Targeting Tumors Using Peptides
Abstract
1. The Need for Low Mw Ligands
2. Identification of Peptides by Biopanning
2.1. Enhancement of Identified Binding Motifs
2.2. Alternative Biopanning Strategies
3. In Silico Identification of Peptides
Protein-Peptide Docking Methods
4. Vascular-Targeting Peptides
5. Extracellular Matrix-Targeting Peptides
6. Tumor Associated Macrophage-Targeting Ligands
7. Non-Peptidic Ligands
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simón-Gracia, L.; Scodeller, P.; Fuentes, S.S.; Vallejo, V.G.; Ríos, X.; San Sebastián, E.; Sidorenko, V.; Di Silvio, D.; Suck, M.; De Lorenzi, F.; et al. Application of polymersomes engineered to target p32 protein for detection of small breast tumors in mice. Oncotarget 2018, 9, 18682–18697. [Google Scholar] [CrossRef] [PubMed]
- Wonder, E.; Simón-Gracia, L.; Scodeller, P.; Majzoub, R.N.; Kotamraju, V.R.; Ewert, K.K.; Teesalu, T.; Safinya, C.R. Competition of charge-mediated and specific binding by peptide-tagged cationic liposome–DNA nanoparticles in vitro and in vivo. Biomaterials 2018. [Google Scholar] [CrossRef] [PubMed]
- Scodeller, P. Hyaluronidase and other Extracellular Matrix Degrading Enzymes for Cancer Therapy: New Uses and Nano-Formulations. J. Carcinog. Mutagen. 2014, 5, 1–5. [Google Scholar] [CrossRef]
- Scodeller, P. Extracellular Matrix Degrading Enzymes for Nanocarrier-Based Anticancer Therapy. Intracell. Deliv. 2016, III, 49–66. [Google Scholar]
- Scodeller, P.; Catalano, P.N.; Salguero, N.; Duran, H.; Wolosiuk, A.; Soler-Illia, G.J.A.A. Hyaluronan degrading silica nanoparticles for skin cancer therapy. Nanoscale 2013, 5, 9690–9698. [Google Scholar] [CrossRef] [PubMed]
- Diop-Frimpong, B.; Chauhan, V.P.; Krane, S.; Boucher, Y.; Jain, R.K. Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 2909–2914. [Google Scholar] [CrossRef]
- Fadnes, H.O.; Reed, R.K.; Aukland, K. Interstitial fluid pressure in rats measured with a modified wick technique. Microvasc. Res. 1977, 14, 27–36. [Google Scholar] [CrossRef]
- Heldin, C.-H.; Rubin, K.; Pietras, K.; Östman, A. High interstitial fluid pressure—An obstacle in cancer therapy. Nat. Rev. Cancer 2004, 4, 806–813. [Google Scholar] [CrossRef]
- Adams, G.P.; Schier, R.; McCall, A.M.; Simmons, H.H.; Horak, E.M.; Alpaugh, R.K.; Marks, J.D.; Weiner, L.M. High affinity restricts the localization and tumor penetration of single-chain Fv antibody molecules. Cancer Res. 2001. [Google Scholar] [CrossRef]
- Hussain, S.; Rodriguez-Fernandez, M.; Braun, G.B.; Doyle, F.J.; Ruoslahti, E. Quantity and accessibility for specific targeting of receptors in tumours. Sci. Rep. 2014. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Scott, J.K. Libraries of Peptides and Proteins Displayed on Filamentous Phage. Methods Enzymol. 1993. [Google Scholar] [CrossRef]
- Clackson, T.; Hoogenboom, H.R.; Griffiths, A.D.; Winter, G. Making antibody fragments using phage display libraries. Nature 1991. [Google Scholar] [CrossRef]
- Koivunen, E.; Gay, D.A.; Ruoslahti, E. Selection of peptides binding to the α5β1 integrin from phage display library. J. Biol. Chem. 1993, 268, 20505–20510. [Google Scholar]
- O’Neil, K.T.; Hoess, R.H.; Jackson, S.A.; Ramachandran, N.S.; Mousa, S.A.; DeGrado, W.F. Identification of novel peptide antagonists for GPIIb/IIIa from a conformationally constrained phage peptide library. Proteins Struct. Funct. Bioinform. 1992. [Google Scholar] [CrossRef]
- Motti, C.; Nuzzo, M.; Meola, A.; Galfré, G.; Felici, F.; Cortese, R.; Nicosia, A.; Monaci, P. Recognition by human sera and immunogenicity of HBsAg mimotopes selected from an M13 phage display library. Gene 1994. [Google Scholar] [CrossRef]
- Pasqualini, R.; Ruoslahti, E. Organ targeting in vivo using phage display peptide libraries. Nature 1996. [Google Scholar] [CrossRef]
- Teesalu, T.; Sugahara, K.N.; Ruoslahti, E. Mapping of vascular ZIP codes by phage display. Methods Enzymol. 2012, 503, 35–56. [Google Scholar]
- Ruoslahti, E. Tumor penetrating peptides for improved drug delivery. Adv. Drug Deliv. Rev. 2017. [Google Scholar] [CrossRef]
- Mann, A.P.; Scodeller, P.; Hussain, S.; Braun, G.B.; Mölder, T.; Toome, K.; Ambasudhan, R.; Teesalu, T.; Lipton, S.A.; Ruoslahti, E. Identification of a peptide recognizing cerebrovascular changes in mouse models of Alzheimer’s disease. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Mann, A.P.; Scodeller, P.; Hussain, S.; Joo, J.; Kwon, E.; Gary, B. A peptide for targeted, systemic delivery of imaging and therapeutic compounds into acute brain injuries. Nat. Commun. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Arap, W.; Kolonin, M.G.; Trepel, M.; Lahdenranta, J.; Cardó-Vila, M.; Giordano, R.J.; Mintz, P.J.; Ardelt, P.U.; Yao, V.J.; Vidal, C.I.; et al. Steps toward mapping the human vasculature by phage display. Nat. Med. 2002. [Google Scholar] [CrossRef]
- Zurita, A.J.; Troncoso, P.; Cardó-Vila, M.; Logothetis, C.J.; Pasqualini, R.; Arap, W. Combinatorial Screenings in Patients: The Interleukin-11 Receptor α as a Candidate Target in the Progression of Human Prostate Cancer. Cancer Res. 2004. [Google Scholar] [CrossRef] [PubMed]
- Lewis, V.O.; Ozawa, M.G.; Deavers, M.T.; Wang, G.; Shintani, T.; Arap, W.; Pasqualini, R. The lnterleukin-11 receptor a as a candidate ligand-directed target in osteosarcoma: Consistent data from cell lines, orthotopic models, and human tumor samples. Cancer Res. 2009. [Google Scholar] [CrossRef] [PubMed]
- Staquicini, F.I.; Cardó-Vila, M.; Kolonin, M.G.; Trepel, M.; Edwards, J.K.; Nunes, D.N.; Sergeeva, A.; Efstathiou, E.; Sun, J.; Almeida, N.F.; et al. Vascular ligand-receptor mapping by direct combinatorial selection in cancer patients. Proc. Natl. Acad. Sci. USA 2011. [Google Scholar] [CrossRef]
- Ngambenjawong, C.; Gustafson, H.H.; Pineda, J.M.; Kacherovsky, N.A.; Cieslewicz, M.; Pun, S.H. Serum stability and affinity optimization of an M2 macrophage-targeting peptide (M2pep). Theranostics 2016. [Google Scholar] [CrossRef]
- Aumailley, M.; Gurrath, M.; Müller, G.; Calvete, J.; Timpl, R.; Kessler, H. Arg-Gly-Asp constrained within cyclic pentapoptides Strong and selective inhibitors of cell adhesion to vitronectin and laminin fragment P1. FEBS Lett. 1991. [Google Scholar] [CrossRef]
- Reichart, F.; Horn, M.; Neundorf, I. Cyclization of a cell-penetrating peptide via click-chemistry increases proteolytic resistance and improves drug delivery. J. Pept. Sci. 2016. [Google Scholar] [CrossRef]
- Molhoek, E.M.; Van Dijk, A.; Veldhuizen, E.J.A.; Haagsman, H.P.; Bikker, F.J. Improved proteolytic stability of chicken cathelicidin-2 derived peptides by d-amino acid substitutions and cyclization. Peptides 2011. [Google Scholar] [CrossRef]
- Walensky, L.D.; Bird, G.H. Hydrocarbon-stapled peptides: Principles, practice, and progress. J. Med. Chem. 2014. [Google Scholar] [CrossRef]
- Bird, G.H.; Mazzola, E.; Opoku-Nsiah, K.; Lammert, M.A.; Godes, M.; Neuberg, D.S.; Walensky, L.D. Biophysical determinants for cellular uptake of hydrocarbon-stapled peptide helices. Nat. Chem. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ngambenjawong, C.; Cieslewicz, M.; Schellinger, J.G.; Pun, S.H. Synthesis and evaluation of multivalent M2pep peptides for targeting alternatively activated M2 macrophages. J. Control. Release 2016, 224, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Duro-Castano, A.; England, R.M.; Razola, D.; Romero, E.; Oteo-Vives, M.; Morcillo, M.A.; Vicent, M.J. Well-defined star-shaped polyglutamates with improved pharmacokinetic profiles as excellent candidates for biomedical applications. Mol. Pharm. 2015. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, N. Multivalent ligand presentation as a central concept to study intricate carbohydrate-protein interactions. Chem. Soc. Rev. 2009. [Google Scholar] [CrossRef] [PubMed]
- Saether, O.; Craik, D.J.; Campbell, I.D.; Sletten, K.; Juul, J.; Norman, D.G. Elucidation of the Primary and Three-Dimensional Structure of the Uterotonic Polypeptide Kalata B1. Biochemistry 1995. [Google Scholar] [CrossRef]
- Wang, C.K.; Craik, D.J. Designing macrocyclic disulfide-rich peptides for biotechnological applications perspective. Nat. Chem. Biol. 2018, 14, 417–427. [Google Scholar] [CrossRef]
- Weidmann, J.; Craik, D.J. Discovery, structure, function, and applications of cyclotides: Circular proteins from plants. J. Exp. Bot. 2016. [Google Scholar] [CrossRef]
- White, A.M.; Craik, D.J. Discovery and optimization of peptide macrocycles. Expert Opin. Drug Discov. 2016. [Google Scholar] [CrossRef]
- Craik, D.J.; Du, J. Cyclotides as drug design scaffolds. Curr. Opin. Chem. Biol. 2017. [Google Scholar] [CrossRef]
- Boy, R.G.; Mier, W.; Nothelfer, E.M.; Altmann, A.; Eisenhut, M.; Kolmar, H.; Tomaszowski, M.; Krämer, S.; Haberkorn, U. Sunflower trypsin inhibitor 1 derivatives as molecular scaffolds for the development of novel peptidic radiopharmaceuticals. Mol. Imaging Biol. 2010. [Google Scholar] [CrossRef]
- Qiu, Y.; Taichi, M.; Wei, N.; Yang, H.; Luo, K.Q.; Tam, J.P. An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor. J. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Durek, T.; Cromm, P.M.; White, A.M.; Schroeder, C.I.; Kaas, Q.; Weidmann, J.; Ahmad Fuaad, A.; Cheneval, O.; Harvey, P.J.; Daly, N.L.; et al. Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1. J. Med. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Baum, R.P.; Kulkarni, H.R.; Schuchardt, C.; Singh, A.; Wirtz, M.; Wiessalla, S.; Schottelius, M.; Mueller, D.; Klette, I.; Wester, H.J. 177Lu-labeled prostate-specific membrane antigen radioligand therapy of metastatic castration-resistant prostate cancer: Safety and efficacy. J. Nucl. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Tak, K.K.; Park, M.O.; Lee, J.T.; Woo, B.H.; Yoo, S.D.; Lee, H.S.; DeLuca, P.P. Preparation and characterization of polyethylene-glycol-modified salmon calcitonins. Pharm. Dev. Technol. 1999. [Google Scholar] [CrossRef]
- DeNardo, S.J.; Yao, Z.; Lam, K.S.; Song, A.; Burke, P.A.; Mirick, G.R.; Lamborn, K.R.; O’Donnell, R.T.; DeNardo, G.L. Effect of molecular size of pegylated peptide on the pharmacokinetics and tumor targeting in lymphoma-bearing mice. Clin. Cancer Res. 2003, 9, 3854s–3864s. [Google Scholar]
- Park, S.I.; Renil, M.; Vikstrom, B.; Amro, N.; Song, L.W.; Xu, B.L.; Lam, K.S. The use of one-bead one-compound combinatorial library method to identify peptide ligands for α4β1 integrin receptor in non-Hodgkin’s lymphoma. Lett. Pept. Sci. 2001. [Google Scholar] [CrossRef]
- Tian, R.; Jacobson, O.; Niu, G.; Kiesewetter, D.O.; Wang, Z.; Zhu, G.; Ma, Y.; Liu, G.; Chen, X. Evans blue attachment enhances somatostatin receptor subtype-2 imaging and radiotherapy. Theranostics 2018. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, X. Simple bioconjugate chemistry serves great clinical advances: Albumin as a versatile platform for diagnosis and precision therapy. Chem. Soc. Rev. 2016. [Google Scholar] [CrossRef]
- Zorzi, A.; Middendorp, S.J.; Wilbs, J.; Deyle, K.; Heinis, C. Acylated heptapeptide binds albumin with high affinity and application as tag furnishes long-acting peptides. Nat. Commun. 2017. [Google Scholar] [CrossRef]
- Zhang, L.; Bulaj, G. Converting Peptides into Drug Leads by Lipidation. Curr. Med. Chem. 2012. [Google Scholar] [CrossRef]
- Levy, O.E.; Jodka, C.M.; Ren, S.S.; Mamedova, L.; Sharma, A.; Samant, M.; D’Souza, L.J.; Soares, C.J.; Yuskin, D.R.; Jin, L.J.; et al. Novel exenatide analogs with peptidic albumin binding domains: Potent anti-diabetic agents with extended duration of action. PLoS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.B.; Braun, G.B.; She, Z.G.; Kotamraju, V.R.; Sugahara, K.N.; Teesalu, T.; Ruoslahti, E. A free cysteine prolongs the half-life of a homing peptide and improves its tumor-penetrating activity. J. Control. Release 2014, 175, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Penchala, S.C.; Miller, M.R.; Pal, A.; Dong, J.; Madadi, N.R.; Xie, J.; Joo, H.; Tsai, J.; Batoon, P.; Samoshin, V.; et al. A biomimetic approach for enhancing the in vivo half-life of peptides. Nat. Chem. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.M.; Mayr, L.M.; Minor, D.L.; Milhollen, M.A.; Burgess, M.W.; Kim, P.S. Identification of D-peptide ligands through mirror-image phage display. Science 1996. [Google Scholar] [CrossRef]
- Heinis, C.; Rutherford, T.; Freund, S.; Winter, G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 2009. [Google Scholar] [CrossRef]
- Borgoño, C.A.; Diamandis, E.P. The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer 2004. [Google Scholar] [CrossRef]
- Panyayai, T.; Sangsawad, P.; Pacharawongsakda, E.; Sawatdichaikul, O.; Tongsima, S.; Choowongkomon, K. The potential peptides against angiotensin-I converting enzyme through a virtual tripeptide-constructing library. Comput. Biol. Chem. 2018, 77, 207–213. [Google Scholar] [CrossRef]
- Pinter, M.; Jain, R.K. Targeting the renin-angiotensin system to improve cancer treatment: Implications for immunotherapy. Sci. Transl. Med. 2017. [Google Scholar] [CrossRef]
- Song, T.; Choi, C.H.; Kim, M.K.; Kim, M.L.; Yun, B.S.; Seong, S.J. The effect of angiotensin system inhibitors (angiotensin-converting enzyme inhibitors or angiotensin receptor blockers) on cancer recurrence and survival: A meta-analysis. Eur. J. Cancer Prev. 2017. [Google Scholar] [CrossRef]
- Mollica, A.; Zengin, G.; Durdagi, S.; Ekhteiari Salmas, R.; Macedonio, G.; Stefanucci, A.; Dimmito, M.P.; Novellino, E. Combinatorial peptide library screening for discovery of diverse α-glucosidase inhibitors using molecular dynamics simulations and binary QSAR models. J. Biomol. Struct. Dyn. 2019, 37, 726–740. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Kevin, J.B.; David, E.C.; Huafeng, X.; Ron, O.D.; Michael, P.E.; Brent, A.G.; John, L.K.; Istvan, K.; Mark, A.M.; Federico, D.S.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Michaeli, A.; Mezan, S.; Kühbacher, A.; Finkelmeier, D.; Elias, M.; Zatsepin, M.; Reed, S.G.; Duthie, M.S.; Rupp, S.; Lerner, I.; et al. Computationally Designed Bispecific MD2/CD14 Binding Peptides Show TLR4 Agonist Activity. J. Immunol. 2018, 201, 3383–3391. [Google Scholar] [CrossRef] [PubMed]
- Lerner, I.; Goldblum, A.; Rayan, A.; Vardi, A.; Michaeli, A. From finance to molecular modeling algorithms: The risk and return heuristic. Curr. Top. Pept. Protein Res. 2017, 18, 117–131. [Google Scholar]
- Volk-Draper, L.; Hall, K.; Griggs, C.; Rajput, S.; Kohio, P.; DeNardo, D.; Ran, S. Paclitaxel therapy promotes breast cancer metastasis in a TLR4-dependent manner. Cancer Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Vukic, V.R.; Vukic, D.V.; Milanovic, S.D.; Ilicic, M.D.; Kanuric, K.G.; Johnson, M.S. In silico identification of milk antihypertensive di- and tripeptides involved in angiotensin I–converting enzyme inhibitory activity. Nutr. Res. 2017, 46, 22–30. [Google Scholar] [CrossRef]
- Cramer, R.D. The inevitable QSAR renaissance. J. Comput. Mol. Des. 2012, 26, 8. [Google Scholar] [CrossRef]
- Klebe, G.; Abraham, U.M.T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J. Med. Chem. 1994, 37, 4130–4146. [Google Scholar] [CrossRef]
- Murumkar, P.R.; Shinde, A.C.; Sharma, M.K.; Yamaguchi, H.; Miniyar, P.B.Y.M. Development of a credible 3D-QSAR CoMSIA model and docking studies for a series of triazoles and tetrazoles containing 11β-HSD1 inhibitors. SAR QSAR Environ. Res. 2016, 27, 265–292. [Google Scholar] [CrossRef]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR Modeling: Where Have You Been? Where Are You Going To? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef]
- Guitard, E.; Parker, F.; Millon, R.; Abecassis, J.; Tocqué, B. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett. 2001. [Google Scholar] [CrossRef]
- Cui, W.; Wei, Z.; Chen, Q.; Cheng, Y.; Geng, L.; Zhang, J.; Chen, J.; Hou, T.; Ji, M. Structure-based design of peptides against G3BP with cytotoxicity on tumor cells. J. Chem. Inf. Model. 2010, 50, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.; Wang, Z.; Yang, X.; Li, D.; Lian, W.; Xiang, Z.; Wang, W.; Bu, X.; Lai, W.; Hu, Z.; et al. Structure-based Design of Peptides with High Affinity and Specificity to HER2 Positive Tumors. Theranostics 2015, 5, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Orlova, A.; Magnusson, M.; Eriksson, T.L.J.; Nilsson, M.; Larsson, B.; Höiden-Guthenberg, I.; Widström, C.; Carlsson, J.; Tolmachev, V.; Ståhl, S.; et al. Tumor imaging using a picomolar affinity HER2 binding Affibody molecule. Cancer Res. 2006. [Google Scholar] [CrossRef]
- Raveh, B.; London, N.; Zimmerman, L.; Schueler-Furman, O. Rosetta FlexPepDock ab-initio: Simultaneous folding, docking and refinement of peptides onto their receptors. PLoS ONE 2011, 6, e18934. [Google Scholar] [CrossRef]
- Donsky, E.; Wolfson, H.J. PepCrawler: A fast RRT-based algorithm for high-resolution refinement and binding affinity estimation of peptide inhibitors. Bioinformatics 2011, 27, 2836–2842. [Google Scholar] [CrossRef]
- Trellet, M.; Melquiond, A.S.; Bonvin, A.M. A unified conformational selection and induced fit approach to protein–peptide docking. PLoS ONE 2013, 8, e58769. [Google Scholar] [CrossRef]
- Hetényi, C.; van der Spoel, D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2009, 11, 1729–1737. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Yan, C.; Xu, X.; Zou, X. Fully blind docking at the atomic level for protein–peptide complex structure prediction. Structure 2016, 24, 1842–1853. [Google Scholar] [CrossRef]
- Schindler, C.E.; de Vries, S.J.; Zacharias, M. Fully blind peptide–protein docking with pepATTRACT. Structure 2015, 23, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, E.; Rajasekaran, R. Rational design of linear tripeptides against the aggregation of human mutant SOD1 protein causing amyotrophic lateral sclerosis. J. Neurol. Sci. 2019, 405, 116425. [Google Scholar] [CrossRef] [PubMed]
- Papa, L.; Manfredi, G.; Germain, D. SOD1, an unexpected novel target for cancer therapy. Genes Cancer 2015. [Google Scholar] [CrossRef]
- Ben-Shimon, A.; Niv, M.Y. AnchorDock: Blind and flexible anchor-driven peptide docking. Structure 2015, 23, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Kurcinski, M.; Pawel Ciemny, M.; Oleniecki, T.; Kuriata, A.; Badaczewska-Dawid, A.E.; Kolinski, A.; Kmiecik, S.; Valencia, A. CABS-dock standalone: A toolbox for flexible protein-peptide docking. Bioinformatics 2019, 35, 4170–4172. [Google Scholar] [CrossRef] [PubMed]
- Blaszczyk, M.; Kurcinski, M.; Kouza, M.; Wieteska, L.; Debinski, A.; Kolinski, A.; Kmiecik, S. Modeling of protein-peptide interactions using the CABS-dock web server for binding site search and flexible docking. Methods 2016, 93, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sanner, M.F. AutoDock CrankPep: Combining folding and docking to predict protein–peptide complexes. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Burkoff, N.S.; Várnai, C.; Wells, S.A.; Wild, D.L. Exploring the energy landscapes of protein folding simulations with bayesian computation. Biophys. J. 2012, 102, 878–886. [Google Scholar] [CrossRef]
- Dundas, J.; Ouyang, Z.; Tseng, J.; Binkowski, A.; Turpaz, Y.L.J. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006, 34, W116–W118. [Google Scholar] [CrossRef]
- Shulman-Peleg, A.; Shatsky, M.; Nussinov, R.; Wolfson, H.J. MultiBind and MAPPIS: Webservers for multiple alignment of protein 3D-binding sites and their interactions. Nucleic Acids Res. 2008, 36, 260–264. [Google Scholar] [CrossRef]
- Gabdoulline, R.R.; Wade, R.C.; Walther, D. MolSurfer: A macromolecular interface navigator. Nucleic Acids Res. 2003, 31, 3349–3351. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saha, R.P.; Bahadur, R.P.; Pal, A.; Mandal, S.C.P. ProFace: A server for the analysis of the physicochemical features of protein-protein interfaces. BMC Struct. Biol. 2006, 6, 11. [Google Scholar]
- Wang, Y.; Guo, H.; Feng, Z.; Wang, S.; Wang, Y.; He, Q.; Li, G.; Lin, W.; Xie, X.Q.; Lin, Z. PD-1-targeted discovery of peptide inhibitors by virtual screening, molecular dynamics simulation, and surface plasmon resonance. Molecules 2019, 24, 3784. [Google Scholar] [CrossRef] [PubMed]
- Ansar, S.; Vetrivel, U. PepVis: An integrated peptide virtual screening pipeline for ensemble and flexible docking protocols. Chem. Biol. Drug Des. 2019. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.I.; Huang, S.-Y. Efficient conformational ensemble generation of protein-bound peptides. J. Cheminform. 2017, 9, 59. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Autodock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient opti- mization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
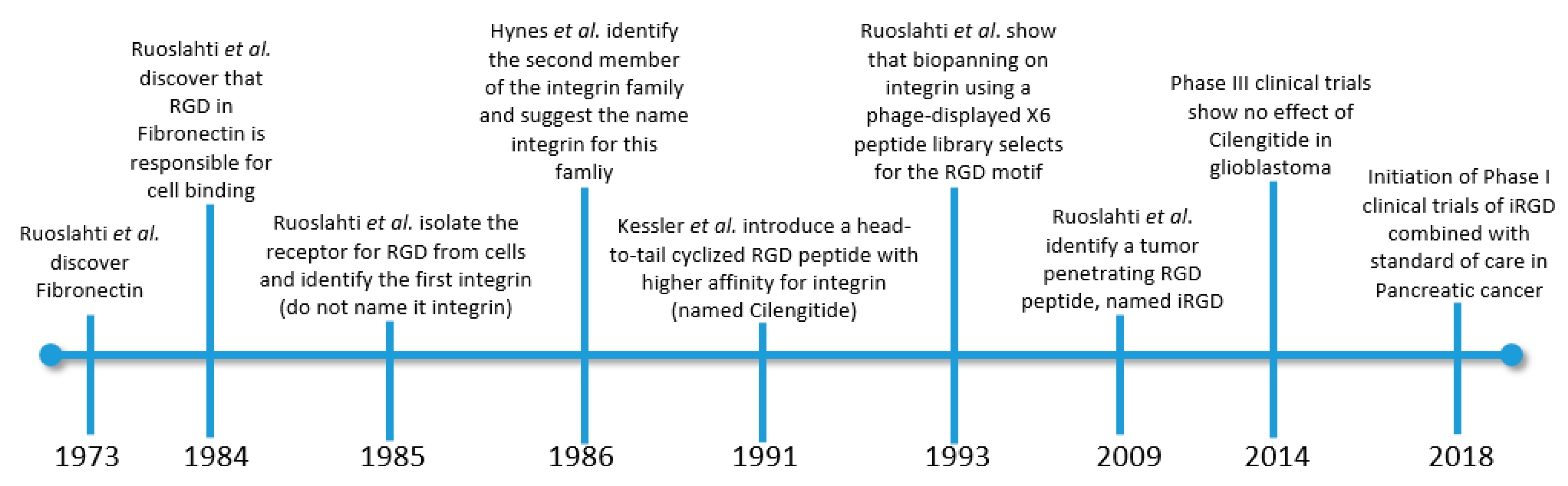
- Ruoslahti, E.; Vaheri, A.; Kuusela, P.; Linder, E. Fibroblast surface antigen: A new serum protein. BBA Protein Struct. 1973. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef]
- Pytela, R.; Pierschbacher, M.D.; Ruoslahti, E. Identification and isolation of a 140 kd cell surface glycoprotein with properties expected of a fibronectin receptor. Cell 1985. [Google Scholar] [CrossRef]
- Tamkun, J.W.; DeSimone, D.W.; Fonda, D.; Patel, R.S.; Buck, C.; Horwitz, A.F.; Hynes, R.O. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 1986. [Google Scholar] [CrossRef]
- Ruoslahti, E.; Pierschbacher, M.D. New perspectives in cell adhesion: RGD and integrins. Science 1987. [Google Scholar] [CrossRef] [PubMed]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-Penetrating Delivery of Compounds and Nanoparticles into Tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Teesalu, T.; Prakash Karmali, P.; Ramana Kotamraju, V.; Agemy, L.; Greenwald, D.R.; Ruoslahti, E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science 2010. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Braun, G.B.; de Mendoza, T.H.; Kotamraju, V.R.; French, R.P.; Lowy, A.M.; Teesalu, T.; Ruoslahti, E. Tumor-Penetrating iRGD Peptide Inhibits Metastasis. Mol. Cancer Ther. 2015, 14, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Akashi, Y.; Oda, T.; Ohara, Y.; Miyamoto, R.; Kurokawa, T.; Hashimoto, S.; Enomoto, T.; Yamada, K.; Satake, M.; Ohkohchi, N. Anticancer effects of gemcitabine are enhanced by co-administered iRGD peptide in murine pancreatic cancer models that overexpressed neuropilin-1. Br. J. Cancer 2014. [Google Scholar] [CrossRef]
- Puig-Saus, C.; Rojas, L.A.; Laborda, E.; Figueras, A.; Alba, R.; Fillat, C.; Alemany, R. IRGD tumor-penetrating peptide-modified oncolytic adenovirus shows enhanced tumor transduction, intratumoral dissemination and antitumor efficacy. Gene Ther. 2014. [Google Scholar] [CrossRef][Green Version]
- Schmithals, C.; Köberle, V.; Korkusuz, H.; Pleli, T.; Kakoschky, B.; Augusto, E.A.; Ibrahim, A.A.; Arencibia, J.M.; Vafaizadeh, V.; Groner, B.; et al. Improving drug penetrability with iRGD leverages the therapeutic response to sorafenib and doxorubicin in hepatocellular carcinoma. Cancer Res. 2015. [Google Scholar] [CrossRef]
- Fadeev, R.; Chekanov, A.; Solovieva, M.; Bezborodova, O.; Nemtsova, E.; Dolgikh, N.; Fadeeva, I.; Senotov, A.; Kobyakova, M.; Evstratova, Y.; et al. Improved anticancer effect of recombinant protein izTRAIL combined with sorafenib and peptide iRGD. Int. J. Mol. Sci. 2019, 20, 525. [Google Scholar] [CrossRef]
- Liu, X.; Lin, P.; Perrett, I.; Lin, J.; Liao, Y.P.; Chang, C.H.; Jiang, J.; Wu, N.; Donahue, T.; Wainberg, Z.; et al. Tumor-penetrating peptide enhances transcytosis of silicasome-based chemotherapy for pancreatic cancer. J. Clin. Investig. 2017. [Google Scholar] [CrossRef]
- Sugahara, K.N.; Scodeller, P.; Braun, G.B.; De Mendoza, T.H.; Yamazaki, C.M.; Kluger, M.D.; Kitayama, J.; Alvarez, E.; Howell, S.B.; Teesalu, T.; et al. A tumor-penetrating peptide enhances circulation-independent targeting of peritoneal carcinomatosis. J. Control. Release 2015, 212, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Mas-Moruno, C.; Rechenmacher, F.; Kessler, H. Cilengitide: The First Anti-Angiogenic Small Molecule Drug Candidate. Design, Synthesis and Clinical Evaluation. Anticancer Agents Med. Chem. 2011. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Bu, X.Y.; Khankaldyyan, V.; Gonzales-Gomez, I.; McComb, J.G.; Laug, W.E. Effect of the angiogenesis inhibitor Cilengitide (EMD 121974) on glioblastoma growth in nude mice. Neurosurgery 2006. [Google Scholar] [CrossRef]
- Stupp, R.; Hegi, M.E.; Neyns, B.; Goldbrunner, R.; Schlegel, U.; Clement, P.M.J.; Grabenbauer, G.G.; Ochsenbein, A.F.; Simon, M.; Dietrich, P.Y.; et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J. Clin. Oncol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Gorlia, T.; Erridge, S.C.; Perry, J.; Hong, Y.K.; Aldape, K.D.; Lhermitte, B.; Pietsch, T.; Grujicic, D.; et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicentre, randomised, open-label, phase 3 trial. Lancet. Oncol. 2014. [Google Scholar] [CrossRef]
- Tucci, M.; Stucci, S.; Silvestris, F. Does cilengitide deserve another chance? Lancet Oncol. 2014. [Google Scholar] [CrossRef]
- Bernhagen, D.; Jungbluth, V.; Quilis, N.G.; Dostalek, J.; White, P.B.; Jalink, K.; Timmerman, P. Bicyclic RGD Peptides with Exquisite Selectivity for the Integrin α v β 3 Receptor Using a ‘random Design’ Approach. ACS Comb. Sci. 2019. [Google Scholar] [CrossRef]
- Altmann, A.; Sauter, M.; Roesch, S.; Mier, W.; Warta, R.; Debus, J.; Dyckhoff, G.; Herold-Mende, C.; Haberkorn, U. Identification of a novel ITGαvβ6-binding peptide using protein separation and phage display. Clin. Cancer Res. 2017. [Google Scholar] [CrossRef]
- Del Gatto, A.; Zaccaro, L.; Grieco, P.; Novellino, E.; Zannetti, A.; Del Vecchio, S.; Iommelli, F.; Salvatore, M.; Pedone, C.; Saviano, M. Novel and selective αvβ3 receptor peptide antagonist: Design, synthesis, and biological behavior. J. Med. Chem. 2006. [Google Scholar] [CrossRef]
- Zannetti, A.; Del Vecchio, S.; Iommelli, F.; Del Gatto, A.; De Luca, S.; Zaccaro, L.; Papaccioli, A.; Sommella, J.; Panico, M.; Speranza, A.; et al. Imaging of αvβ3 expression by a bifunctional chimeric RGD peptide not cross-reacting with α vβ5. Clin. Cancer Res. 2009. [Google Scholar] [CrossRef]
- Farina, B.; De Paola, I.; Russo, L.; Capasso, D.; Liguoro, A.; Del Gatto, A.; Saviano, M.; Pedone, P.V.; Di Gaetano, S.; Malgieri, G.; et al. A Combined NMR and Computational Approach to Determine the RGDechi-hCit-αvβ3 Integrin Recognition Mode in Isolated Cell Membranes. Chem. A Eur. J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Chen, X. Anti-angiogenic cancer therapy based on integrin alphavbeta3 antagonism. Anticancer. Agents Med. Chem. 2006, 6, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Sloan, E.K.; Pouliot, N.; Stanley, K.L.; Chia, J.; Moseley, J.M.; Hards, D.K.; Anderson, R.L. Tumor-specific expression of αvβ3 integrin promotes spontaneous metastasis of breast cancer to bone. Breast Cancer Res. 2006. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.A.; Pasqualini, R.; Arap, W.; Ruoslahti, E.; Stallcup, W.B. NG2 proteoglycan-binding peptides target tumor neovasculature. Cancer Res. 1999, 59, 2869–2874. [Google Scholar]
- Guan, Y.Y.; Luan, X.; Xu, J.R.; Liu, Y.R.; Lu, Q.; Wang, C.; Liu, H.J.; Gao, Y.G.; Chen, H.Z.; Fang, C. Selective eradication of tumor vascular pericytes by peptide-conjugated nanoparticles for antiangiogenic therapy of melanoma lung metastasis. Biomaterials 2014. [Google Scholar] [CrossRef]
- Svensen, N.; Walton, J.G.A.; Bradley, M. Peptides for cell-selective drug delivery. Trends Pharmacol. Sci. 2012. [Google Scholar] [CrossRef]
- Pasqualini, R.; Koivunen, E.; Kain, R.; Lahdenranta, J.; Sakamoto, M.; Stryhn, A.; Ashmun, R.A.; Shapiro, L.H.; Arap, W.; Ruoslahti, E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000, 60, 722–727. [Google Scholar]
- Suwiwat, S.; Ricciardelli, C.; Tammi, R.; Tammi, M.; Auvinen, P.; Kosma, V.; Lebaron, R.G.; Raymond, W.A.; Tilley, W.D.; Horsfall, D.J. Expression of Extracellular Matrix Components Versican, Chondroitin Sulfate, Tenascin, and Hyaluronan and Their Association with Disease Outcome in Node-Negative Breast Cancer. Clin. Cancer Res. 2004, 10, 2491–2498. [Google Scholar] [CrossRef]
- Asimakopoulou, A.P.; Theocharis, A.D.; Tzanakakis, G.N.; Karamanos, N.K. The biological role of chondroitin sulfate in cancer and chondroitin-based anticancer agents. In Vivo 2008, 22, 385–390. [Google Scholar]
- Kim, Y.; Lee, H.G.; Dmitrieva, N.; Kim, J.; Kaur, B.; Friedman, A. Choindroitinase ABC I-mediated enhancement of oncolytic virus spread and anti tumor efficacy: A mathematical model. PLoS ONE 2014, 9, e102499. [Google Scholar] [CrossRef]
- Carnemolla, B.; Castellani, P.; Ponassi, M.; Borsi, L.; Urbini, S.; Nicolo, G.; Dorcaratto, A.; Viale, G.; Winter, G.; Neri, D.; et al. Identification of a glioblastoma-associated Tenascin-C isoform by a high affinity recombinant antibody. Am. J. Pathol. 1999. [Google Scholar] [CrossRef]
- Park, J.; Kim, S.; Saw, P.E.; Lee, I.H.; Yu, M.K.; Kim, M.; Lee, K.; Kim, Y.C.; Jeong, Y.Y.; Jon, S. Fibronectin extra domain B-specific aptide conjugated nanoparticles for targeted cancer imaging. J. Control. Release 2012. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.H.; Beppu, T.; Kurose, A.; Yamauchi, K.; Sugawara, A.; Suzuki, M.; Ogawa, A.; Sawai, T. Neoplastic cells and proliferating endothelial cells express connective tissue growth factor (CTGF) in glioblastoma. Neurol. Res. 2002. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Chen, W.; O’Kelly, J.; Lu, D.; Ham, M.; Doan, N.B.; Xie, D.; Wang, C.; Vadgama, J.; Said, J.W.; et al. Connective tissue growth factor associated with oncogenic activities and drug resistance in glioblastoma multiforme. Int. J. Cancer 2010, 127, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Campos, B.; Meier, J.; Devens, F.; Liesenberg, F.; Wolter, M.; Reifenberger, G.; Herold-Mende, C.; Lichter, P.; Radlwimmer, B. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 2010. [Google Scholar] [CrossRef] [PubMed]
- Röhrich, M.; Zhang, M.; Capper, D.; Wirkner, U.; Trinh, T.; Debus, J.; Timke, C.; Jenne, J.; Huber, P. An Anti-CTGF-antibody Attenuates Tumor Invasion, Delays Tumor-growth, and Prolongs Survival Alone and in Combination with Radiation in an Orthotopic Glioma. Int. J. Radiat. Oncol. 2011. [Google Scholar] [CrossRef]
- Lingasamy, P.; Tobi, A.; Haugas, M.; Hunt, H.; Paiste, P.; Asser, T.; Rätsep, T.; Kotamraju, V.R.; Bjerkvig, R.; Teesalu, T. Bi-specific tenascin-C and fibronectin targeted peptide for solid tumor delivery. Biomaterials 2019. [Google Scholar] [CrossRef]
- Han, Z.; Zhou, Z.; Shi, X.; Wang, J.; Wu, X.; Sun, D.; Chen, Y.; Zhu, H.; Magi-Galluzzi, C.; Lu, Z.R. EDB fibronectin specific peptide for prostate cancer targeting. Bioconjug. Chem. 2015. [Google Scholar] [CrossRef]
- Yeow, Y.L.; Kotamraju, V.R.; Wang, X.; Chopra, M.; Azme, N.; Wu, J.; Schoep, T.D.; Delaney, D.S.; Feindel, K.; Li, J.; et al. Immune-mediated ECM depletion improves tumour perfusion and payload delivery. EMBO Mol. Med. 2019, 11. [Google Scholar] [CrossRef]
- Golombek, S.K.; May, J.N.; Theek, B.; Appold, L.; Drude, N.; Kiessling, F.; Lammers, T. Tumor targeting via EPR: Strategies to enhance patient responses. Adv. Drug Deliv. Rev. 2018, 130, 17–38. [Google Scholar] [CrossRef]
- Danhier, F. To exploit the tumor microenvironment: Since the EPR effect fails in the clinic, what is the future of nanomedicine? J. Control. Release 2016. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, M.; Güç, E.; Park, A.J.; Julier, Z.; Briquez, P.S.; Kuhn, G.A.; Müller, R.; Swartz, M.A.; Hubbell, J.A.; Martino, M.M. Growth factors with enhanced syndecan binding generate tonic signalling and promote tissue healing. Nat. Biomed. Eng. 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Katsumata, K.; Ishihara, J.; Mansurov, A.; Ishihara, A.; Raczy, M.M.; Yuba, E.; Hubbell, J.A. Targeting inflammatory sites through collagen affinity enhances the therapeutic efficacy of anti-inflammatory antibodies. Sci. Adv. 2019, 5, eaay1971. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Ishihara, J.; Ishihara, A.; Miura, R.; Mansurov, A.; Fukunaga, K.; Hubbell, J.A. Engineered collagen-binding serum albumin as a drug conjugate carrier for cancer therapy. Sci. Adv. 2019. [Google Scholar] [CrossRef]
- Ishihara, J.; Ishihara, A.; Sasaki, K.; Lee, S.S.Y.; Williford, J.M.; Yasui, M.; Abe, H.; Potin, L.; Hosseinchi, P.; Fukunaga, K.; et al. Targeted antibody and cytokine cancer immunotherapies through collagen affinity. Sci. Transl. Med. 2019. [Google Scholar] [CrossRef]
- Ishihara, J.; Fukunaga, K.; Ishihara, A.; Larsson, H.M.; Potin, L.; Hosseinchi, P.; Galliverti, G.; Swartz, M.A.; Hubbell, J.A. Matrix-binding checkpoint immunotherapies enhance antitumor efficacy and reduce adverse events. Sci. Transl. Med. 2017. [Google Scholar] [CrossRef]
- González, L.O.; Pidal, I.; Junquera, S.; Corte, M.D.; Vázquez, J.; Rodríguez, J.C.; Lamelas, M.L.; Merino, A.M.; García-Mũiz, J.L.; Vizoso, F.J. Overexpression of matrix metalloproteinases and their inhibitors in mononuclear inflammatory cells in breast cancer correlates with metastasis-relapse. Br. J. Cancer 2007. [Google Scholar] [CrossRef]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Lewis, C.E.; Harney, A.S.; Pollard, J.W. The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell 2016, 30, 18–25. [Google Scholar] [CrossRef]
- Hiratsuka, S.; Nakamura, K.; Iwai, S.; Murakami, M.; Itoh, T.; Kijima, H.; Shipley, J.M.; Senior, R.M.; Shibuya, M. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell 2002. [Google Scholar] [CrossRef]
- Madsen, D.H.; Bugge, T.H. Imaging collagen degradation in vivo highlights a key role for M2-polarized macrophages in extracellular matrix degradation. Oncoimmunology 2013, 2, e27127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hughes, R.; Qian, B.Z.; Rowan, C.; Muthana, M.; Keklikoglou, I.; Olson, O.C.; Tazzyman, S.; Danson, S.; Addison, C.; Clemons, M.; et al. Perivascular M2 macrophages stimulate tumor relapse after chemotherapy. Cancer Res. 2015, 75, 3479–3491. [Google Scholar] [CrossRef] [PubMed]
- Wyckoff, J.B.; Wang, Y.; Lin, E.Y.; Li, J.F.; Goswami, S.; Stanley, E.R.; Segall, J.E.; Pollard, J.W.; Condeelis, J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007, 67, 2649–2656. [Google Scholar] [CrossRef] [PubMed]
- Sable, R.; Jaynes, J.; Ronzetti, M.; Guzman, W.; Knotts, Z.; Val, N.; de Morgan, J.; Yates, C.; Bolormaa, B.; Rudloff, U. Abstract B49: Precision targeting of M2-like macrophages by the innate defense regulator RP-182 in pancreatic cancer and noncancerous diseases. Cancer Res. 2019, 79, B49. [Google Scholar]
- Cieslewicz, M.; Tang, J.; Yu, J.L.; Cao, H.; Zavaljevski, M.; Motoyama, K.; Lieber, A.; Raines, E.W.; Pun, S.H. Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival. Proc. Natl. Acad. Sci. USA 2013, 110, 15919–15924. [Google Scholar] [CrossRef] [PubMed]
- Scodeller, P.; Simón-Gracia, L.; Kopanchuk, S.; Tobi, A.; Kilk, K.; Säälik, P.; Kurm, K.; Squadrito, M.L.; Kotamraju, V.R.; Rinken, A.; et al. Precision Targeting of Tumor Macrophages with a CD206 Binding Peptide. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Asciutto, E.K.; Kopanchuk, S.; Lepland, A.; Simón-Gracia, L.; Aleman, C.; Teesalu, T.; Scodeller, P. Phage-Display-Derived Peptide Binds to Human CD206 and Modeling Reveals a New Binding Site on the Receptor. J. Phys. Chem. B 2019. [Google Scholar] [CrossRef]
- Ehlers, S. DC-SIGN and mannosylated surface structures of Mycobacterium tuberculosis: A deceptive liaison. Eur. J. Cell Biol. 2010. [Google Scholar] [CrossRef]
- Lasala, F.; Arce, E.; Otero, J.R.; Rojo, J.; Delgado, R. Mannosyl Glycodendritic Structure Inhibits DC-SIGN-Mediated Ebola Virus Infection in cis and in trans. Antimicrob. Agents Chemother. 2003. [Google Scholar] [CrossRef]
- Leber, N.; Kaps, L.; Yang, A.; Aslam, M.; Giardino, M.; Klefenz, A.; Choteschovsky, N.; Rosigkeit, S.; Mostafa, A.; Nuhn, L.; et al. α-Mannosyl-Functionalized Cationic Nanohydrogel Particles for Targeted Gene Knockdown in Immunosuppressive Macrophages. Macromol. Biosci. 2019. [Google Scholar] [CrossRef]
- Leamon, C.P.; Low, P.S. Delivery of macromolecules into living cells: A method that exploits folate receptor endocytosis. Proc. Natl. Acad. Sci. USA 1991. [Google Scholar] [CrossRef] [PubMed]
- Hilgenbrink, A.R.; Low, P.S. Folate receptor-mediated drug targeting: From therapeutics to diagnostics. J. Pharm. Sci. 2005. [Google Scholar] [CrossRef] [PubMed]
- Paulos, C.M.; Turk, M.J.; Breur, G.J.; Low, P.S. Folate receptor-mediated targeting of therapeutic and imaging agents to activated macrophages in rheumatoid arthritis. Adv. Drug Deliv. Rev. 2004. [Google Scholar] [CrossRef] [PubMed]
- Low, P.S.; Kularatne, S.A. Folate-targeted therapeutic and imaging agents for cancer. Curr. Opin. Chem. Biol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Puig-Kröger, A.; Sierra-Filardi, E.; Domínguez-Soto, A.; Samaniego, R.; Corcuera, M.T.; Gómez-Aguado, F.; Ratnam, M.; Sánchez-Mateos, P.; Corbí, A. Folate receptor β is expressed by tumor-associated macrophages and constitutes a marker for M2 anti-inflammatory/regulatory Macrophages. Cancer Res. 2009. [Google Scholar] [CrossRef] [PubMed]
- Guertin, A.D.; O’Neil, J.; Stoeck, A.; Reddy, J.A.; Cristescu, R.; Haines, B.B.; Hinton, M.C.; Dorton, R.; Bloomfield, A.; Nelson, M.; et al. High Levels of Expression of P-glycoprotein/Multidrug Resistance Protein Result in Resistance to Vintafolide. Mol. Cancer Ther. 2016. [Google Scholar] [CrossRef]
- Lee, Y.G.; Chu, H.; Lu, Y.; Leamon, C.P.; Srinivasarao, M.; Putt, K.S.; Low, P.S. Regulation of CAR T cell-mediated cytokine release syndrome-like toxicity using low molecular weight adapters. Nat. Commun. 2019. [Google Scholar] [CrossRef]
- Lee, Y.G.; Marks, I.; Srinivasarao, M.; Kanduluru, A.K.; Mahalingam, S.M.; Liu, X.; Chu, H.; Low, P.S. Use of a single CAR T cell and several bispecific adapters facilitates eradication of multiple antigenically different solid tumors. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Silver, D.A.; Pellicer, I.; Fair, W.R.; Heston, W.D.W.; Cordon-Cardo, C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997, 3, 81–85. [Google Scholar]
- Perner, S.; Hofer, M.D.; Kim, R.; Shah, R.B.; Li, H.; Möller, P.; Hautmann, R.E.; Gschwend, J.E.; Kuefer, R.; Rubin, M.A. Prostate-specific membrane antigen expression as a predictor of prostate cancer progression. Hum. Pathol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Choy, C.J.; Ling, X.; Geruntho, J.J.; Beyer, S.K.; Latoche, J.D.; Langton-Webster, B.; Anderson, C.J.; Berkman, C.E. 177Lu-labeled phosphoramidate-based PSMA inhibitors: The effect of an albumin binder on biodistribution and therapeutic efficacy in prostate tumor-bearing mice. Theranostics 2017. [Google Scholar] [CrossRef] [PubMed]
- Locke, L.W.; Mayo, M.W.; Yoo, A.D.; Williams, M.B.; Berr, S.S. PET imaging of tumor associated macrophages using mannose coated 64Cu liposomes. Biomaterials 2012, 33, 7785–7793. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Cai, H.; Peng, X.; Zhang, P.; Wu, X.; Tian, R. Targeted imaging of tumor-associated macrophages by cyanine 7-labeled mannose in xenograft tumors. Mol. Imaging 2017. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Rajaram, M.V.S.; Metz, W.L.; Cope, F.O.; Blue, M.S.; Vera, D.R.; Schlesinger, L.S. Tilmanocept, a New Radiopharmaceutical Tracer for Cancer Sentinel Lymph Nodes, Binds to the Mannose Receptor (CD206). J. Immunol. 2015, 195, 2019–2029. [Google Scholar] [CrossRef]
- Wallace, A.M.; Hoh, C.K.; Vera, D.R.; Darrah, D.D.; Schulteis, G. Lymphoseek: A molecular radiopharmaceutical for sentinel node detection. Ann. Surg. Oncol. 2003. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Zupancic, E.; Vandermeulen, G.; Oliveira, V.G.; Salgado, A.; Videira, M.; Gaspar, M.; Graca, L.; Préat, V.; Florindo, H.F. In vivo delivery of peptides and Toll-like receptor ligands by mannose-functionalized polymeric nanoparticles induces prophylactic and therapeutic anti-tumor immune responses in a melanoma model. J. Control. Release 2015. [Google Scholar] [CrossRef]
- Conniot, J.; Scomparin, A.; Peres, C.; Yeini, E.; Pozzi, S.; Matos, A.I.; Kleiner, R.; Moura, L.I.F.; Zupančič, E.; Viana, A.S.; et al. Immunization with mannosylated nanovaccines and inhibition of the immune-suppressing microenvironment sensitizes melanoma to immune checkpoint modulators. Nat. Nanotechnol. 2019. [Google Scholar] [CrossRef]
- Di Matteo, P.; Curnis, F.; Longhi, R.; Colombo, G.; Sacchi, A.; Crippa, L.; Protti, M.P.; Ponzoni, M.; Toma, S.; Corti, A. Immunogenic and structural properties of the Asn-Gly-Arg (NGR) tumor neovasculature-homing motif. Mol. Immunol. 2006. [Google Scholar] [CrossRef]
- De, G.; Ko, J.K.; Tan, T.; Zhu, H.; Li, H.; Ma, J. Amphipathic tail-anchoring peptide is a promising therapeutic agent for prostate cancer treatment. Oncotarget 2014. [Google Scholar] [CrossRef][Green Version]
- Suhorutsenko, J.; Oskolkov, N.; Arukuusk, P.; Kurrikoff, K.; Eriste, E.; Copolovici, D.M.; Langel, Ü. Cell-penetrating peptides, PepFects, show no evidence of toxicity and immunogenicity in vitro and in vivo. Bioconjug. Chem. 2011. [Google Scholar] [CrossRef]
- Reardon, D.A.; Fink, K.L.; Mikkelsen, T.; Cloughesy, T.F.; O’Neill, A.; Plotkin, S.; Glantz, M.; Ravin, P.; Raizer, J.J.; Rich, K.M.; et al. Randomized phase II study of cilengitide, an integrin-targeting arginine-glycine-aspartic acid peptide, in recurrent glioblastoma multiforme. J. Clin. Oncol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Krüger, D.M.; Glas, A.; Bier, D.; Pospiech, N.; Wallraven, K.; Dietrich, L.; Ottmann, C.; Koch, O.; Hennig, S.; Grossmann, T.N. Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions. J. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Cryo-electron microscopy shapes up. Nature 2018. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, D.; Sasaki, A.; Ikeya, T.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; Wälchli, M.; et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 2009. [Google Scholar] [CrossRef]
| Docking Protocol | Description | URL |
|---|---|---|
| AutoDock | A parameter set based on the AMBER force field (Cornell et al. 1995) is used. Flexible or fixed torsions for the ligands can be used | http://autodock.scripps.edu/ |
| HPEPDOCK | Peptide flexibility is included as an ensemble of peptide conformations | http://huanglab.phys.hust.edu.cn/hpepdock/ |
| AnchorDock | Uses prior identification of anchoring spots on the protein surface and the peptide is simulated around the predicted spots. | http://dx.doi.org/10.1016/j.str.2015.03.010 |
| CABS-dock | Multiscale procedure using coarse-grained protein model and a Monte Carlo scheme. | http://biocomp.chem.uw.edu.pl/CABSdock |
| AutoDock CrankPeP | Folds the peptide in the potential energy landscape created by the receptor. | http://adcp.scripps.edu/ |
| PepVis | Pipeline for automated script generation | https://github.com/inpacdb/PepVis |
| Peptide | Sequence/Structure | Ref | Identification | Advantages |
|---|---|---|---|---|
| iRGD | CRGDKGPDC (disulfide cyclized) | [103] | In vitro phage display | * Tumor-penetrating peptide * Medium-high affinity for αvβ5 integrin (KD: 62nM) and αvβ3 integrin (KD: 18 nM) * Extensively validated in different tumor models * Proven non-immunogenicity in mice * In phase I clinical trials |
| Cilengitide | RGDfV (head to tail cyclized) | [27] | By rational design based on RGD motif | * High affinity for αvβ3 integrin (KD: 0.58nM) * Safe in humans |
| SFITGv6 | GRCTFRGDLMQLCYPD (head-to-tail cyclized, disulfide bonded) | [119] | In vitro phage display | * High affinity for ανβ6 integrin (KD: 15nM) * Validated applicability in cancer patients |
| CNGRC | CNGRC (disulfide cyclized) | [128] | In vitro phage display | * Validated in vitro as peptide-drug conjugate * Proven non-immungenicity in mice |
| RGDechi | Cyclic RGD/echistatin hybrid PubChem CID: 91936353 | [120] | By rational design based on cyclic RGD and echistatin | * Validated applicability in PET imaging * Can discriminate between αvβ3 and αvβ5 integrin |
| Peptide Name | Sequence/Structure | Ref | Identification | Advantages |
|---|---|---|---|---|
| DAG | CDAGRKQKC (disulfide cyclized) | [20] | In vivo phage display in Alzheimer mice | * Robust homing in patient-derived glioblastoma * Known receptor (CTGF) |
| ZD2 | CTVRTSADC (disulfide cyclized) | [139] | In vitro phage display on recombinant protein | * Known receptor and affinity (Fibronectin extra domain B, KD:11µM) * PET and MRI active conjugates of ZD2 are being commercially developed by the company Molecular Theranostics |
| CSG | CSGRRSSKC (disulfide cyclized) | [140] | In vitro phage display on Matrigel TM | * Robust homing in different mouse models * Known receptor (laminin-nidogen complex) |
| PlGF-2122-144 | RRRPKGRGKRRREKQRPTDCHL | [147] | Imitation of natural ligand | * High affinity (low nM KD) binding to multiple ECM proteins * Proven therapeutic value |
| BT1718 (Peptide-Drug Conjugate) | Undisclosed (bicyclic peptide coupled to maytansinoid) | [140] | In vitro phage display | * High affinity (low nM KD) to MMP-14 * In Phase I/IIa clinical trials |
| Ligand Name | Sequence | Reference | Identification | Advantages | Disadvantages/Question Marks |
|---|---|---|---|---|---|
| RP-182 | KFRKAFKRFF | [155] | In silico |
|
|
| M2pep | YEQDPWGVKWWY | [156] | In vitro phage display |
|
|
| cyclic M2pep(RY) | CGYEQDPWGVRYWYGCkkk | [26] | Rational modification of M2pep |
|
|
| Mannose | [161] | Natural ligand of CD206 |
|
| |
| mUNO | CSPGAK | [157,158] | In vivo phage display |
|
|
| Identification | Advantages | Disadvantages/Question Marks |
|---|---|---|
| Phage Display-Cyclic Library (CXiC) |
|
|
| Phage Display-Bicyclic Library (CXiCXjC) |
|
|
| Phage Display-Sunflower Trypsin Inhibitor I Library |
|
|
| Mirror Image Phage Display |
|
|
| Imitation of Natural Binder |
|
|
| In silico Identification |
|
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scodeller, P.; Asciutto, E.K. Targeting Tumors Using Peptides. Molecules 2020, 25, 808. https://doi.org/10.3390/molecules25040808
Scodeller P, Asciutto EK. Targeting Tumors Using Peptides. Molecules. 2020; 25(4):808. https://doi.org/10.3390/molecules25040808
Chicago/Turabian StyleScodeller, Pablo, and Eliana K. Asciutto. 2020. "Targeting Tumors Using Peptides" Molecules 25, no. 4: 808. https://doi.org/10.3390/molecules25040808
APA StyleScodeller, P., & Asciutto, E. K. (2020). Targeting Tumors Using Peptides. Molecules, 25(4), 808. https://doi.org/10.3390/molecules25040808





