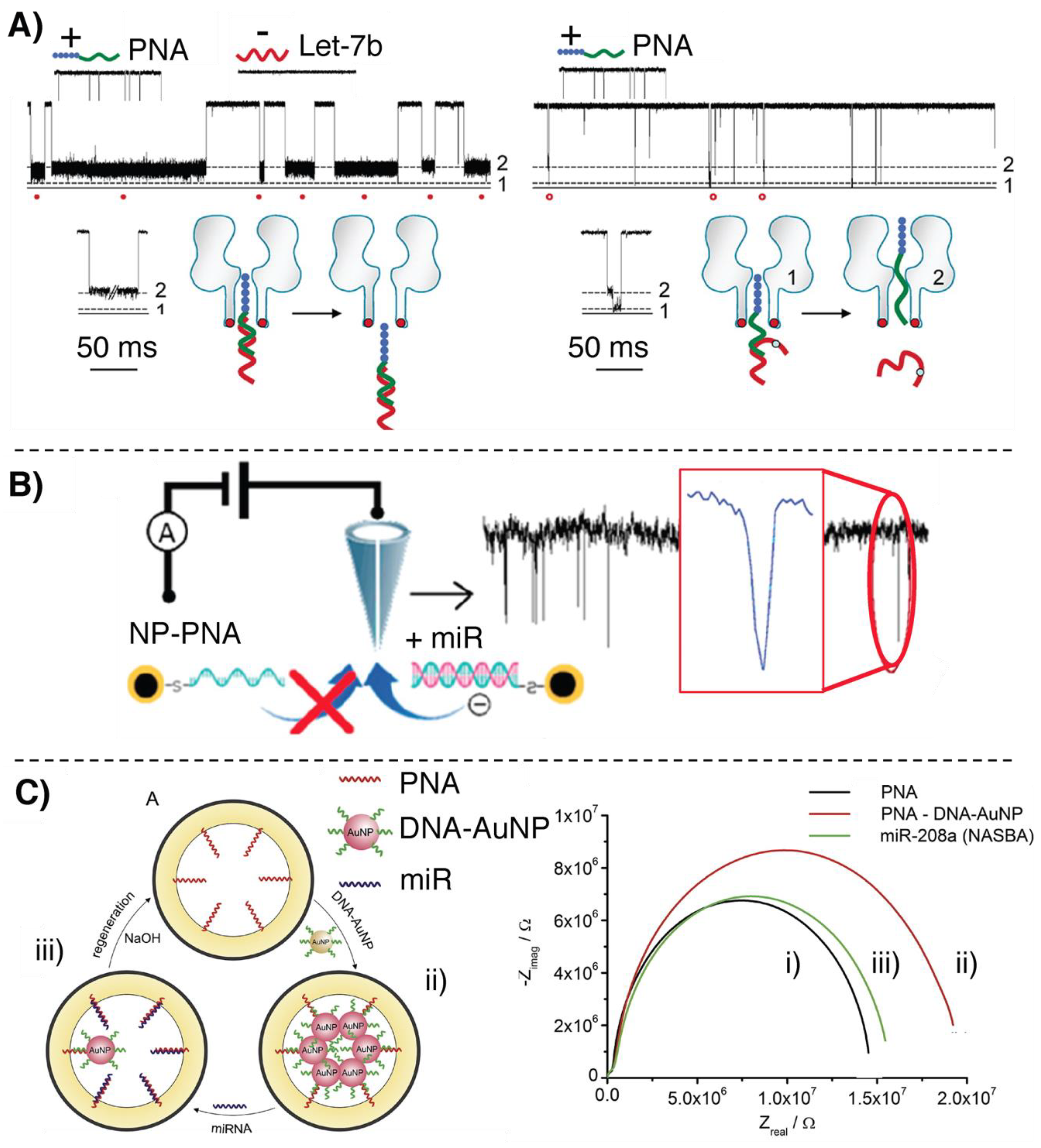
2.2. Nanopore-Based Methodologies
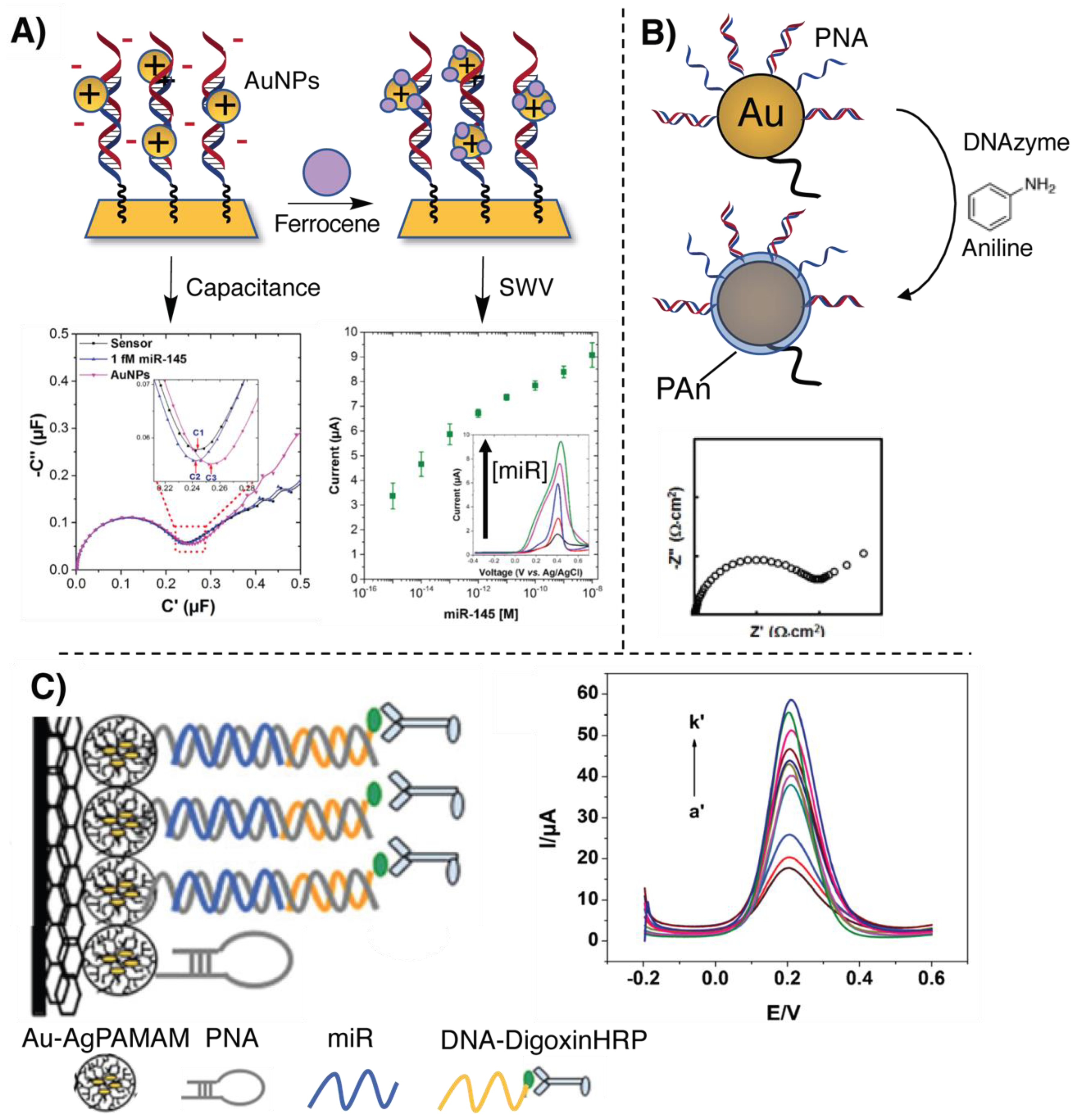
2.3. Signal Amplification Methodologies
3. Fluorescence-Based Methodologies
3.1. Fluorescence-Based Detection Methodologies Based on Hybridization
3.2. Templated Reactions
3.2.1. Hybridization-Triggered Templated Reactions
3.2.2. Light-Triggered Templated Reactions
3.3. Fluorescence-Based Detection Methodologies Featuring Signal Amplification
4. Colorimetric Detection Methodologies
4.1. Colorimetric Assays Based on Hybridization
4.2. Colorimetric Assays Based on Templated Reactions
5. Other Methodologies
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Search Strategy and Selection Criteria
References
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Gao, F.; Kataoka, M.; Liu, N.; Liang, T.; Huang, Z.P.; Gu, F.; Ding, J.; Liu, J.; Zhang, F.; Ma, Q.; et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Eskildsen, T.; Taipaleenmäki, H.; Stenvang, J.; Abdallah, B.M.; Ditzel, N.; Nossent, A.Y.; Bak, M.; Kauppinen, S.; Kassem, M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6139–6144. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-Resolution structure of the pre-microrna nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2018, 20. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Miller, B.H.; Wahlestedt, C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010, 1338, 89–99. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. 2019, 99, 452–469. [Google Scholar] [CrossRef]
- Shi, H.; Yang, F.; Li, W.; Zhao, W.; Nie, K.; Dong, B.; Liu, Z. A review: Fabrications, detections and applications of peptide nucleic acids (PNAs) microarray. Biosens. Bioelectron. 2015, 66, 481–489. [Google Scholar] [CrossRef]
- Sassolas, A.; Leca-Bouvier, B.D.; Blum, L.J. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Creighton, C.J.; Fountain, M.D.; Yu, Z.; Nagaraja, A.K.; Zhu, H.; Khan, M.; Olokpa, E.; Zariff, A.; Gunaratne, P.H.; Matzuk, M.M.; et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010, 70, 1906–1915. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.C.N.; Xie, W.; Yang, M.; Hsieh, C.L.; Drouin, S.; Lee, G.S.M.; Kantoff, P.W. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate 2013, 73, 346–354. [Google Scholar] [CrossRef]
- Eades, G.; Wolfson, B.; Zhang, Y.; Li, Q.; Yao, Y.; Zhou, Q. LincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting Arf6. Mol. Cancer Res. 2015, 13, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 2013, 532, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raitoharju, E.; Lyytikäinen, L.P.; Levula, M.; Oksala, N.; Mennander, A.; Tarkka, M.; Klopp, N.; Illig, T.; Kähönen, M.; Karhunen, P.J.; et al. MiR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011, 219, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Nielsen, P.; Egholm, M.; Berg, R.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Nielsen, P.E. PNA Technology. Mol. Biotechnol. 2004, 26, 233–248. [Google Scholar] [CrossRef]
- Demidov, V.; Frank-kamenetskii, M.D.; Egholm, M.; Buchardt, O.; Nielsen, P.E. Sequence selective double strand DNA cleavage by peptide nucleic acid (PNA) targeting using nuclease S1. Nucleic Acids Res. 1993, 21, 2103–2107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Appella, D.H. Advantages of Peptide Nucleic Acids as Diagnostic Platforms for Detection of Nucleic Acids in Resource-Limited Settings. J. Infect. Dis. 2010, 201, S42–S45. [Google Scholar] [CrossRef] [PubMed]
- Anstaett, P.; Zheng, Y.; Thai, T.; Funston, A.M.; Bach, U.; Gasser, G. Synthesis of stable peptide nucleic acid-modified gold nanoparticles and their assembly onto gold surfaces. Angew. Chem.-Int. Ed. 2013, 52, 4217–4220. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Lautner, G.; Gyurcsányi, R.E. Reliable microspotting methodology for peptide-nucleic acid layers with high hybridization efficiency on gold SPR imaging chips. Anal. Methods 2015, 7, 6077–6082. [Google Scholar] [CrossRef]
- Cadoni, E.; Rosa-Gastaldo, D.; Manicardi, A.; Mancin, F.; Madder, A. Exploiting Double Exchange Diels-Alder Cycloadditions for Immobilization of Peptide Nucleic Acids on Gold Nanoparticles. Front. Chem. 2020, 8, 1–7. [Google Scholar] [CrossRef]
- Fabbri, E.; Manicardi, A.; Tedeschi, T.; Sforza, S.; Bianchi, N.; Brognara, E.; Finotti, A.; Breveglieri, G.; Borgatti, M.; Corradini, R.; et al. Modulation of the Biological Activity of microRNA-210 with Peptide Nucleic Acids (PNAs). ChemMedChem 2011, 6, 2192–2202. [Google Scholar] [CrossRef]
- Brognara, E.; Fabbri, E.; Aimi, F.; Manicardi, A.; Bianchi, N.; Finotti, A.; Breveglieri, G.; Borgatti, M.; Corradini, R.; Marchelli, R.; et al. Peptide nucleic acids targeting miR-221 modulate p27Kip1 expression in breast cancer MDA-MB-231 cells. Int. J. Oncol. 2012, 41, 2119–2127. [Google Scholar] [CrossRef]
- Brandén, L.J.; Mohamed, A.J.; Smith, C.I. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999, 17, 784–787. [Google Scholar] [CrossRef]
- Gambari, R. Peptide nucleic acids: A review on recent patents and technology transfer. Expert Opin. Ther. Pat. 2014, 24, 267–294. [Google Scholar] [CrossRef]
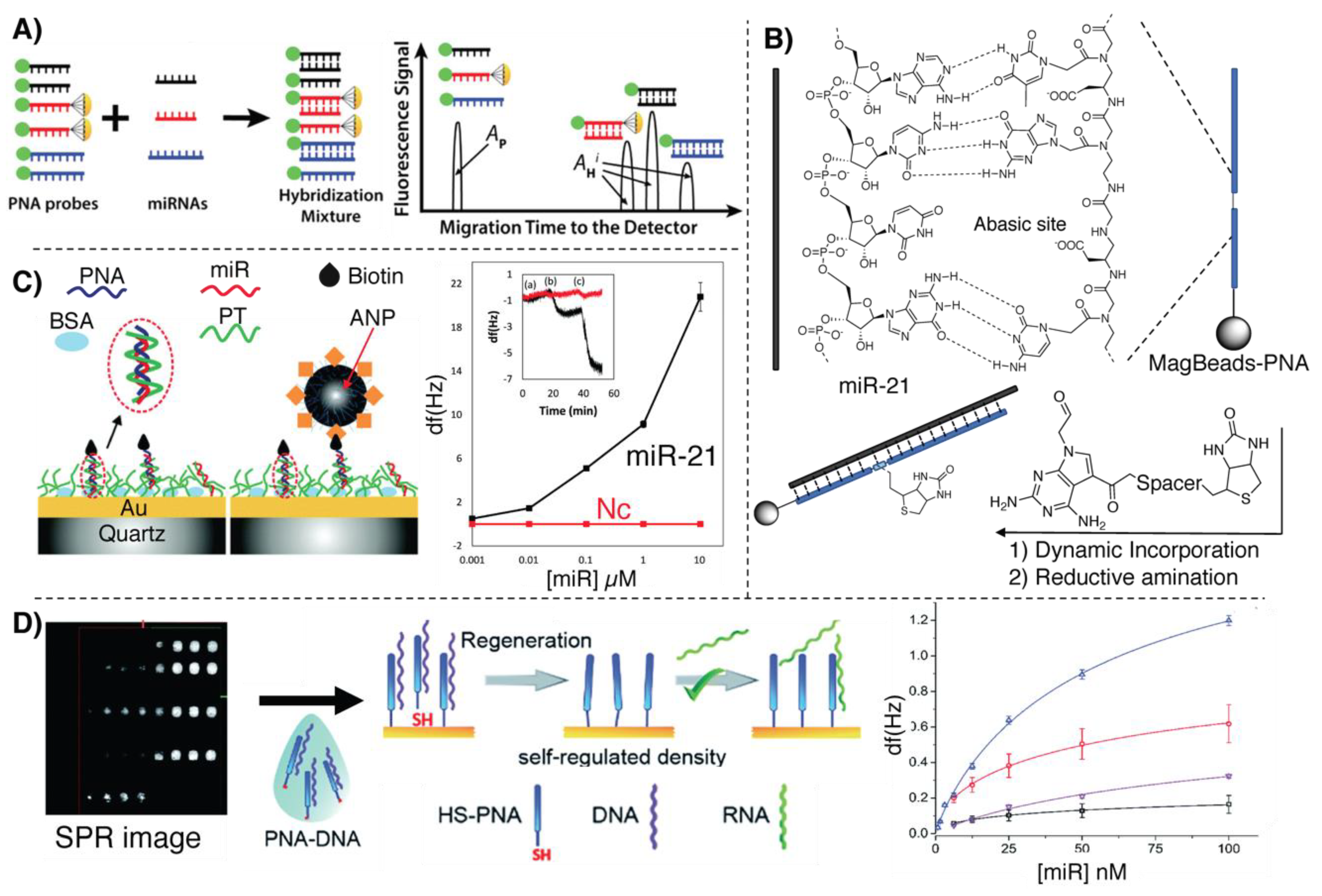
- Gillespie, P.; Ladame, S.; O’Hare, D. Molecular methods in electrochemical microRNA detection. Analyst 2019, 144, 114–129. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Labib, M.; Sargent, E.H.; Kelley, S.O. Electrochemical Methods for the Analysis of Clinically Relevant Biomolecules. Chem. Rev. 2016, 116, 9001–9090. [Google Scholar] [CrossRef] [PubMed]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Giuffrida, M.C.; Spoto, G. Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis. Molecules 2017, 22, 1951. [Google Scholar] [CrossRef]
- Singh, R.P.; Oh, B.K.; Choi, J.W. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry 2010, 79, 153–161. [Google Scholar] [CrossRef]
- Willner, I.; Katz, E. Integration of Layered Redox Proteins and Conductive Supports for Bioelectronic Applications. Angew. Chemie Int. Ed. 2000, 39, 1180–1218. [Google Scholar] [CrossRef]
- Schwierz, F. Graphene transistors. Nat. Nanotechnol. 2010, 5, 487–496. [Google Scholar] [CrossRef]
- Cai, B.; Wang, S.; Huang, L.; Ning, Y.; Zhang, Z.; Zhang, G.-J. Ultrasensitive Label-Free Detection of PNA–DNA Hybridization by Reduced Graphene Oxide Field-Effect Transistor Biosensor. ACS Nano 2014, 8, 2632–2638. [Google Scholar] [CrossRef]
- Cai, B.; Huang, L.; Zhang, H.; Sun, Z.; Zhang, Z.; Zhang, G.-J. Gold nanoparticles-decorated graphene field-effect transistor biosensor for femtomolar MicroRNA detection. Biosens. Bioelectron. 2015, 74, 329–334. [Google Scholar] [CrossRef]
- Kangkamano, T.; Numnuam, A.; Limbut, W.; Kanatharana, P.; Vilaivan, T.; Thavarungkul, P. Pyrrolidinyl PNA polypyrrole/silver nanofoam electrode as a novel label-free electrochemical miRNA-21 biosensor. Biosens. Bioelectron. 2018, 102, 217–225. [Google Scholar] [CrossRef]
- Suparpprom, C.; Srisuwannaket, C.; Sangvanich, P.; Vilaivan, T. Synthesis and oligodeoxynucleotide binding properties of pyrrolidinyl peptide nucleic acids bearing prolyl-2-aminocyclopentanecarboxylic acid (ACPC) backbones. Tetrahedron Lett. 2005, 46, 2833–2837. [Google Scholar] [CrossRef]
- Aoki, H.; Torimura, M.; Nakazato, T. 384-Channel electrochemical sensor array chips based on hybridization-triggered switching for simultaneous oligonucleotide detection. Biosens. Bioelectron. 2019, 136, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 2011, 6, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, B.M.; Bashir, R. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011, 6, 615–624. [Google Scholar] [CrossRef]
- Tian, K.; He, Z.; Wang, Y.; Chen, S.J.; Gu, L.Q. Designing a polycationic probe for simultaneous enrichment and detection of microRNAs in a nanopore. ACS Nano 2013, 7, 3962–3969. [Google Scholar] [CrossRef]
- Wang, H.; Tang, H.; Yang, C.; Li, Y. Selective Single Molecule Nanopore Sensing of microRNA Using PNA Functionalized Magnetic Core–Shell Fe 3 O 4 –Au Nanoparticles. Anal. Chem. 2019, 91, 7965–7970. [Google Scholar] [CrossRef]
- Zhang, Y.; Rana, A.; Stratton, Y.; Czyzyk-Krzeska, M.F.; Esfandiari, L. Sequence-Specific Detection of MicroRNAs Related to Clear Cell Renal Cell Carcinoma at fM Concentration by an Electroosmotically Driven Nanopore-Based Device. Anal. Chem. 2017, 89, 9201–9208. [Google Scholar] [CrossRef]
- Lautner, G.; Plesz, M.; Jágerszki, G.; Fürjes, P.; Gyurcsányi, R.E. Nanoparticle displacement assay with electrochemical nanopore-based sensors. Electrochem. commun. 2016, 71, 13–17. [Google Scholar] [CrossRef][Green Version]
- Makra, I.; Brajnovits, A.; Jágerszki, G.; Fürjes, P.; Gyurcsányi, R.E. Potentiometric sensing of nucleic acids using chemically modified nanopores. Nanoscale 2017, 9, 739–747. [Google Scholar] [CrossRef]
- Fozooni, T.; Ravan, H.; Sasan, H. Signal Amplification Technologies for the Detection of Nucleic Acids: From Cell-Free Analysis to Live-Cell Imaging. Appl. Biochem. Biotechnol. 2017, 183, 1224–1253. [Google Scholar] [CrossRef]
- Jolly, P.; Batistuti, M.R.; Miodek, A.; Zhurauski, P.; Mulato, M.; Lindsay, M.A.; Estrela, P. Highly sensitive dual mode electrochemical platform for microRNA detection. Sci. Rep. 2016, 6, 36719. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Shen, W.; Ren, Y.; Gao, Z. A highly sensitive microRNA biosensor based on hybridized microRNA-guided deposition of polyaniline. Biosens. Bioelectron. 2014, 60, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jiang, S.; Wang, L.; Zhang, Z.; Xie, G. Direct detection of microRNA-126 at a femtomolar level using a glassy carbon electrode modified with chitosan, graphene sheets, and a poly(amidoamine) dendrimer composite with gold and silver nanoclusters. Microchim. Acta 2015, 182, 77–84. [Google Scholar] [CrossRef]
- Huang, X.; Song, J.; Yung, B.C.; Huang, X.; Xiong, Y.; Chen, X. Ratiometric optical nanoprobes enable accurate molecular detection and imaging. Chem. Soc. Rev. 2018, 47, 2873–2920. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular beacon probes that fluoresce on hybridiztion. Nat. Publ. Gr. 1996, 14, 303–308. [Google Scholar]
- Vilaivan, T. Fluorogenic PNA probes. Beilstein J. Org. Chem. 2018, 14, 253–281. [Google Scholar] [CrossRef]
- Croci, S.; Manicardi, A.; Rubagotti, S.; Bonacini, M.; Iori, M.; Capponi, P.C.; Cicoria, G.; Parmeggiani, M.; Salvarani, C.; Versari, A.; et al. 64 Cu and fluorescein labeled anti-miRNA peptide nucleic acids for the detection of miRNA expression in living cells. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Hwang, D.W.; Kim, H.Y.; Li, F.; Park, J.Y.; Kim, D.; Park, J.H.; Han, H.S.; Byun, J.W.; Lee, Y.S.; Jeong, J.M.; et al. In vivo visualization of endogenous miR-21 using hyaluronic acid-coated graphene oxide for targeted cancer therapy. Biomaterials 2017, 121, 144–154. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, S.; Na, H.K.; Min, D.H. MicroRNA-Responsive Drug Release System for Selective Fluorescence Imaging and Photodynamic Therapy In Vivo. Adv. Healthc. Mater. 2016, 5, 2386–2395. [Google Scholar] [CrossRef]
- Tian, F.; Lyu, J.; Shi, J.; Yang, M. Graphene and graphene-like two-denominational materials based fluorescence resonance energy transfer (FRET) assays for biological applications. Biosens. Bioelectron. 2017, 89, 123–135. [Google Scholar] [CrossRef]
- Oh, H.J.; Kim, J.; Park, H.; Chung, S.; Hwang, D.W.; Lee, D.S. Graphene-oxide quenching-based molecular beacon imaging of exosome-mediated transfer of neurogenic miR-193a on microfluidic platform. Biosens. Bioelectron. 2019, 126, 647–656. [Google Scholar] [CrossRef] [PubMed]
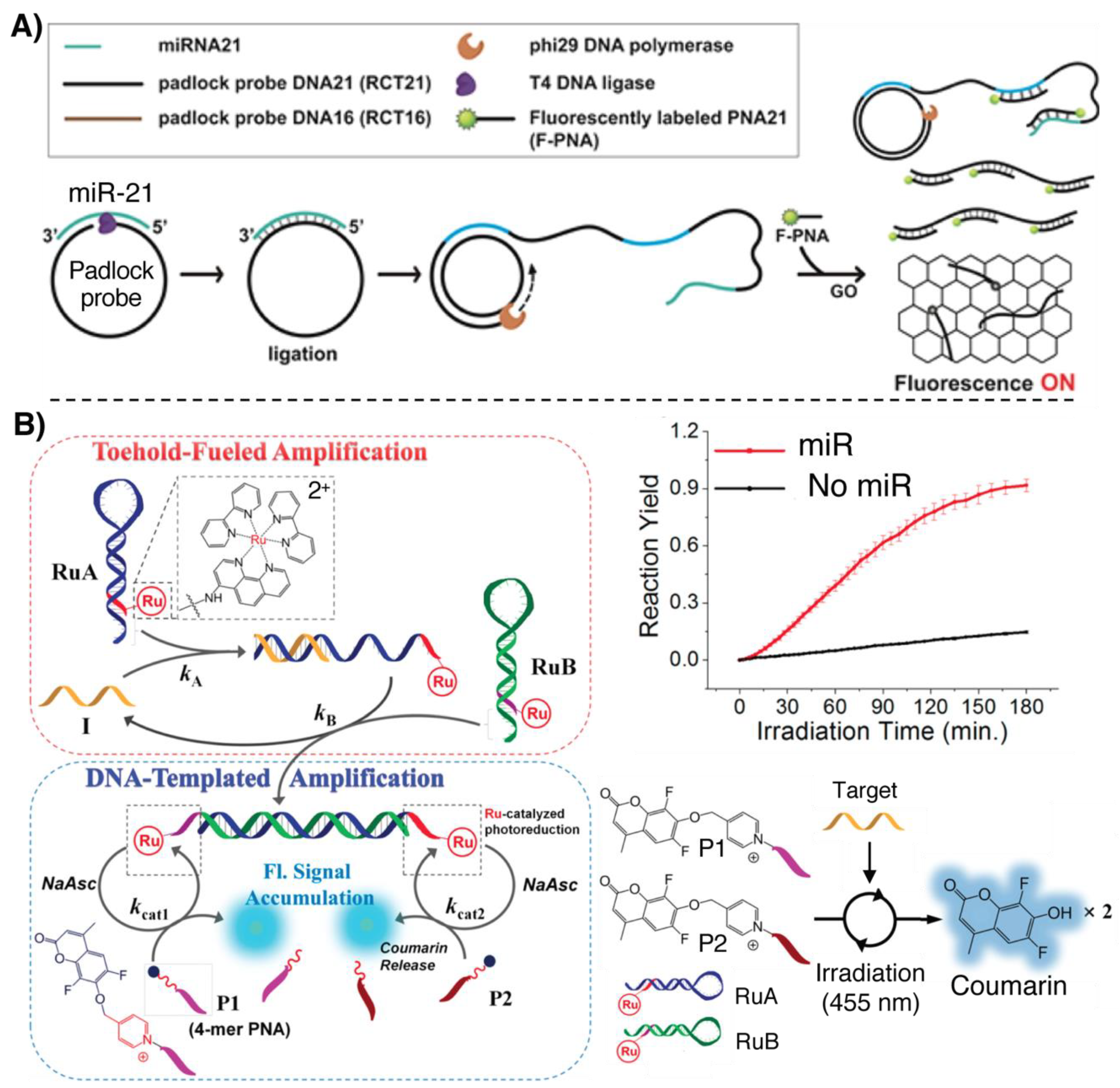
- Ryoo, S.R.; Lee, J.; Yeo, J.; Na, H.K.; Kim, Y.K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N.; et al. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 2013, 7, 5882–5891. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Han, J.; Xue, P.; Xu, R.; Kang, Y. Nano metal-organic framework (NMOF)-based strategies for multiplexed microRNA detection in solution and living cancer cells. Nanoscale 2015, 7, 1753–1759. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, Q.; Ju, H. A peptide nucleic acid-functionalized carbon nitride nanosheet as a probe for in situ monitoring of intracellular microRNA. Analyst 2015, 140, 4245–4252. [Google Scholar] [CrossRef]
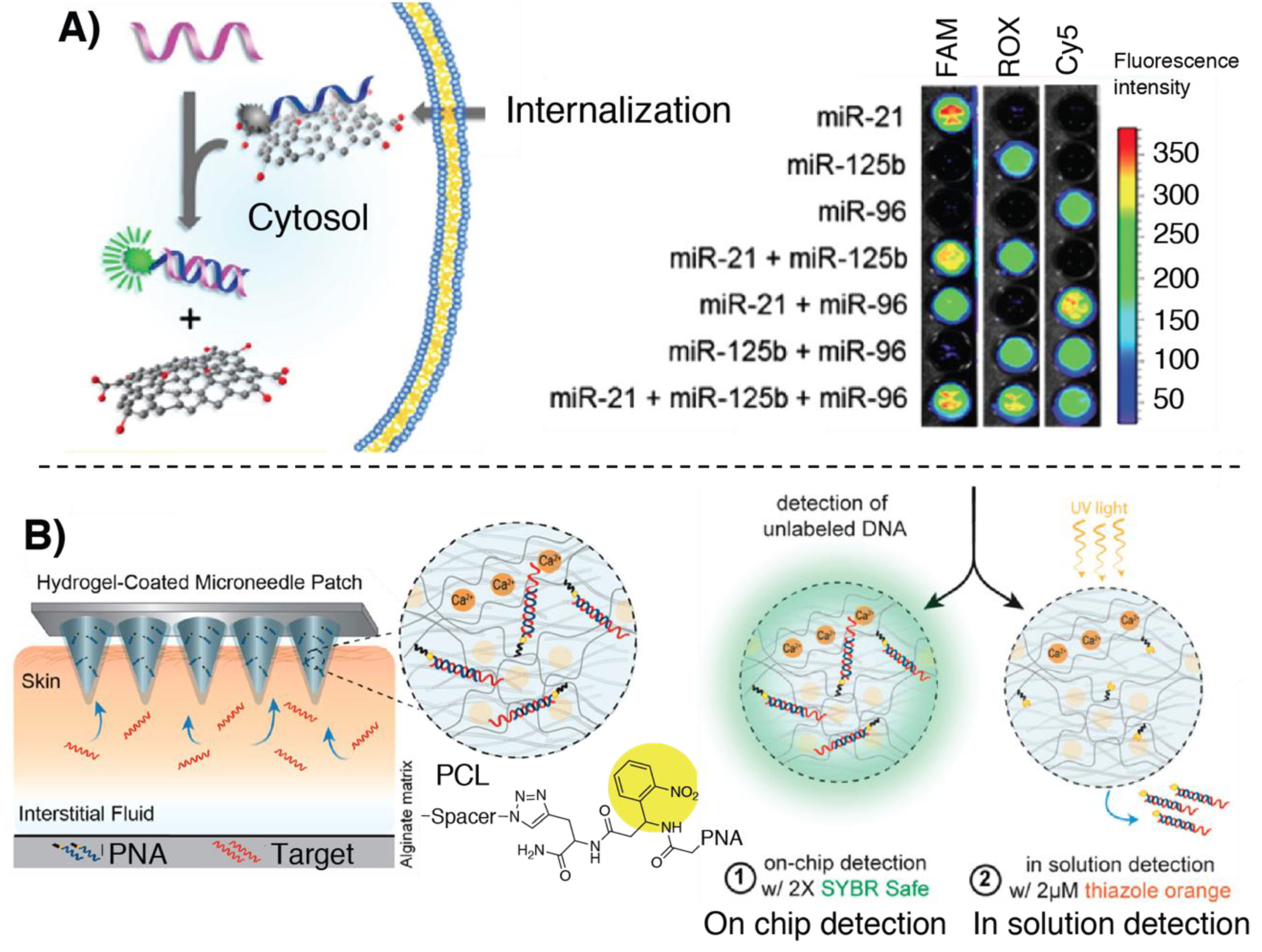
- Al Sulaiman, D.; Chang, J.Y.H.; Bennett, N.R.; Topouzi, H.; Higgins, C.A.; Irvine, D.J.; Ladame, S. Hydrogel-Coated Microneedle Arrays for Minimally Invasive Sampling and Sensing of Specific Circulating Nucleic Acids from Skin Interstitial Fluid. ACS Nano 2019, 13, 9620–9628. [Google Scholar] [CrossRef]
- Di Pisa, M.; Seitz, O. Nucleic Acid Templated Reactions for Chemical Biology. ChemMedChem 2017, 12, 872–882. [Google Scholar] [CrossRef]
- Michaelis, J.; Roloff, A.; Seitz, O. Amplification by nucleic acid-templated reactions. Org. Biomol. Chem. 2014, 12, 2821–2833. [Google Scholar] [CrossRef]
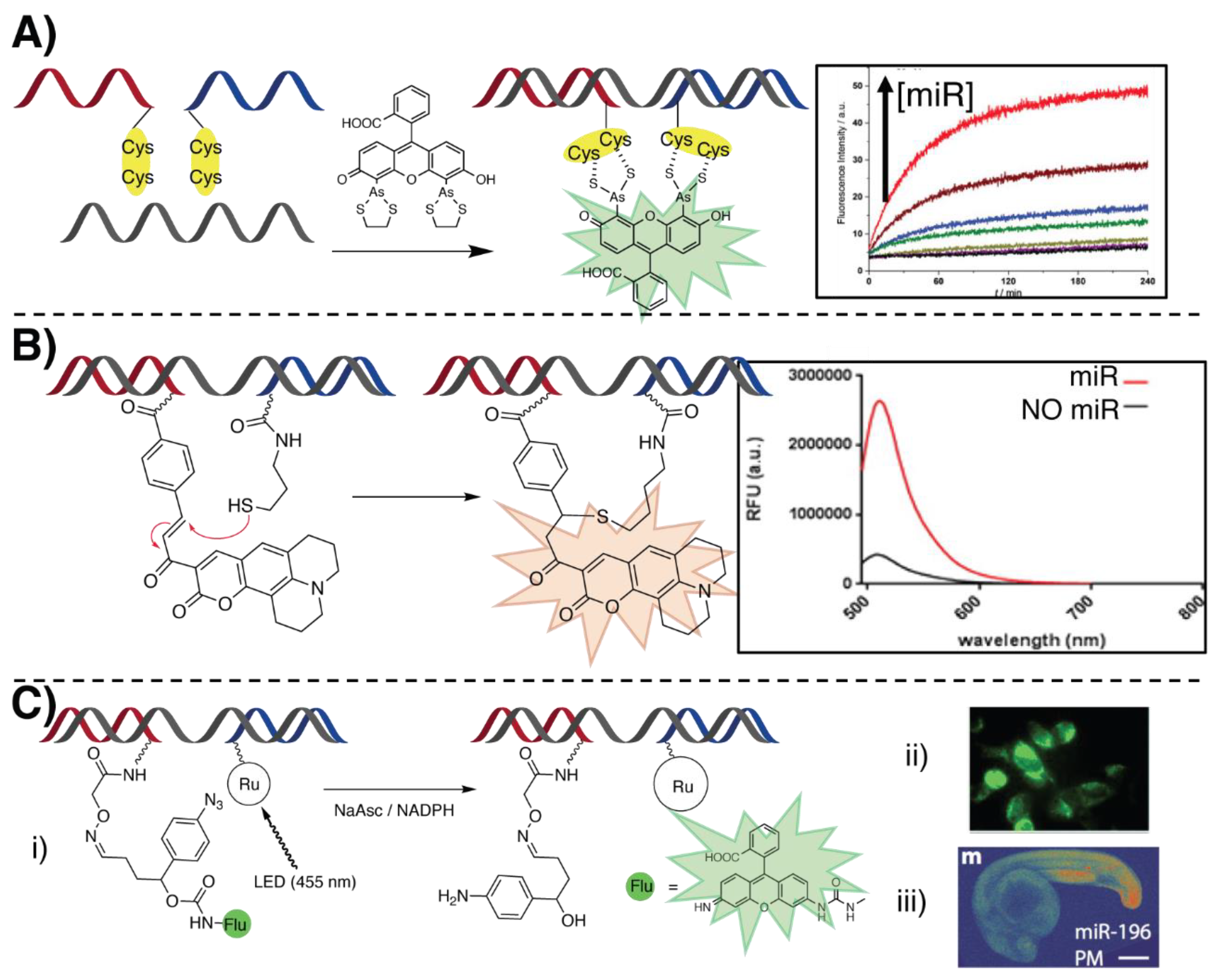
- Fang, G.M.; Seitz, O. Bivalent Display of Dicysteine on Peptide Nucleic Acids for Homogenous DNA/RNA Detection through in Situ Fluorescence Labelling. ChemBioChem 2017, 18, 189–194. [Google Scholar] [CrossRef]
- Piras, L.; Avitabile, C.; D’Andrea, L.D.; Saviano, M.; Romanelli, A. Detection of oligonucleotides by PNA-peptide conjugates recognizing the biarsenical fluorescein complex FlAsH-EDT2. Biochem. Biophys. Res. Commun. 2017, 493, 126–131. [Google Scholar] [CrossRef]
- Metcalf, G.A.D.; Shibakawa, A.; Patel, H.; Sita-Lumsden, A.; Zivi, A.; Rama, N.; Bevan, C.L.; Ladame, S. Amplification-free detection of circulating microRNA biomarkers from body fluids based on fluorogenic oligonucleotide-templated reaction between engineered peptide nucleic acid probes: Application to prostate cancer diagnosis. Anal. Chem. 2016, 88, 8091–8098. [Google Scholar] [CrossRef]
- Al Sulaiman, D.; Chang, J.Y.H.; Ladame, S. Subnanomolar Detection of Oligonucleotides through Templated Fluorogenic Reaction in Hydrogels: Controlling Diffusion to Improve Sensitivity. Angew. Chem.-Int. Ed. 2017, 56, 5247–5251. [Google Scholar] [CrossRef] [PubMed]
- Pavagada, S.; Channon, R.B.; Chang, J.Y.H.; Kim, S.H.; MacIntyre, D.; Bennett, P.R.; Terzidou, V.; Ladame, S. Oligonucleotide-templated lateral flow assays for amplification-free sensing of circulating microRNAs. Chem. Commun. 2019, 55, 12451–12454. [Google Scholar] [CrossRef] [PubMed]
- Saarbach, J.; Lindberg, E.; Winssinger, N. Ruthenium-based Photocatalysis in Templated Reactions. Chim. Int. J. Chem. 2018, 72, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, K.K.; Winssinger, N. Detection of miRNA in live cells by using templated RuII- catalyzed unmasking of a fluorophore. Chem.-A Eur. J. 2013, 19, 8182–8189. [Google Scholar] [CrossRef]
- Holtzer, L.; Oleinich, I.; Anzola, M.; Lindberg, E.; Sadhu, K.K.; Gonzalez-Gaitan, M.; Winssinger, N. Nucleic acid templated chemical reaction in a live vertebrate. ACS Cent. Sci. 2016, 2, 394–400. [Google Scholar] [CrossRef]
- Kim, K.T.; Chang, D.; Winssinger, N. Double-Stranded RNA-Specific Templated Reaction with Triplex Forming PNA. Helv. Chim. Acta 2018, 101, e1700295. [Google Scholar] [CrossRef]
- Chang, D.; Lindberg, E.; Winssinger, N. Critical analysis of rate constants and turnover frequency in nucleic acid-templated reactions: Reaching terminal velocity. J. Am. Chem. Soc. 2017, 139, 1444–1447. [Google Scholar] [CrossRef]
- Hong, C.; Baek, A.; Hah, S.S.; Jung, W.; Kim, D.E. Fluorometric Detection of MicroRNA Using Isothermal Gene Amplification and Graphene Oxide. Anal. Chem. 2016, 88, 2999–3003. [Google Scholar] [CrossRef]
- Kim, K.T.; Angerani, S.; Chang, D.; Winssinger, N. Coupling of DNA Circuit and Templated Reactions for Quadratic Amplification and Release of Functional Molecules. J. Am. Chem. Soc. 2019, 141, 16288–16295. [Google Scholar] [CrossRef]
- Dykman, L.; Khlebtsov, N. Gold nanoparticles in biomedical applications: Recent advances and perspectives. Chem. Soc. Rev. 2012, 41, 2256–2282. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2018, 10, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Erdem, A.; Ozsoz, M.; Carrara, S. microRNA biosensors: Opportunities and challenges among conventional and commercially available techniques. Biosens. Bioelectron. 2018, 99, 525–546. [Google Scholar] [CrossRef] [PubMed]
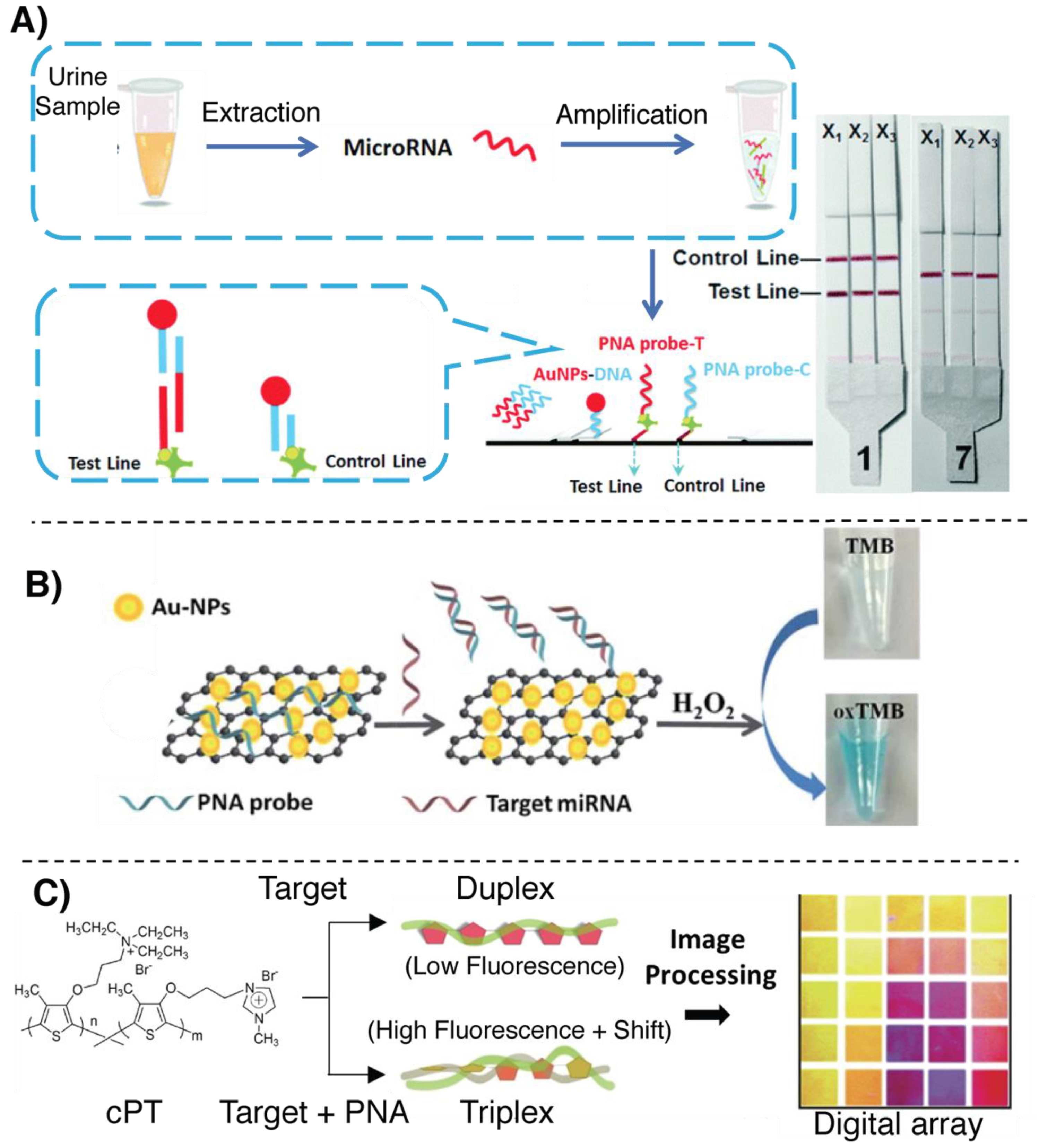
- Cheng, N.; Xu, Y.; Luo, Y.; Zhu, L.; Zhang, Y.; Huang, K.; Xu, W. Specific and relative detection of urinary microRNA signatures in bladder cancer for point-of-care diagnostics. Chem. Commun. 2017, 53, 4222–4225. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Shang, Y.; Xu, Y.; Zhang, L.; Luo, Y.; Huang, K.; Xu, W. On-site detection of stacked genetically modified soybean based on event-specific TM-LAMP and a DNAzyme-lateral flow biosensor. Biosens. Bioelectron. 2017, 91, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Duan, D.; Cao, R.; Zhang, L.; Zheng, K.; Li, J. A reverse transcription-free real-time PCR assay for rapid miRNAs quantification based on effects of base stacking. Chem. Commun. 2011, 47, 7452–7454. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, Z.; Liu, C.; Cheng, Y. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew. Chem.-Int. Ed. 2010, 49, 5498–5501. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Qu, Y.; Yuan, F.; Quan, X. A visible and label-free colorimetric sensor for miRNA-21 detection based on peroxidase-like activity of graphene/gold-nanoparticle hybrids. Anal. Methods 2016, 8, 2005–2012. [Google Scholar] [CrossRef]
- Yildiz, U.H.; Alagappan, P.; Liedberg, B. Naked eye detection of lung cancer associated miRNA by paper based biosensing platform. Anal. Chem. 2013, 85, 820–824. [Google Scholar] [CrossRef]
- Kaloni, T.P.; Giesbrecht, P.K.; Schreckenbach, G.; Freund, M.S. Polythiophene: From Fundamental Perspectives to Applications. Chem. Mater. 2017, 29, 10248–10283. [Google Scholar] [CrossRef]
- Rajwar, D.; Ammanath, G.; Cheema, J.A.; Palaniappan, A.; Yildiz, U.H.; Liedberg, B. Tailoring Conformation-Induced Chromism of Polythiophene Copolymers for Nucleic Acid Assay at Resource Limited Settings. ACS Appl. Mater. Interfaces 2016, 8, 8349–8357. [Google Scholar] [CrossRef]
- Ammanath, G.; Yeasmin, S.; Srinivasulu, Y.; Vats, M.; Cheema, J.A.; Nabilah, F.; Srivastava, R.; Yildiz, U.H.; Alagappan, P.; Liedberg, B. Flow-through colorimetric assay for detection of nucleic acids in plasma. Anal. Chim. Acta 2019, 1066, 102–111. [Google Scholar] [CrossRef]
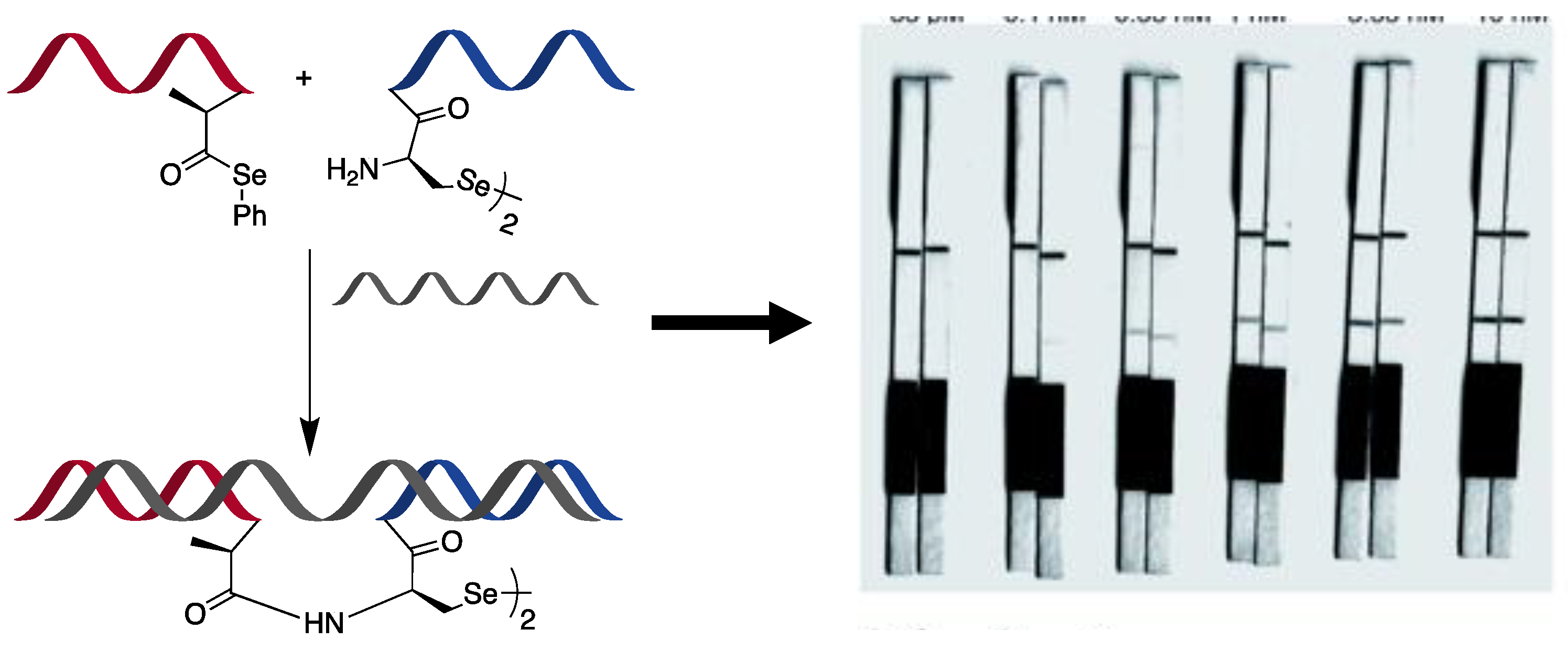
- Sayers, J.; Payne, R.J.; Winssinger, N. Peptide nucleic acid-templated selenocystine–selenoester ligation enables rapid miRNA detection. Chem. Sci. 2018, 9, 896–903. [Google Scholar] [CrossRef]
- Mitchell, N.J.; Malins, L.R.; Liu, X.; Thompson, R.E.; Chan, B.; Radom, L.; Payne, R.J. Rapid Additive-Free Selenocystine-Selenoester Peptide Ligation. J. Am. Chem. Soc. 2015, 137, 14011–14014. [Google Scholar] [CrossRef]
- Hu, L.; Anand, M.; Krylova, S.M.; Yang, B.B.; Liu, S.K.; Yousef, G.M.; Krylov, S.N. Direct Quantitative Analysis of Multiple microRNAs (DQAMmiR) with Peptide Nucleic Acid Hybridization Probes. Anal. Chem. 2018, 90, 14610–14615. [Google Scholar] [CrossRef]
- Delgado-Gonzalez, A.; Robles-Remacho, A.; Marin-Romero, A.; Detassis, S.; Lopez-Longarela, B.; Lopez-Delgado, F.J.; de Miguel-Perez, D.; Guardia-Monteagudo, J.J.; Fara, M.A.; Tabraue-Chavez, M.; et al. PCR-free and chemistry-based technology for miR-21 rapid detection directly from tumour cells. Talanta 2019, 200, 51–56. [Google Scholar] [CrossRef]
- Palaniappan, A.; Cheema, J.A.; Rajwar, D.; Ammanath, G.; Xiaohu, L.; Seng Koon, L.; Yi, W.; Yildiz, U.H.; Liedberg, B. Polythiophene derivative on quartz resonators for miRNA capture and assay. Analyst 2015, 140, 7912–7917. [Google Scholar] [CrossRef]
| hsa-miR | Sequence (5′→3′) | Cellular Effect * | Phenotypical Effect | Ref. |
|---|---|---|---|---|
| miR-15a | UAGCAGCACAUAAUGGUUUGUG | Regulation of antiapoptotic BCL2 gene (D) | Onset of chronic lymphocytic leukemia (CLL) | [29] |
| miR-16 | UAGCAGCACGUAAAUAUUGGCG | |||
| miR-17 | CAAAGUGCUUACAGUGCAGGUAG | E2F1 expression (U) | Cell proliferation | [5] |
| miR-20 | UAAAGUGCUUAUAGUGCAGGUAG | |||
| miR-19a | UGUGCAAAUCUAUGCAAAACUGA | upregulation of genes related to the immune response, T-cell activation, extracellular matrix and collagen network (U) | Regeneration of infarcted myocardium | [6] |
| miR-19b | UGUGCAAAUCCAUGCAAAACUGA | |||
| miR-21 | UAGCUUAUCAGACUGAUGUUGA | Antiapoptotic action via cell growth regulation. No effects on cell proliferation (U) | Glioblastoma, breast cancer onset | [30,31] |
| miR-31 | UGCUAUGCCAACAUAUUGCCAU | Defects in protein p53 pathways (D) | Found in ovarian cancer | [32] |
| miR-33 | GUGCAUUGUAGUUGCAUUGCA | Upregulated expression of cholesterol efflux transporters ABCA1 in liver (D) | Increased levels of HDL in plasma | [33] |
| miR-138 | GCUACUUCACAACACCAGGGCC | Negative regulation of osteogenic differentiation of human mesenchymal cells (U) | Bone formation reduction | [8] |
| miR-141 | UAACACUGUCUGGUAAAGAUGG | Upregulation of Androgen receptor transcriptional activity (U) | Prostate cancer onset | [34] |
| miR-375 | UUUGUUCGUUCGGCUCGCGUGA | |||
| miR-145 | GUCCAGUUUUCCCAGGAAUCCCU | ARF6 overexpression (D) | Triple negative breast cancer onset | [35] |
| miR-155 | UUAAUGCUAAUCGUGAUAGGGGUU | MYC overexpression (U) | CLL, Burkitt’s lymphoma, lung and colon cancer onset | [36] |
| miR-210 | CUGUGCGUGUGACAGCGGCUGA | EFNA3 (VEGF signaling and angiogenesis) downregulation (U) | Upregulated in atherosclerotic plaques | [37] |
| miR-221 | AGCUACAUUGUCUGCUGGGUUUC | KIT downregulation (U) | Modulation of erythropoiesis (CD34+) | [38] |
| miR-222 | AGCUACAUCUGGCUACUGGGU | |||
| let-7 | miR family (miRs let-7a-2, let-7c and let-7g involved) | RAS protein upregulation (D) | lung cancer onset | [7] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadoni, E.; Manicardi, A.; Madder, A. PNA-Based MicroRNA Detection Methodologies. Molecules 2020, 25, 1296. https://doi.org/10.3390/molecules25061296
Cadoni E, Manicardi A, Madder A. PNA-Based MicroRNA Detection Methodologies. Molecules. 2020; 25(6):1296. https://doi.org/10.3390/molecules25061296
Chicago/Turabian StyleCadoni, Enrico, Alex Manicardi, and Annemieke Madder. 2020. "PNA-Based MicroRNA Detection Methodologies" Molecules 25, no. 6: 1296. https://doi.org/10.3390/molecules25061296
APA StyleCadoni, E., Manicardi, A., & Madder, A. (2020). PNA-Based MicroRNA Detection Methodologies. Molecules, 25(6), 1296. https://doi.org/10.3390/molecules25061296






