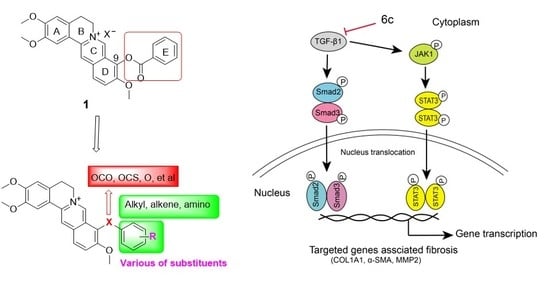
Discovery of 9O-Substituted Palmatine Derivatives as a New Class of Anti-COL1A1 Agents Via Repressing TGF-β1/Smads and JAK1/STAT3 Pathways
Abstract
1. Introduction
2. Results and Discussion
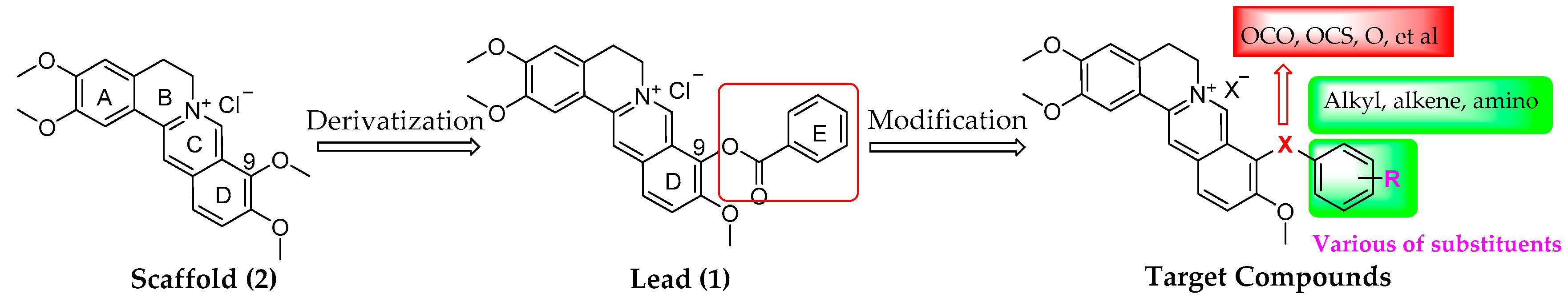
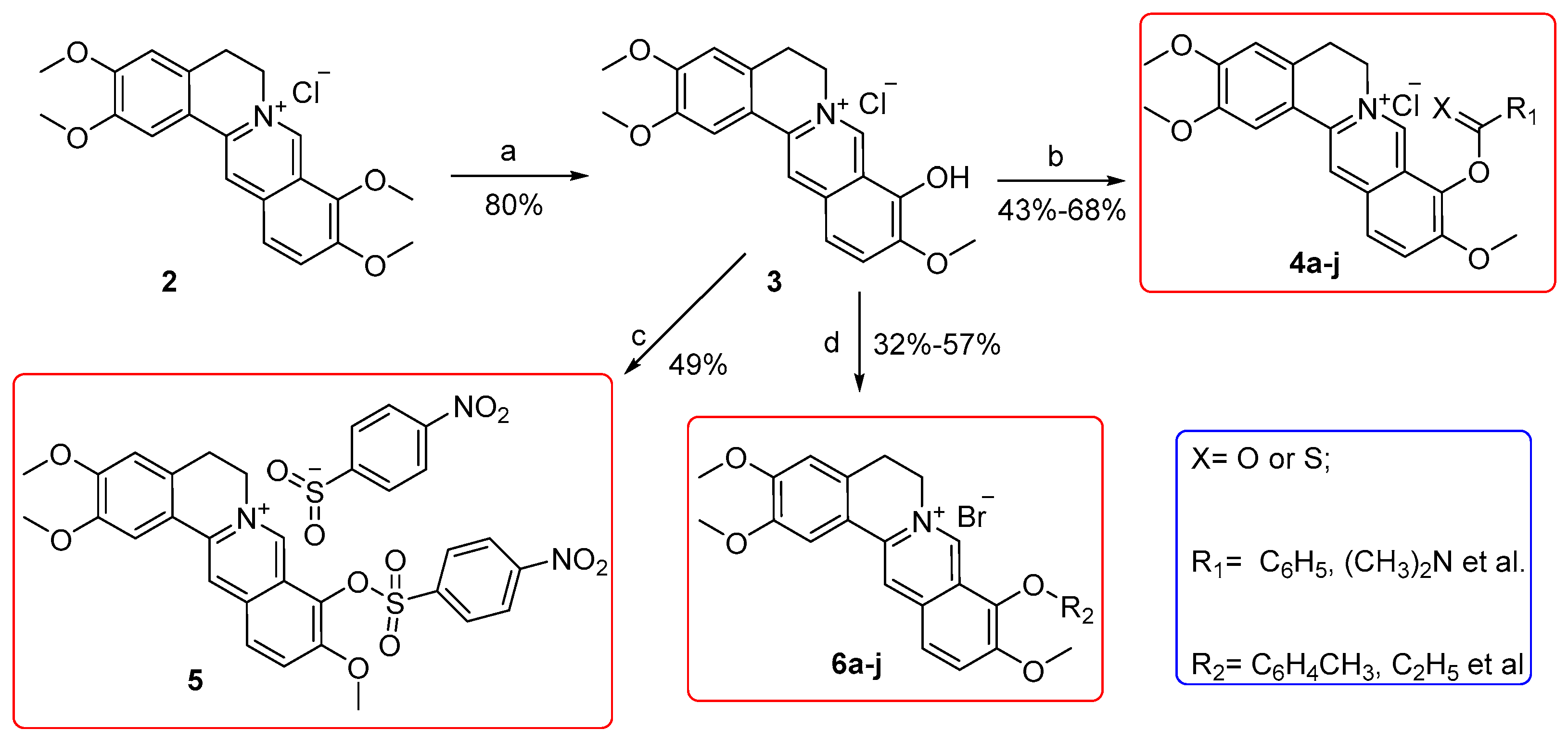
2.1. Chemistry
2.2. Target Compounds Inhibited the Activity of COL1A1 Promotor in Human Hepatic Stellate LX-2 Cells
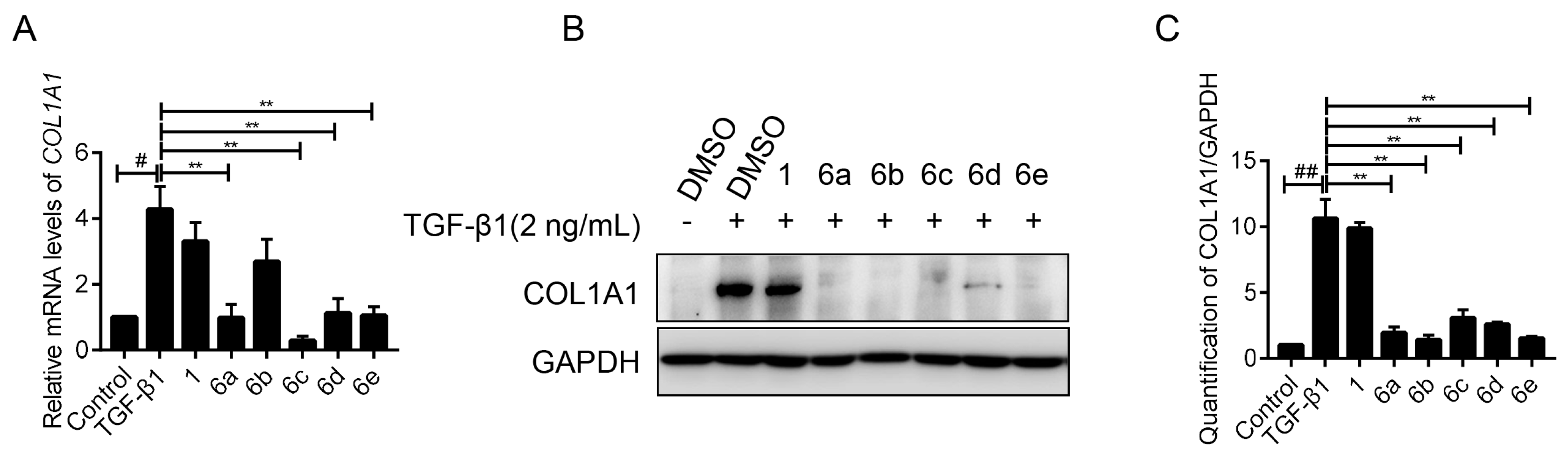
2.3. Key Compounds Inhibited the Expression of COL1A1 in mRNA and Protein Levels
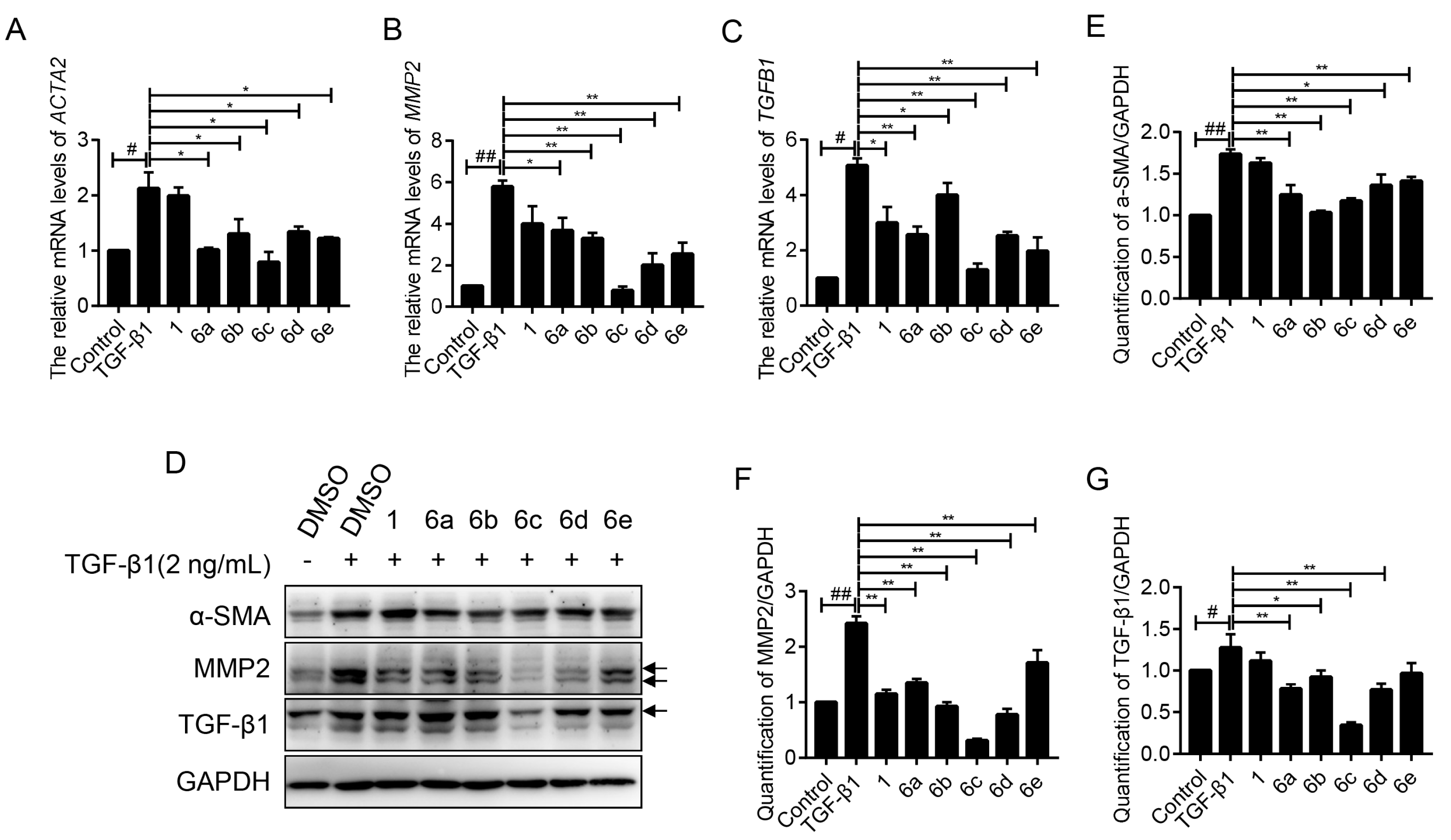
2.4. Key Compounds Inhibited the Expressions of Fibrogenic Genes
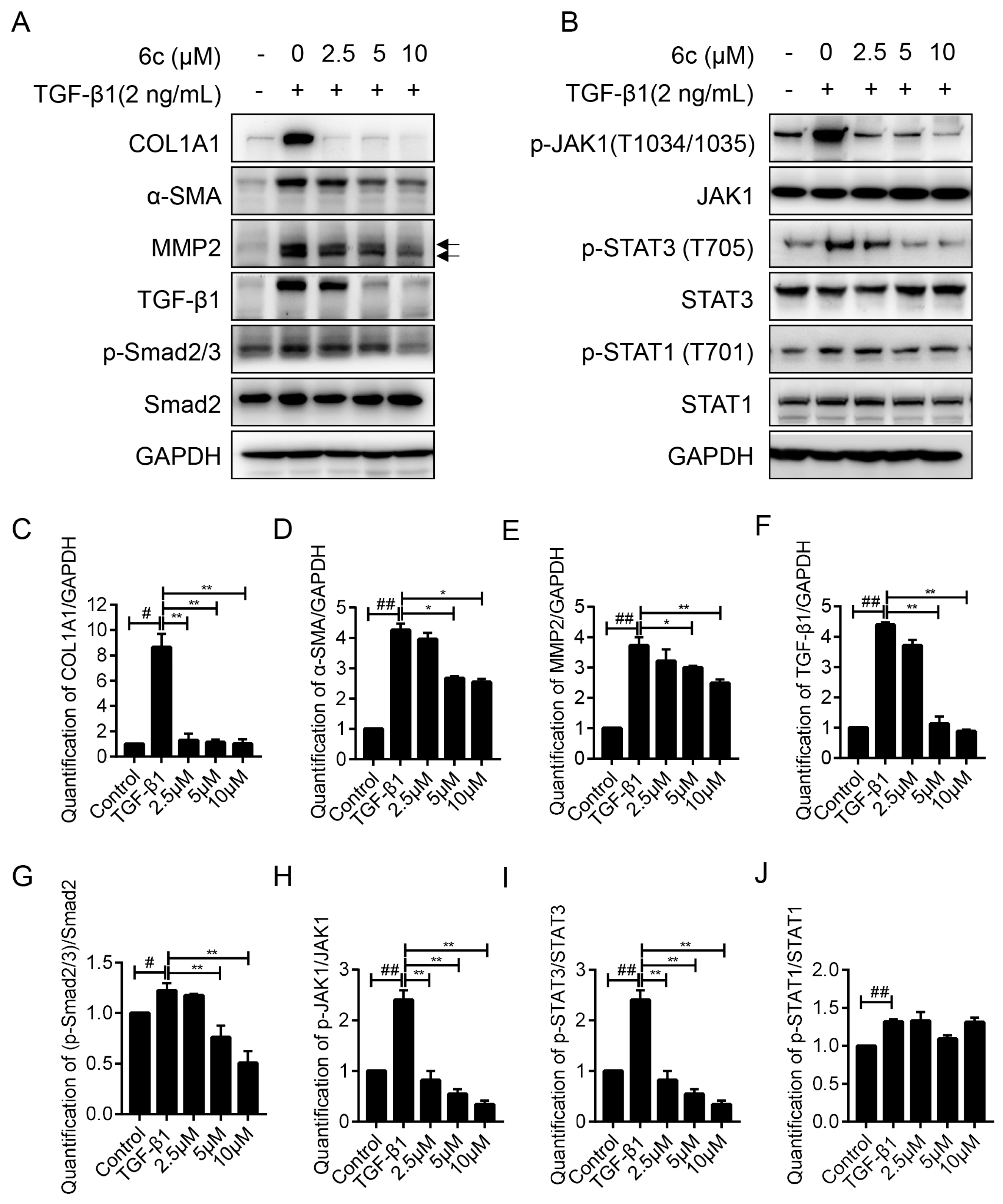
2.5. 6c Inhibited the Expressions of Fibrogentic Proteins in a Dose-Dependent Manner
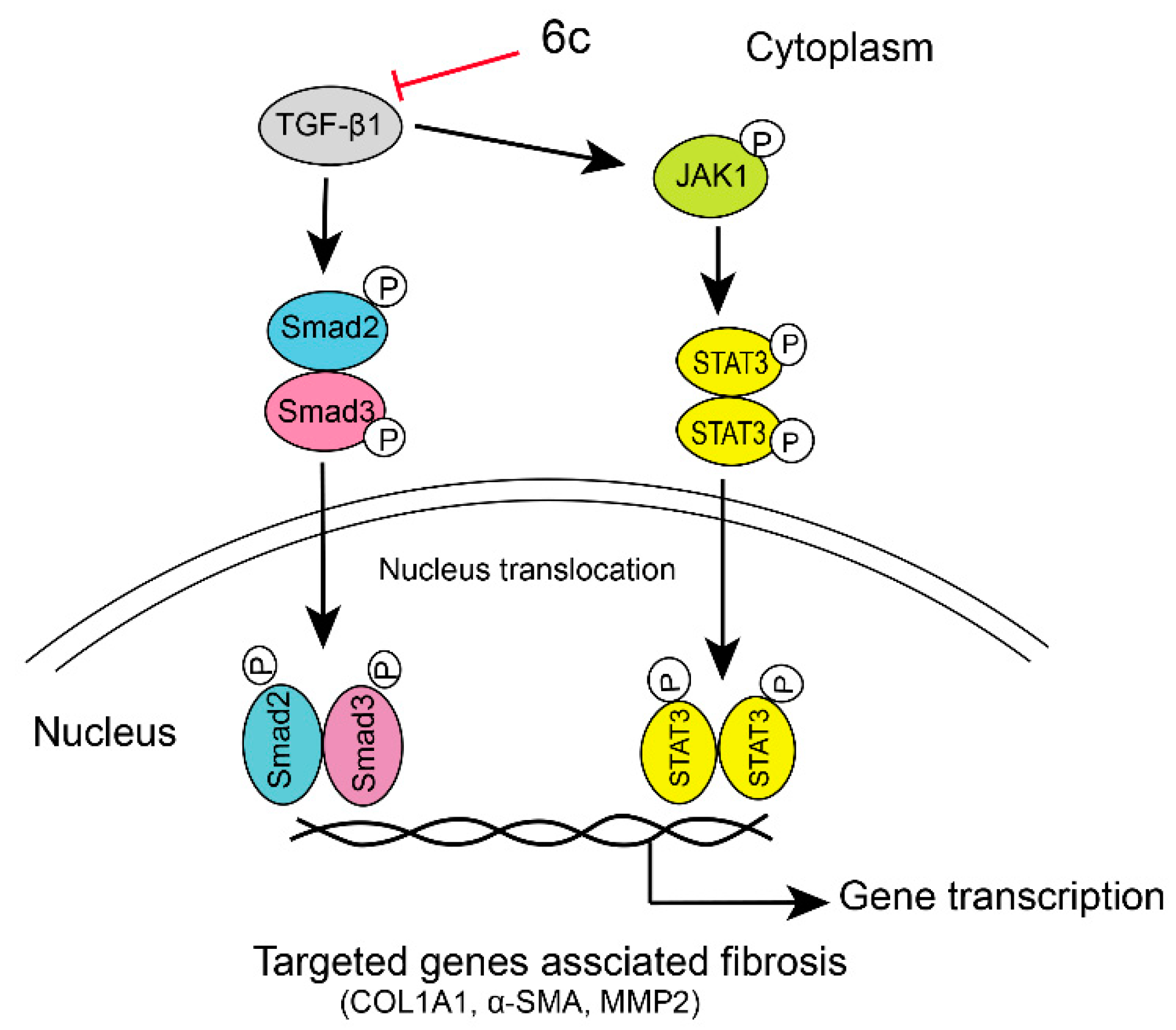
2.6. 6c Supressed the JAK1/STAT3 Signaling Pathway
2.7. Safety Profile of 6c
2.8. Druglike Property Prediction of 6c
3. Materials and Methods
3.1. Apparatus, Materials, and Analysis Reagents
3.2. Chemistry
3.2.1. General Procedure for the Synthesis of Compounds 4a–i and 5
3.2.2. General Procedure for the Synthesis of 6a–j
3.3. Biology Assay
3.3.1. Cell Culture and Screening of Compounds
3.3.2. Cell Survival Assay
3.3.3. RT-PCR Assay
3.3.4. Western blot
3.3.5. Acute Toxicity
3.3.6. Statistics
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lachiondo-Ortega, S.; Mercado-Gómez, M.; Serrano-Maciá, M.; Lopitz-Otsoa, F.; Salas-Villalobos, T.B.; Varela-Rey, M.; Delgado, T.C.; Martínez-Chantar, M.L. Ubiquitin-Like Post-Translational Modifications (Ubl-PTMs): Small Peptides with Huge Impact in Liver Fibrosis. Cells 2019, 8, 1575. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, C.T.; Wang, Y.; Nyberg, S.L. Cell therapy in chronic liver disease. Curr. Opin. Gastroenterol. 2016, 32, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.J.; Friedman, S.L.; Lee, Y.A. Antifibrotic Therapies: Where Are We Now? Semin. Liver Dis. 2016, 36, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Blachier, M.; Leleu, H.; Peck-Radosavljevic, M.; Valla, D.C.; Roudot-Thoraval, F. The burden of liver disease in Europe: A review of available epidemiological data. J. Hepatol. 2013, 58, 593–608. [Google Scholar] [CrossRef]
- Zoubek, M.E.; Trautwein, C.; Strnad, P. Reversal of liver fibrosis: From fiction to reality. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 129–141. [Google Scholar] [CrossRef]
- Starkel, P.; Sempoux, C.; Leclercq, I.; Herin, M.; Deby, C.; Desager, J.P.; Horsmans, Y. Oxidative stress, KLF6 and transforming growth factor-beta up-regulation differentiate non-alcoholic steatohepatitis progressing to fibrosis from uncomplicated steatosis in rats. J. Hepatol. 2003, 39, 538–546. [Google Scholar] [CrossRef]
- Huang, Y.; Deng, X.; Liang, J. Modulation of hepatic stellate cells and reversibility of hepatic fibrosis. Exp. Cell Res. 2017, 352, 420–426. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef]
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar]
- Zhao, S.S.; Wang, J.X.; Wang, Y.C.; Shao, R.G.; He, H.W. Establishment and application of a high-throughput drug screening model based on COL1A1 promoter for anti-liver fibrosis. Acta Pharmacol. Sin. 2015, 50, 169–173. [Google Scholar]
- Ge, M.; Liu, H.; Zhang, Y.; Li, N.; Zhao, S.; Zhao, W.; Zhen, Y.; Yu, J.; He, H.; Shao, R.G. The anti-hepatic fibrosis effects of dihydrotanshinone I are mediated by disrupting the yes-associated protein and transcriptional enhancer factor D2 complex and stimulating autophagy. Br. J. Pharmacol. 2017, 174, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Niu, T.; Niu, W.; Bao, Y.; Liu, T.; Song, D.; Li, Y.; He, H. Discovery of matrinic thiadiazole derivatives as a novel family of anti-liver fibrosis agents via repression of the TGFβ/Smad pathway. Molecules 2018, 23, 1644. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Li, Y.; Bao, Y.; Dai, Z.; Niu, T.; Wang, K.; He, H.; Song, D. Novel cytisine derivatives exert anti-liver fibrosis effect via PI3K/Akt/Smad pathway. Bioorg. Chem. 2019, 90, 103032. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Pang, Y.; Tang, S.; Niu, T.; Guo, Z.; He, H.; Li, Y.; Song, D. 12N-Substituted matrinol derivatives inhibited the expression of fibrogenic genes via repressing Integrin/FAK/PI3K/Akt pathway in hepatic stellate cells. Molecules 2019, 24, 3748. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Song, D.Q.; Li, Y.H.; Kong, W.J.; Wang, Y.X.; Gao, L.M.; Liu, S.Y.; Cao, R.Q.; Jiang, J.D. Synthesis and structure-activity relationships of berberine analogues as a novel class of low-density-lipoprotein receptor up-regulators. Bioorg. Med. Chem. Lett. 2008, 18, 4675–4677. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Yang, P.; Kong, W.J.; Wang, Y.X.; Hu, C.Q.; Zuo, Z.Y.; Wang, Y.M.; Gao, H.; Gao, L.M.; Feng, Y.C.; et al. Berberine analogues as a novel class of the low-density-lipoprotein receptor up-regulators: Synthesis, structure-activity relationships, and cholesterol-lowering efficacy. J. Med. Chem. 2009, 52, 492–501. [Google Scholar] [CrossRef]
- Fan, T.Y.; Wang, Y.X.; Tang, S.; Hu, X.X.; Zeng, Q.X.; Pang, J.; Yang, Y.S.; You, X.F.; Song, D.Q. Synthesis and antibacterial evaluation of 13-substituted cycloberberine derivatives as a novel class of anti-MRSA agents. Eur. J. Med. Chem. 2018, 157, 877–886. [Google Scholar] [CrossRef]
- Zeng, Q.X.; Wang, H.Q.; Wei, W.; Guo, T.T.; Yu, L.; Wang, Y.X.; Li, Y.H.; Song, D.Q. Synthesis and biological evaluation of berberine derivatives as a new class of broad-spectrum antiviral agents against Coxsackievirus B. Bioorg. Chem. 2020, 95, 103490. [Google Scholar] [CrossRef]
- Wang, Y.X.; Yang, L.; Wang, H.Q.; Zhao, X.Q.; Liu, T.; Li, Y.H.; Zeng, Q.X.; Li, Y.H.; Song, D.Q. Synthesis and evolution of berberine derivatives as a new class of antiviral agents against enterovirus 71 through the MEK/ERK pathway and autophagy. Molecules 2018, 23, 2084. [Google Scholar] [CrossRef]
- Yang, Y.S.; Wei, W.; Hu, X.X.; Tang, S.; Pang, J.; You, X.F.; Fan, T.Y.; Wang, Y.X.; Song, D.Q. Evolution and antibacterial evaluation of 8-hydroxy-cycloberberine derivatives as a novel family of antibacterial agents against MRSA. Molecules 2019, 24, 984. [Google Scholar] [CrossRef]
- Fan, T.Y.; Hu, X.X.; Wang, Y.X.; Tang, S.; You, X.F.; Song, D.Q. Design, synthesis and anti-MRSA activities of cycloberberine derivatives with a novel chemical scaffold. Acta Pharm. Sin. 2018, 8, 629–638. [Google Scholar]
- Fan, T.Y.; Hu, X.X.; Tang, S.; Liu, X.J.; Wang, Y.X.; Deng, H.B.; You, X.F.; Jiang, J.D.; Li, Y.H.; Song, D.Q. Discovery and development of 8-substituted cycloberberine derivatives as novel antibacterial agents against MRSA. ACS Med. Chem. Lett. 2018, 9, 484–489. [Google Scholar] [CrossRef]
- Yu, D.K.; Zhang, C.X.; Zhao, S.S.; Zhang, S.H.; Zhang, H.; Cai, S.Y.; Shao, R.G.; He, H.W. The anti-fibrotic effects of epigallocatechin-3-gallate in bile duct-ligated cholestatic rats and human hepatic stellate LX-2 cells are mediated by the PI3K/Akt/Smad pathway. Acta Pharmacol. Sin. 2015, 36, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.J. Fibrogenesis II. Metalloproteinases and their inhibitors in liver fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G245–G249. [Google Scholar] [CrossRef] [PubMed]
- Iredale, J.P. Hepatic stellate cell behavior during resolution of liver injury. Semin. Liver Dis. 2001, 21, 427–436. [Google Scholar] [CrossRef]
- Choi, S.; Jung, H.J.; Kim, M.W.; Kang, J.H.; Shin, D.; Jang, Y.S.; Yoon, Y.S.; Oh, S.H. A novel STAT3 inhibitor, STX-0119, attenuates liver fibrosis by inactivating hepatic stellate cells in mice. Biochem. Biophys. Res. Commun. 2019, 513, 49–55. [Google Scholar] [CrossRef]
- Tang, L.Y.; Heller, M.; Meng, Z.J.; Yu, L.R.; Zhang, Y.E. TGF-β directly activates the JAK1-STAT3 axis to induce hepatic fibrosis in coordination with SMAD pathway. J. Biol. Chem. 2017, 292, 4302–4312. [Google Scholar] [CrossRef]
- Chakraborty, D.; Šumová, B.; Mallano, T.; Chen, C.W.; Distler, A.; Bergmann, C.; Ludolph, I.; Horch, R.E.; Gelse, K.; Ramming, A.; et al. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat. Commun. 2017, 24, 1130–1145. [Google Scholar] [CrossRef]
- Cao, S.; Cao, R.; Liu, X.; Luo, X.; Zhong, W. Design, Synthesis and Biological Evaluation of Novel Benzothiazole Derivatives as Selective PI3Kβ Inhibitors. Molecules 2016, 21, 876. [Google Scholar] [CrossRef]
- Ken, Y.; Michio, T.; Tetsuro, I.; Fumitake, S.; Mieko, T.; Shigeki, N. Relative inhibitory activity of berberine-type alkaloids against 12-O-tetradecanoylphorbol-13-acetate-induced inflammation in mice. Chem. Pharm. Bull. 1991, 39, 1462–1465. [Google Scholar]
- Lu, Z.M.; Zhao, S.H.; Liang, J.Q.; Tu, P.F. Synthesis and characterization of 9-O-alkyl substituted palmatine derivatives. Zhong Guo Zhong Yao Za Zhi 2014, 39, 699–703. [Google Scholar]
Sample Availability: Samples of all targeted compounds are available from the authors. |
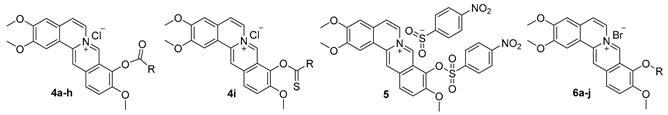
| Code | R | Inhibition Rate a | CC50 b | IC50 c | SI d |
|---|---|---|---|---|---|
| 1 | - | 24.73 ± 3.27% | NTe | NT | NT |
| 4a |  | −5.12 ± 0.86% | NT | NT | NT |
| 4b |  | 30.57 ± 7.77% | NT | NT | NT |
| 4c | CH2CH3 | 13.12 ± 2.38% | NT | NT | NT |
| 4d | (CH2)2CH3 | 6.42 ± 3.63% | NT | NT | NT |
| 4e | (CH2)3CH3 | 16.05 ± 4.09% | NT | NT | NT |
| 4f |  | 7.14 ± 1.79% | NT | NT | NT |
| 4g | N(CH3)2 | 14.83 ± 1.42% | NT | NT | NT |
| 4h |  | 2.86 ± 2.69% | NT | NT | NT |
| 4i | N(CH3)2 | 5.67 ± 4.61% | NT | NT | NT |
| 5 | - | 26.78 ± 3.61% | NT | NT | NT |
| 6a | CH2C6H5 | 38.16 ± 3.81% | NT | NT | NT |
| 6b | 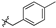 | 77.60 ± 2.93% | NT | NT | NT |
| 6c | 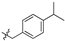 | 96.77 ± 5.64% | 39.30 ± 2.09 | 3.98 ± 0.67 | 9.9 |
| 6d | 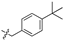 | 84.10 ± 7.91% | 21.43 ± 3.98 | 5.56 ± 0.98 | 3.8 |
| 6e | 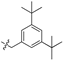 | 96.53 ± 3.00% | 20.66 ± 1.54 | 4.47 ± 0.61 | 4.6 |
| 6f | 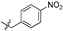 | 25.37 ± 4.40% | NT | NT | NT |
| 6g |  | 33.39 ± 3.71% | NT | NT | NT |
| 6h | CH2CH3 | 26.19 ± 3.63% | NT | NT | NT |
| 6i | (CH2)3CH3 | 26.74 ± 4.00% | NT | NT | NT |
| 6j | CH2CH=CH | 5.26 ± 1.78% | NT | NT | NT |
| EGCG | - | 25.5 ± 7.90% | NT | NT | NT |
| DMSO | - | 2.90 ± 0.00% | NT | NT | NT |
| Code | Absn Risk a | CYP Risk b | TOX Risk c | ADMET Risk d |
|---|---|---|---|---|
| 6c | 0.994 | 1.8 | 1 | 3.825 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, T.; Ge, M.; Guo, Z.; He, H.; Zhang, N.; Li, Y.; Song, D. Discovery of 9O-Substituted Palmatine Derivatives as a New Class of Anti-COL1A1 Agents Via Repressing TGF-β1/Smads and JAK1/STAT3 Pathways. Molecules 2020, 25, 773. https://doi.org/10.3390/molecules25040773
Fan T, Ge M, Guo Z, He H, Zhang N, Li Y, Song D. Discovery of 9O-Substituted Palmatine Derivatives as a New Class of Anti-COL1A1 Agents Via Repressing TGF-β1/Smads and JAK1/STAT3 Pathways. Molecules. 2020; 25(4):773. https://doi.org/10.3390/molecules25040773
Chicago/Turabian StyleFan, Tianyun, Maoxu Ge, Zhihao Guo, Hongwei He, Na Zhang, Yinghong Li, and Danqing Song. 2020. "Discovery of 9O-Substituted Palmatine Derivatives as a New Class of Anti-COL1A1 Agents Via Repressing TGF-β1/Smads and JAK1/STAT3 Pathways" Molecules 25, no. 4: 773. https://doi.org/10.3390/molecules25040773
APA StyleFan, T., Ge, M., Guo, Z., He, H., Zhang, N., Li, Y., & Song, D. (2020). Discovery of 9O-Substituted Palmatine Derivatives as a New Class of Anti-COL1A1 Agents Via Repressing TGF-β1/Smads and JAK1/STAT3 Pathways. Molecules, 25(4), 773. https://doi.org/10.3390/molecules25040773