The Interaction of Cyclic Naphthalene Diimide with G-Quadruplex under Molecular Crowding Condition
Abstract
1. Introduction
2. Results
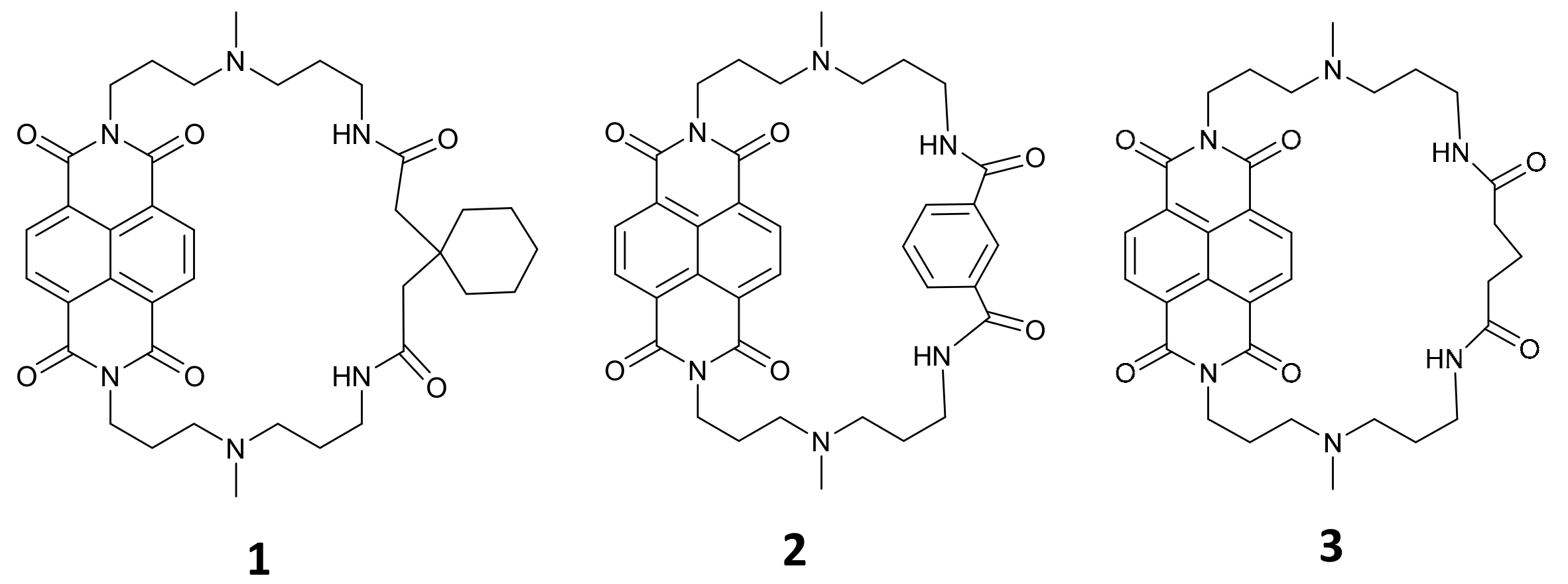
2.1. Synthesis of Cyclic Naphthalene Diimide Derivatives
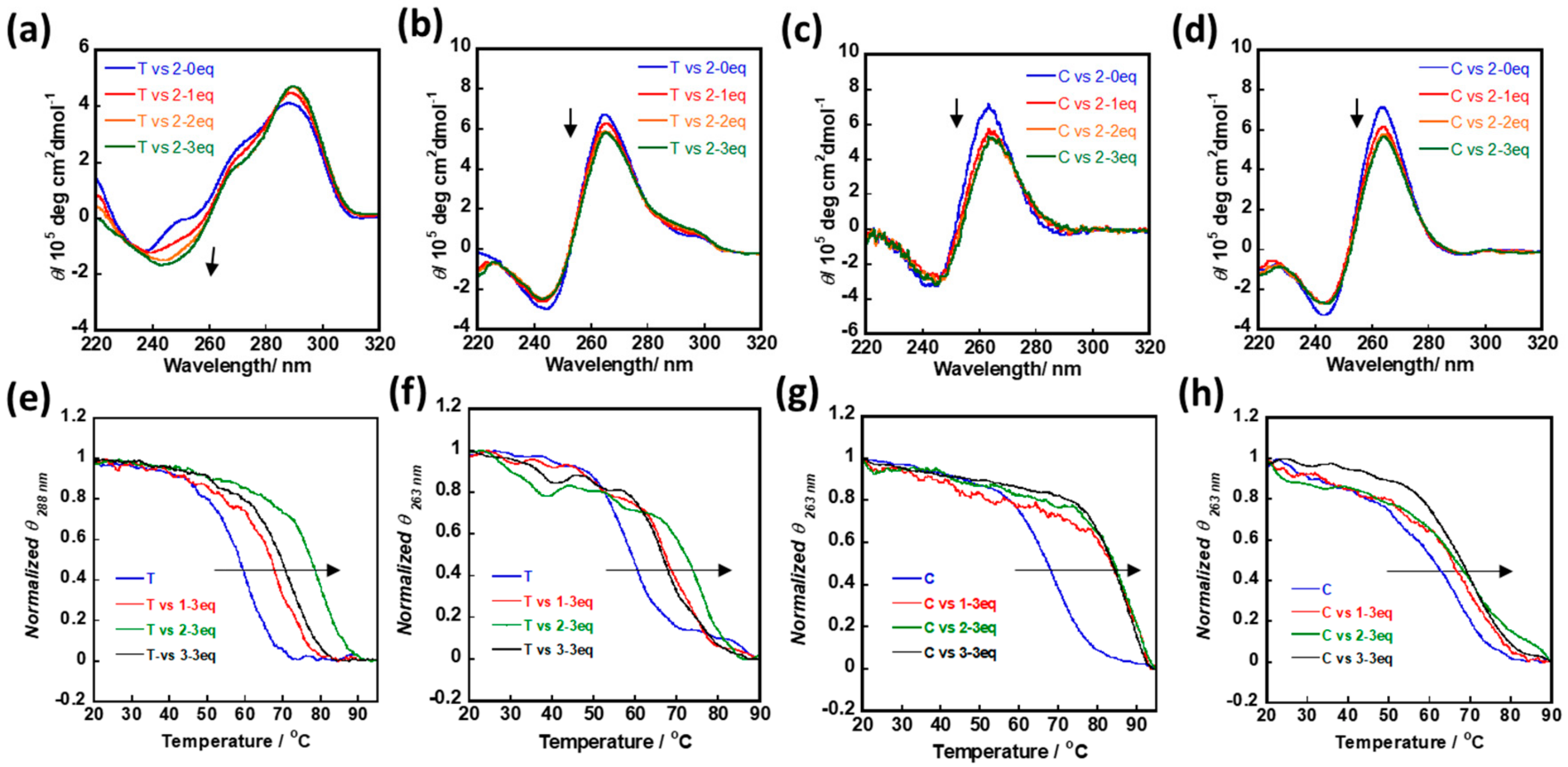
2.2. cNDI Derivatives Recognize and Stabilize G-Quadruplex under Molecular Crowding Condition
2.3. cNDI Derivatives Bind to G-quadruplex under Molecular Crowding Condition
2.4. cNDI Derivatives Induced the Formation of Telomere Hybrid G-quadruplex under Cation-Deficient Molecular Crowding Condition
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of cNDI Derivatives
4.3. Circular Dichroism (CD) Measurement
4.4. Melting Temperature (Tm) Detection
4.5. Isothermal Titration Calorimetry (ITC) Measurements
4.6. UV-Vis Absorption Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Kang, H.J.; Luevano, L.A.; Gokhale, V.; Wu, K.; Pandey, R.; Sherry Chow, H.H.; Hurley, L.H.; Kraft, A.S. Small-Molecule-Targeting Hairpin Loop of hTERT Promoter G-Quadruplex Induces Cancer Cell Death. Cell. Chem. Biol. 2019, 26, 1110–1121.e4. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Fenk, K.D.; Huang, D.; Sen, S.; Cowan, J.A. Rapid Telomere Reduction in Cancer Cells Induced by G-Quadruplex-Targeting Copper Complexes. J. Med. Chem. 2019, 62, 5040–5048. [Google Scholar] [CrossRef]
- Neidle, S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125. [Google Scholar] [CrossRef]
- Oganesian, L.; Bryan, T.M. Physiological relevance of telomeric G-quadruplex formation: A potential drug target. Bioessays 2007, 29, 155–165. [Google Scholar] [CrossRef]
- Cimino-Reale, G.; Zaffaroni, N.; Folini, M. Emerging Role of G-quadruplex DNA as Target in Anticancer Therapy. Curr. Pharm. Des. 2016, 22, 6612–6624. [Google Scholar] [CrossRef]
- Tan, J.H.; Gu, L.Q.; Wu, J.Y. Design of selective G-quadruplex ligands as potential anticancer agents. Mini. Rev. Med. Chem. 2008, 8, 1163–1178. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef]
- Muller, S.; Kumari, S.; Rodriguez, R.; Balasubramanian, S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010, 2, 1095–1098. [Google Scholar] [CrossRef]
- Micco, M.; Collie, G.W.; Dale, A.G.; Ohnmacht, S.A.; Pazitna, I.; Gunaratnam, M.; Reszka, A.P.; Neidle, S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013, 56, 2959–2974. [Google Scholar] [CrossRef]
- Esaki, Y.; Islam, M.M.; Fujii, S.; Sato, S.; Takenaka, S. Design of tetraplex specific ligands: Cyclic naphthalene diimide. Chem. Commun. 2014, 50, 5967–5969. [Google Scholar] [CrossRef]
- Islam, M.M.; Sato, S.; Shinozaki, S.; Takenaka, S. Cyclic ferrocenylnaphthalene diimide derivative as a new class of G-quadruplex DNA binding ligand. Bioorg. Med. Chem. Lett. 2017, 27, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Yaku, H.; Murashima, T.; Tateishi-Karimata, H.; Nakano, S.; Miyoshi, D.; Sugimoto, N. Study on effects of molecular crowding on G-quadruplex-ligand binding and ligand-mediated telomerase inhibition. Methods 2013, 64, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Sugimoto, N. Molecular crowding effects on structure and stability of DNA. Biochimie 2008, 90, 1040–1051. [Google Scholar] [CrossRef]
- Zhu, L.N.; Wu, B.; Kong, D.M. Specific recognition and stabilization of monomeric and multimeric G-quadruplexes by cationic porphyrin TMPipEOPP under molecular crowding conditions. Nucleic Acids Res. 2013, 41, 4324–4335. [Google Scholar] [CrossRef]
- Wang, Z.F.; Chang, T.C. Molecular engineering of G-quadruplex ligands based on solvent effect of polyethylene glycol. Nucleic Acids Res. 2012, 40, 8711–8720. [Google Scholar] [CrossRef][Green Version]
- Xue, Y.; Kan, Z.-Y.; Wang, Q.; Yao, Y.; Liu, J.; Hao, Y.H.; Tan, Z. Human Telomeric DNA Forms Parallel-Stranded Intramolecular G-Quadruplex in K+ Solution under Molecular Crowding Condition. J. AM. Chem. Soc. 2007, 129, 11185–11191. [Google Scholar] [CrossRef]
- Viglasky, V.; Tluckova, K.; Bauer, L. The first derivative of a function of circular dichroism spectra: Biophysical study of human telomeric G-quadruplex. Eur. Biophys. J. 2011, 40, 29–37. [Google Scholar] [CrossRef]
- Petraccone, L.; Fotticchia, I.; Cummaro, A.; Pagano, B.; Ginnari-Satriani, L.; Haider, S.; Randazzo, A.; Novellino, E.; Neidle, S.; Giancola, C. The triazatruxene derivative azatrux binds to the parallel form of the human telomeric G-quadruplex under molecular crowding conditions: Biophysical and molecular modeling studies. Biochimie 2011, 93, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Martino, L.; Pagano, B.; Fotticchia, I.; Neidle, S.; Giancola, C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B 2009, 113, 14779–14786. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 2001, 26, 597–604. [Google Scholar] [CrossRef]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Wu, L.; Oka, H.; Karimata, H.T.; Kirihata, T.; Sato, Y.; Fujii, S.; Sakai, H.; Kuwahara, M.; Sawai, H.; et al. Conformation and the sodium ion condensation on DNA and RNA structures in the presence of a neutral cosolute as a mimic of the intracellular media. Mol. Biosyst. 2008, 4, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Nagatoishi, S.; Tanaka, Y.; Tsumoto, K. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007, 352, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
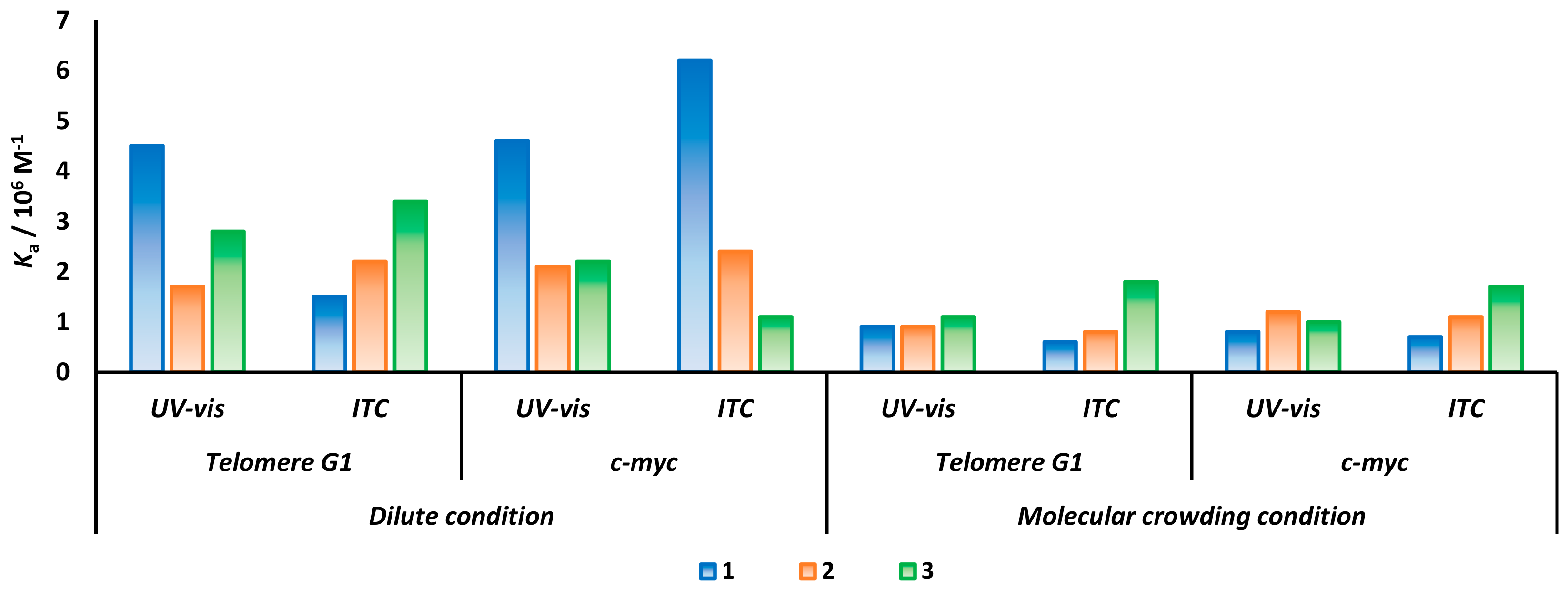

| Dilute Condition | Molecular Crowding Condition | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Telomere G1 | c-myc | Telomere G1 | c-myc | |||||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| 10−6; Ka/M−1 | 1.5 | 2.2 | 3.4 | 6.2 | 2.4 | 1.1 | 0.6 | 0.7 | 1.9 | 0.7 | 0.8 | 1.6 |
| n | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ∆H/kcal mol−1 | −12.8 | −9 | −8.5 | −13.9 | −10 | −11 | −43 | −35 | −22 | −20 | −53 | −20 |
| −T∆S/kcal mol−1 | 4.4 | 0.36 | −0.4 | 4.7 | 1.3 | 2.9 | 35 | 27 | 14 | 12 | 45 | 12 |
| ∆G/kcal mol−1 | −8.4 | −8.7 | −8.9 | −9.2 | −8.7 | −8.1 | −7.9 | −8.0 | −8.6 | −8.0 | −8.0 | −8.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, T.; Sato, S.; Yasukawa, R.; Takeuchi, R.; Ozaki, S.; Fujii, S.; Takenaka, S. The Interaction of Cyclic Naphthalene Diimide with G-Quadruplex under Molecular Crowding Condition. Molecules 2020, 25, 668. https://doi.org/10.3390/molecules25030668
Zou T, Sato S, Yasukawa R, Takeuchi R, Ozaki S, Fujii S, Takenaka S. The Interaction of Cyclic Naphthalene Diimide with G-Quadruplex under Molecular Crowding Condition. Molecules. 2020; 25(3):668. https://doi.org/10.3390/molecules25030668
Chicago/Turabian StyleZou, Tingting, Shinobu Sato, Rui Yasukawa, Ryusuke Takeuchi, Shunsuke Ozaki, Satoshi Fujii, and Shigeori Takenaka. 2020. "The Interaction of Cyclic Naphthalene Diimide with G-Quadruplex under Molecular Crowding Condition" Molecules 25, no. 3: 668. https://doi.org/10.3390/molecules25030668
APA StyleZou, T., Sato, S., Yasukawa, R., Takeuchi, R., Ozaki, S., Fujii, S., & Takenaka, S. (2020). The Interaction of Cyclic Naphthalene Diimide with G-Quadruplex under Molecular Crowding Condition. Molecules, 25(3), 668. https://doi.org/10.3390/molecules25030668






