Biological Evaluation and In Silico Study of Benzoic Acid Derivatives from Bjerkandera adusta Targeting Proteostasis Network Modules
Abstract
1. Introduction
2. Results and Discussion
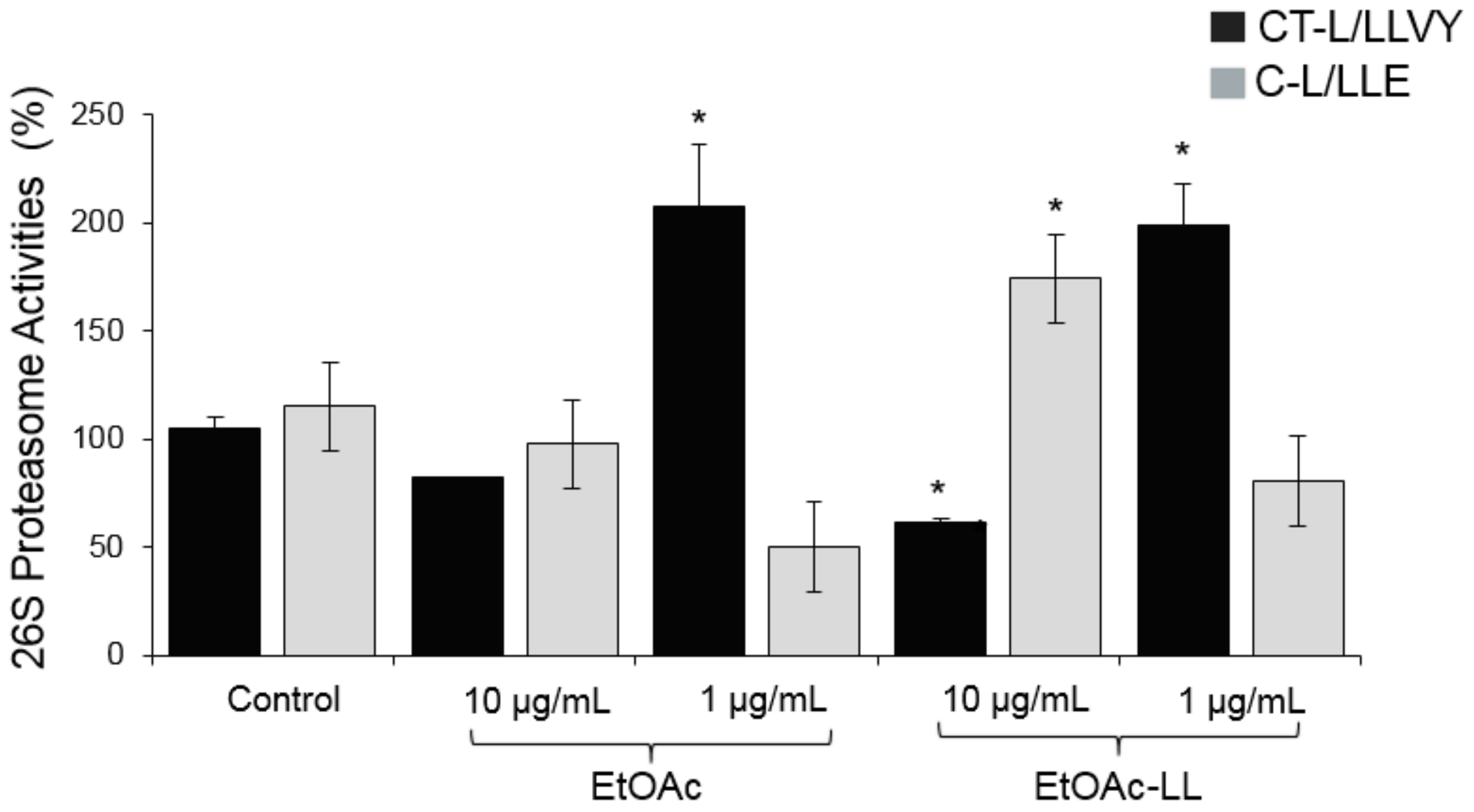
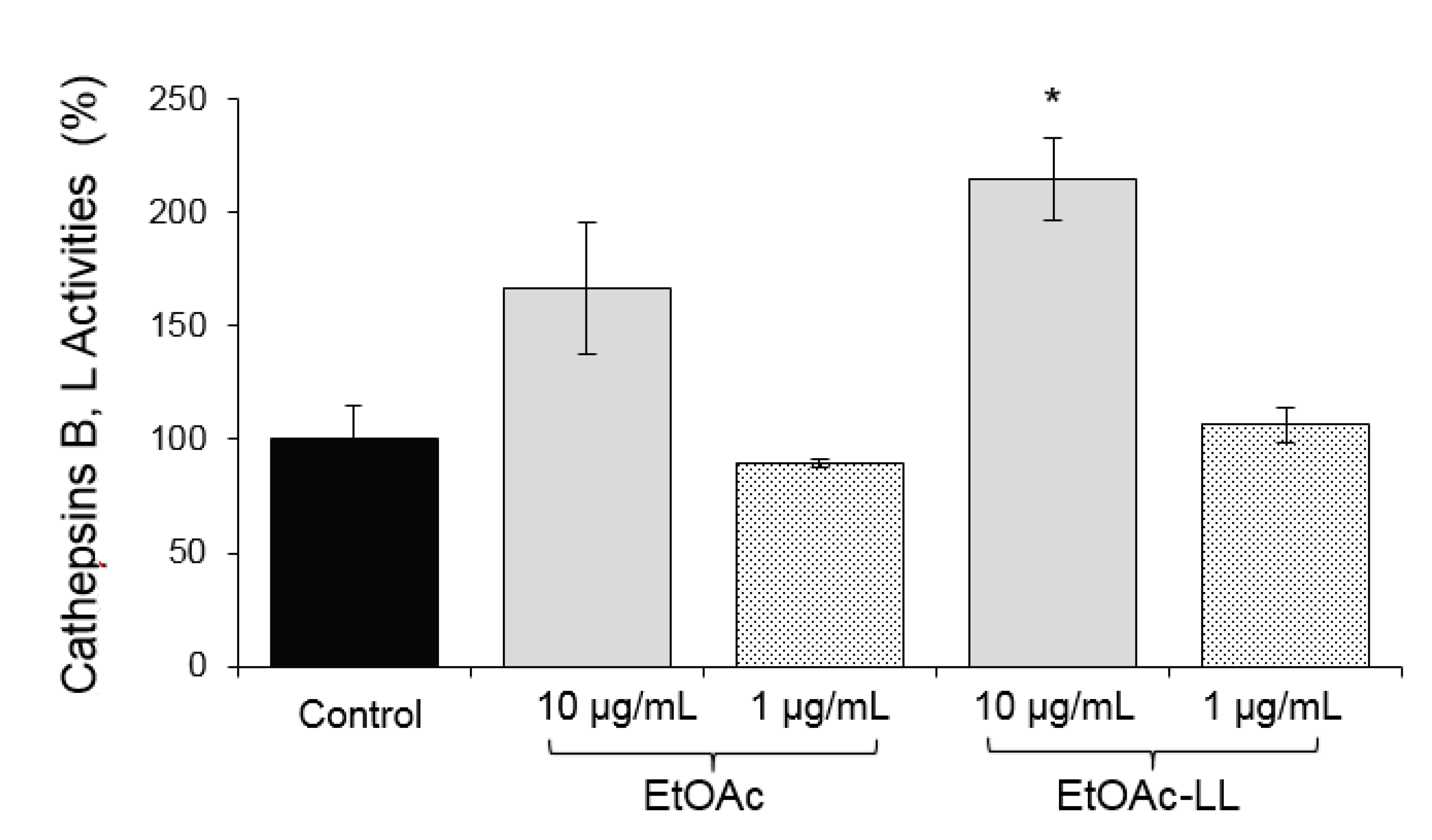
2.1. Extraction and Bioevaluation
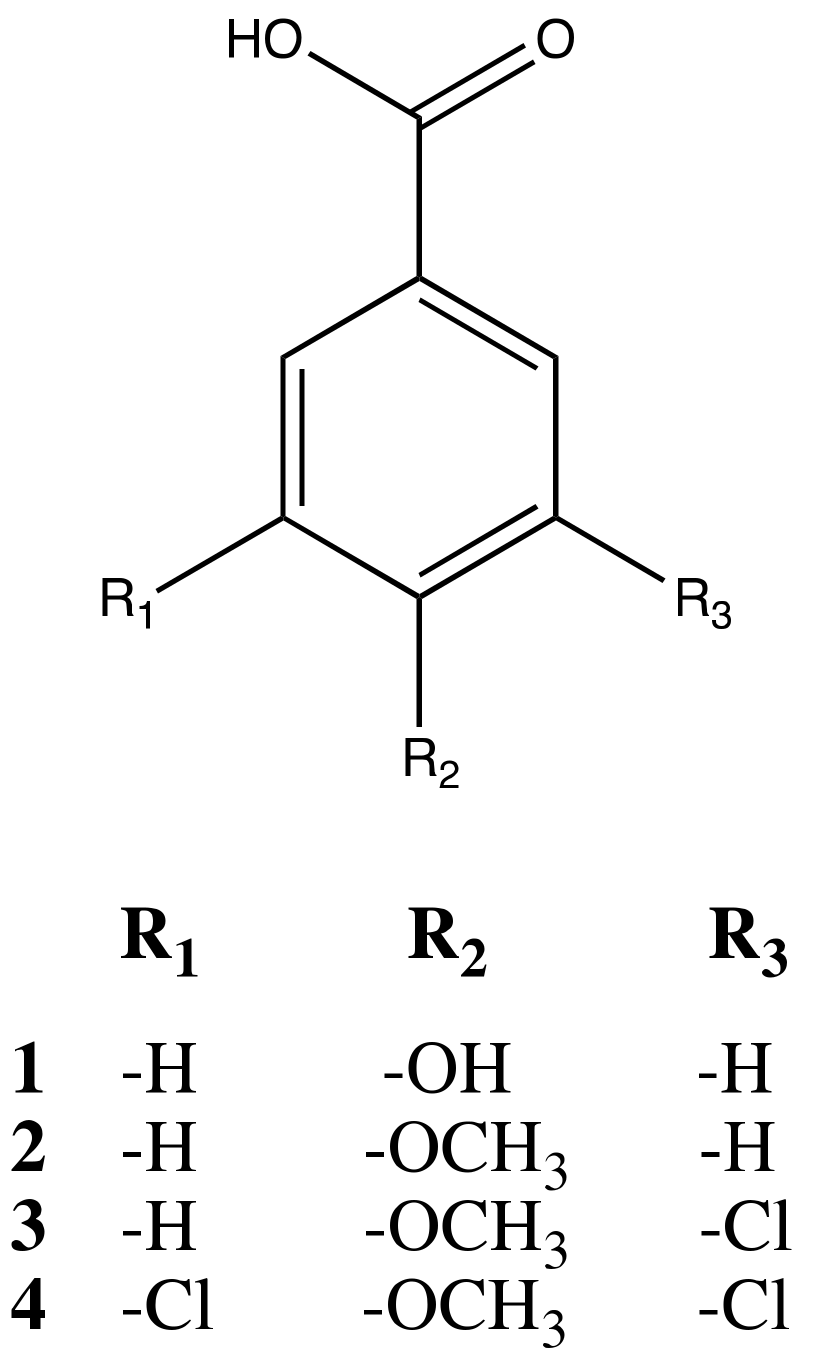
2.2. Isolation
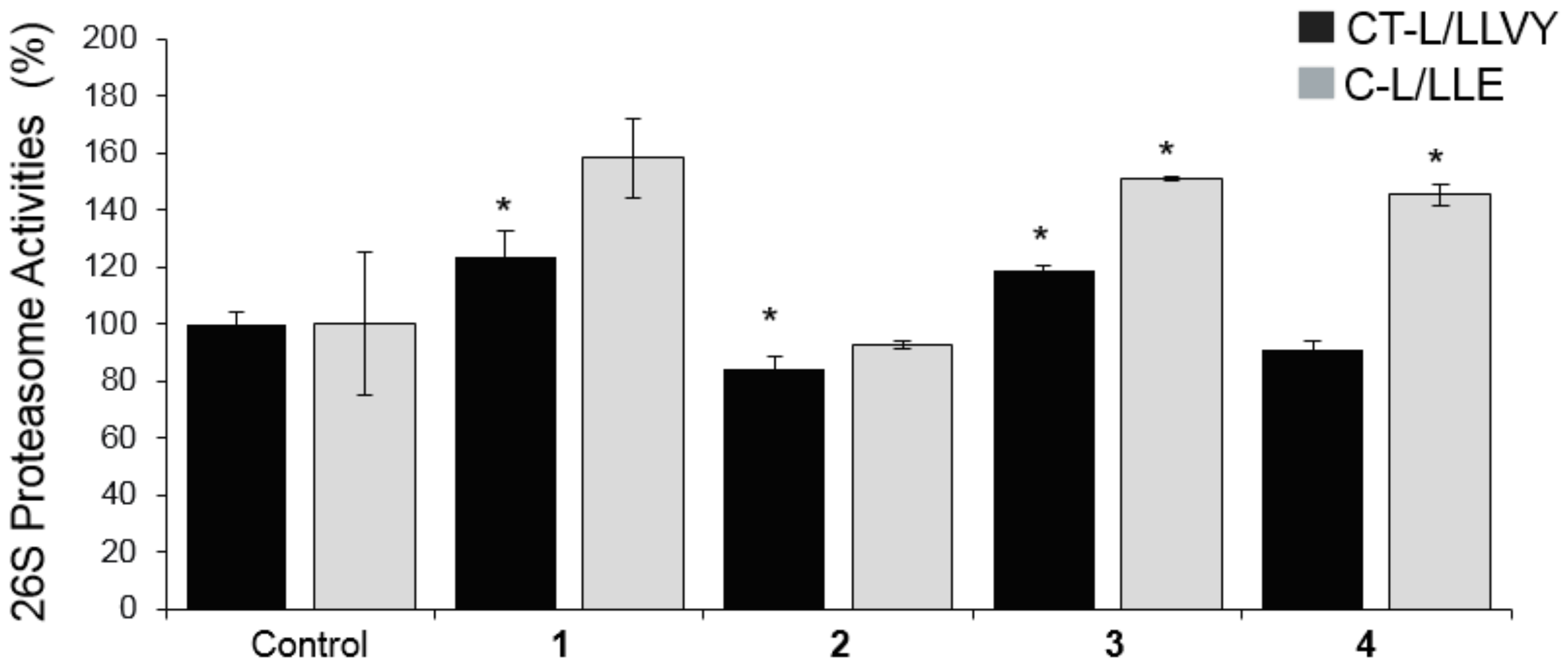
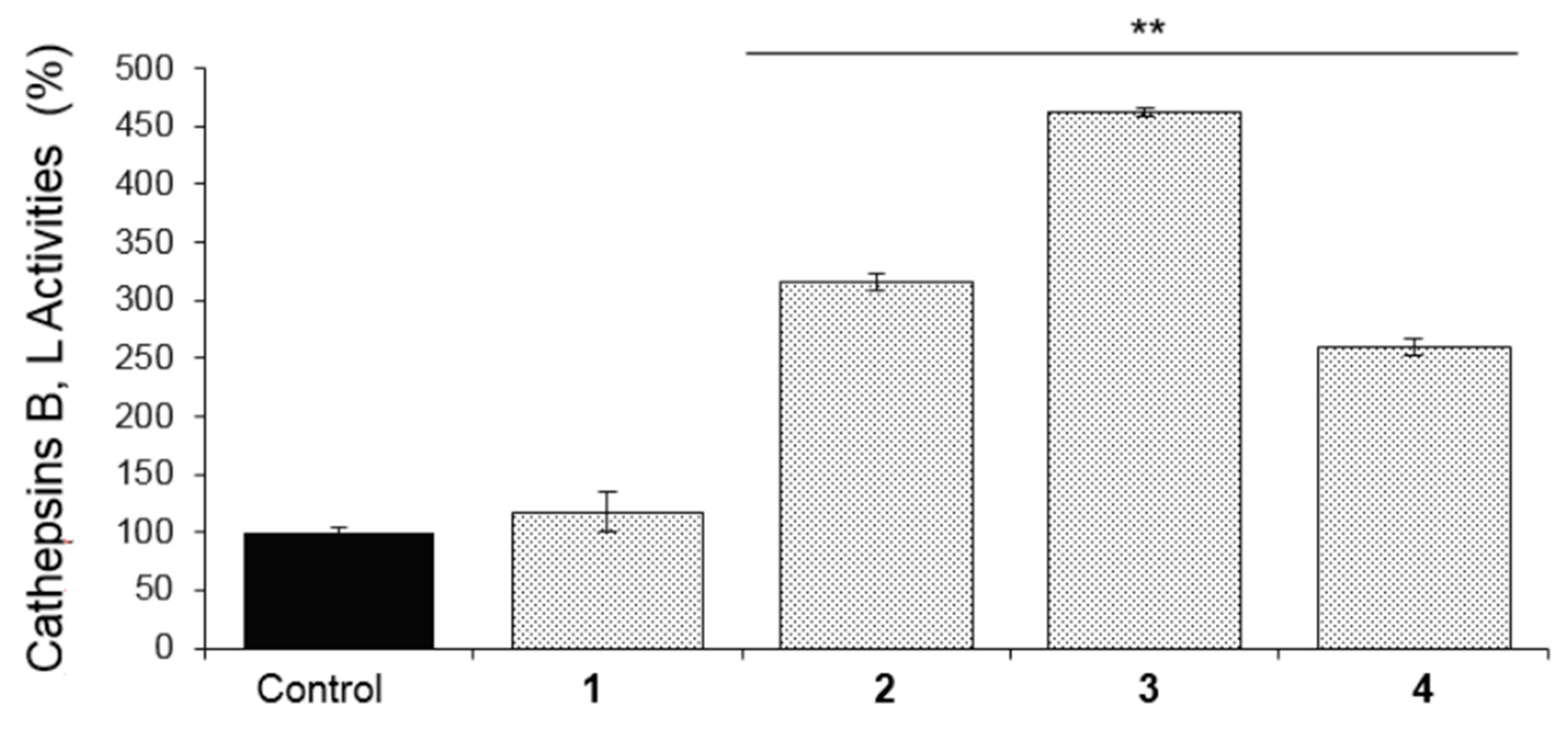
2.3. Biological Evaluation of Isolated Compounds
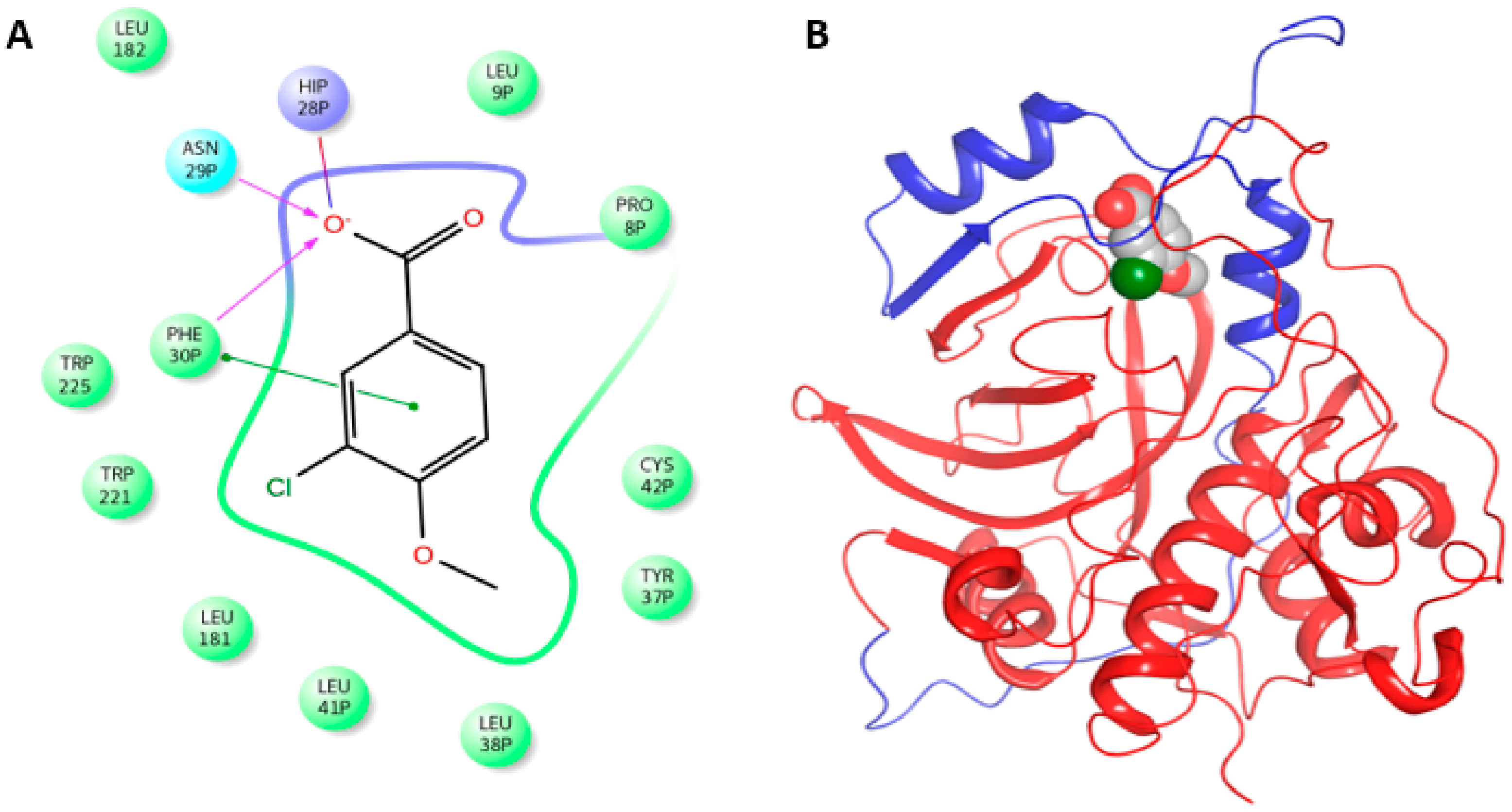
2.4. Molecular Docking Simulation of the Binding Mode of the Isolated Compounds with Procathepsins B and L
3. Materials and Methods
3.1. Collection and Characterization of the Strain CF-092983
3.2. Fermentation and Extraction
3.3. Isolation of Compounds
3.4. HPLC Profiling of the Extracts and Quantification of Compound 1
3.5. Cell lines and Cell Culture Conditions
3.6. Measurement of Cathepsin B, L Enzymatic Activity
3.7. Proteasome Activity
3.8. Cytotoxicity
3.9. Molecular Simulations
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALP | autophagy-lysosome pathway |
| ATP | adenosine triphosphate |
| CAM | chlorinated anisyl metabolites |
| CH3CN | acetonitrile |
| C-L/LLE | caspase-like activity of the proteasome |
| CT-L/LLVY | chymotrypsin-like activity of the proteasome |
| DNA | deoxyribonucleic acid |
| EDTA | ethylenediaminetetraacetic acid |
| EtOAc | ethyl acetate |
| EtOH | ethanol |
| FBS | fetal bovine serum |
| GAGs | glycosaminoglycans |
| H2O | water |
| Hex | hexane |
| HPLC | high-performance liquid chromatography |
| UPLC-HRMS | ultra-high performance liquid chromatography coupled high resolution mass spectrometry |
| MeOH | methanol |
| NMR | nuclear magnetic resonance |
| PCR | polymerase chain reaction |
| PN | proteostasis network |
| TFA | trifluoroacetic acid |
| UPP | ubiquitin-proteasome pathway |
References
- Nakanishi, H.; Tominaga, K.; Amano, T.; Hirotsu, I.; Inoue, T.; Yamamoto, K. Age-related changes in activities and localizations of cathepsins D, E, B, and L in the rat brain tissues. Exp. Neurol. 1994, 126, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Labbadia, J.; Morimoto, R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015, 84, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Tsakiri, E.N.; Gumeni, S.; Iliaki, K.K.; Benaki, D.; Vougas, K.; Sykiotis, G.P.; Gorgoulis, V.G.; Mikros, E.; Scorrano, L.; Trougakos, I.P. Hyperactivation of Nrf2 increases stress tolerance at the cost of aging acceleration due to metabolic deregulation. Aging Cell 2019, 18, e12845. [Google Scholar] [PubMed]
- Koga, H.; Kaushik, S.; Cuervo, A.M. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res. Rev. 2011, 10, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Gumeni, S.; Trougakos, I.P. Cross Talk of Proteostasis and Mitostasis in Cellular Homeodynamics, Ageing, and Disease. Available online: https://www.hindawi.com/journals/omcl/2016/4587691/ (accessed on 10 March 2019).
- Gumeni, S.; Evangelakou, Z.; Gorgoulis, V.G.; Trougakos, I.P. Proteome Stability as a Key Factor of Genome Integrity. Int. J. Mol. Sci. 2017, 18, 2036. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef]
- Wang, Y.; Vazquez-Duhalt, R.; Pickard, M.A. Manganese-lignin peroxidase hybrid from Bjerkandera adusta oxidizes polycyclic aromatic hydrocarbons more actively in the absence of manganese. Can. J. Microbiol. 2003, 49, 675–682. [Google Scholar] [CrossRef]
- Higson, F.K. Degradation of xenobiotics by white rot fungi. Rev. Env. Contam Toxicol 1991, 122, 111–152. [Google Scholar]
- Reddy, C. The potential for white-rot fungi in the treatment of pollutants. Curr. Opin. Biotechnol. 1995, 6, 320–328. [Google Scholar] [CrossRef]
- Aro, N.; Pakula, T.; Penttilä, M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. Fems Microbiol Rev. 2005, 29, 719–739. [Google Scholar] [CrossRef]
- De Jong, E.; Field, J.A.; Dings, J.A.; Wijnberg, J.B.; de Bont, J.A. De-novo biosynthesis of chlorinated aromatics by the white-rot fungus Bjerkandera sp. BOS55. Formation of 3-chloro-anisaldehyde from glucose. Febs Lett. 1992, 305, 220–224. [Google Scholar] [CrossRef]
- Lauritsen, F.R.; Kotiaho, T.; Lloyd, D. Rapid and direct monitoring of volatile fermentation products in the fungus Bjerkandera adusta by membrane inlet tandem mass spectrometry. Biol. Mass Spectrom. 1993, 22, 585–589. [Google Scholar] [CrossRef]
- Pfefferle, W.; Anke, H.; Bross, M.; Steglich, W. Inhibition of Solubilized Chitin Synthase by Chlorinated Aromatic Compounds Isolated from Mushroom Cultures. Agric. Biol. Chem. 1990, 54, 1381–1384. [Google Scholar]
- De Jong, E.; Cazemier, A.E.; Field, J.A.; Bont, J.A.M. de Physiological Role of Chlorinated Aryl Alcohols Biosynthesized De Novo by the White Rot Fungus Bjerkandera sp. Strain BOS55. Appl. Environ. Microbiol. 1994, 60, 271. [Google Scholar] [CrossRef] [PubMed]
- Swarts, H.J.; Verhagen, F.J.M.; Field, J.A.; Wijnberg, J.B.P.A. Novel chlorometabolites produced by Bjerkandera species. Phytochemistry 1996, 42, 1699–1701. [Google Scholar] [CrossRef]
- Lauritsen, F.R.; Lunding, A. A Study of the Bioconversion Potential of the Fungus Bjerkandera Adusta with Respect to a Production of Chlorinated Aromatic Compounds. Enzym. Microb. Technol. 1998, 22, 459–465. [Google Scholar] [CrossRef]
- Silk, P.J.; Aubry, C.; Lonergan, G.C.; Macaulay, J.B. Chlorometabolite production by the ecologically important white rot fungus Bjerkandera adusta. Chemosphere 2001, 44, 1603–1616. [Google Scholar] [CrossRef]
- Tapsell, L.C.; Hemphill, I.; Cobiac, L.; Patch, C.S.; Sullivan, D.R.; Fenech, M.; Roodenrys, S.; Keogh, J.B.; Clifton, P.M.; Williams, P.G.; et al. Health benefits of herbs and spices: the past, the present, the future. Med. J. Aust. 2006, 185, S4–S24. [Google Scholar] [CrossRef]
- Sklirou, A.; Papanagnou, E.-D.; Fokialakis, N.; Trougakos, I.P. Cancer chemoprevention via activation of proteostatic modules. Cancer Lett. 2018, 413, 110–121. [Google Scholar] [CrossRef]
- Peyrat, L.-A.; Tsafantakis, N.; Georgousaki, K.; Ouazzani, J.; Genilloud, O.; Trougakos, I.P.; Fokialakis, N. Terrestrial Microorganisms: Cell Factories of Bioactive Molecules with Skin Protecting Applications. Molecules 2019, 24, 1836. [Google Scholar] [CrossRef]
- Georgousaki, K.; Tsafantakis, N.; Gumeni, S.; González-Menéndez, V.; de Pedro, N.; Tormo, J.R.; Almeida, C.; Lambert, C.; Genilloud, O.; Trougakos, I.P.; et al. Cercospora sp. as a source of anti-aging polyketides targeting 26S proteasome and scale-up production in submerged bioreactor. J. Biotechnol. 2019, 301, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Silk, P.J.; Macaulay, J.B. Stereoselective biosynthesis of chloroarylpropane diols by the basidiomycete Bjerkandera adusta. Chemosphere 2003, 52, 503–512. [Google Scholar] [CrossRef]
- Wang, F.W.; Hou, Z.M.; Wang, C.R.; Li, P.; Shi, D.H. Bioactive metabolites from Penicillium sp., an endophytic fungus residing in Hopea hainanensis. World J. Microbiol. Biotechnol. 2008, 24, 2143–2147. [Google Scholar] [CrossRef]
- El-Gamal, M.I.; Oh, C.-H. Design and Synthesis of 3-(3-Chloro-4-substituted phenyl)-4-(pyridin-4-yl)-1Hpyrazole- 1-carboxamide Derivatives and Their Antiproliferative Activity Against Melanoma Cell Line. Bull. Korean Chem. Soc. 2011, 32, 821–828. [Google Scholar] [CrossRef]
- Uzor, P.F.; Odimegwu, D.C.; Ebrahim, W.; Osadebe, P.O.; Nwodo, N.J.; Okoye, F.B.C.; Liu, Z.; Proksch, P. Anti-Respiratory Syncytial Virus Compounds from Two Endophytic Fungi Isolated from Nigerian Medicinal Plants. Drug Res. (Stuttg) 2016, 66, 527–531. [Google Scholar] [CrossRef]
- Fotso, S.; Maskey, R.P.; Schröder, D.; Ferrer, A.S.; Grün-Wollny, I.; Laatsch, H. Furan oligomers and beta-carbolines from terrestrial streptomycetes. J. Nat. Prod. 2008, 71, 1630–1633. [Google Scholar] [CrossRef]
- Wang, G.; Dai, S.; Chen, M.; Wu, H.; Xie, L.; Luo, X.; Li, X. Two diketopiperazine cyclo(pro-phe) isomers from marine bacteria Bacillus subtilis sp. 13-2. Chem Nat. Compd. 2010, 46, 583–585. [Google Scholar] [CrossRef]
- Tian, S.-Z.; Pu, X.; Luo, G.; Zhao, L.-X.; Xu, L.-H.; Li, W.-J.; Luo, Y. Isolation and characterization of new p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agric. Food Chem. 2013, 61, 3006–3012. [Google Scholar] [CrossRef]
- El Euch, I.Z.; Frese, M.; Sewald, N.; Smaoui, S.; Shaaban, M.; Mellouli, L. Bioactive secondary metabolites from new terrestrial Streptomyces sp TN82 strain: Isolation, structure elucidation and biological activity. Med. Chem. Res. 2018, 27. [Google Scholar] [CrossRef]
- Wang, J.; Lu, Z.; Liu, P.; Wang, Y.; Li, J.; Hong, K.; Zhu, W. Cytotoxic polyphenols from the fungus Penicillium expansum 091 006 endogenous with the mangrove plant Excoecaria agallocha. Planta Med. 2012, 78, 1861–1866. [Google Scholar] [CrossRef]
- Olennikov, D.N.; Penzina, T.A. 2-Methoxy-3,4-Dihydroxybenzoic Acid and other Compounds from Ramaria aurea AND Clavariadelphus ligula. Chem Nat. Compd 2014, 50, 391–393. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Cheng, M.-J.; Hsiao, Y.; Chan, H.-Y.; Hsieh, S.-Y.; Chang, C.-W.; Liu, T.-W.; Chang, H.-S.; Chen, I.-S. Chemical Constituents of the Endophytic Fungus Hypoxylon sp. 12F0687 Isolated from Taiwanese Ilex formosana. Helv. Chim. Acta. 2015, 98, 1167–1176. [Google Scholar] [CrossRef]
- Song, Q.-Y.; Nan, Z.-B.; Gao, K.; Song, H.; Tian, P.; Zhang, X.-X.; Li, C.-J.; Xu, W.-B.; Li, X.-Z. Antifungal, Phytotoxic, and Cytotoxic Activities of Metabolites from Epichloë bromicola, a Fungus Obtained from Elymus tangutorum Grass. J. Agric. Food Chem. 2015, 63, 8787–8792. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Yu, L.; Wang, Q.; Ding, W.; Chen, Z.; Ma, Z. New brefeldins and penialidins from marine fungus Penicillium janthinellum DT-F29. Nat. Prod. Res. 2018, 32, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Hjelm, O.; Borén, H.; Öberg, G. Analysis of halogenated organic compounds in coniferous forest soil from a Lepista nuda (wood blewitt) fairy ring. Chemosphere 1996, 32, 1719–1728. [Google Scholar] [CrossRef]
- Thines, E.; Daußmann, T.; Sterner, O.; Semar, M.; Anke, H. Fungal Melanin Biosynthesis Inhibitors: Introduction of a Test System Based on the Production of Dihydroxynaphthalene (DHN) Melanin in Agar Cultures. Z. Für Nat. C 2014, 50, 813–819. [Google Scholar] [CrossRef]
- Shibata, H.; Kondo, K.; Katsuyama, R.; Kawazoe, K.; Sato, Y.; Murakami, K.; Takaishi, Y.; Arakaki, N.; Higuti, T. Alkyl gallates, intensifiers of beta-lactam susceptibility in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 549–555. [Google Scholar] [CrossRef]
- Eerdunbayaer; Orabi, M.A.A.; Aoyama, H.; Kuroda, T.; Hatano, T. Structures of two new flavonoids and effects of licorice phenolics on vancomycin-resistant Enterococcus species. Molecules 2014, 19, 3883–3897. [Google Scholar] [CrossRef]
- Kubo, I.; Chen, Q.-X.; Nihei, K.-I.; Calderón, J.S.; Céspedes, C.L. Tyrosinase inhibition kinetics of anisic acid. Z. Naturforsch. Cj. Biosci. 2003, 58, 713–718. [Google Scholar] [CrossRef]
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Hassan, H.M.; Zaki, M.A.; Rateb, M.E.; Mohammed, T.A.; Amin, E.; Abdelmohsen, U.R. Epigenetic Modifiers Induce Bioactive Phenolic Metabolites in the Marine-Derived Fungus Penicillium brevicompactum. Mar. Drugs 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Azizuddin; Khan, A.M.; Choudhary, M.I. Tyrosinase inhibitory potential of natural products isolated from various medicinal plants. Nat. Prod. Res. 2011, 25, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wen, H.; Cui, Y.; Fan, M.; Liu, Z.; Mei, L.; Shao, Y.; Wang, Y.; Tao, Y. Phenolics from Lagotis brevituba Maxim. Nat. Prod. Res. 2017, 31, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, B.; Riaz, N.; Saleem, M.; Naveed, M.A.; Ashraf, M.; Alam, U.; Rafiq, H.M.; Tareen, R.B.; Jabbar, A. Isolation of natural compounds from Phlomis stewartii showing α-glucosidase inhibitory activity. Phytochemistry 2013, 96, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Vad, N.M.; Shaik, I.H.; Mehvar, R.; Moridani, M.Y. Metabolic bioactivation and toxicity of ethyl 4-hydroxybenzoate in human SK-MEL-28 melanoma cells. J. Pharm. Sci. 2008, 97, 1934–1945. [Google Scholar] [CrossRef]
- Awale, S.; Miyamoto, T.; Linn, T.Z.; Li, F.; Win, N.N.; Tezuka, Y.; Esumi, H.; Kadota, S. Cytotoxic constituents of Soymida febrifuga from Myanmar. J. Nat. Prod. 2009, 72, 1631–1636. [Google Scholar] [CrossRef]
- Stadler, M.; Hellwig, V.; Mayer-Bartschmid, A.; Denzer, D.; Wiese, B.; Burkhardt, N. Novel analgesic triglycerides from cultures of Agaricus macrosporus and other basidiomycetes as selective inhibitors of neurolysin. J. Antibiot. 2005, 58, 775–786. [Google Scholar] [CrossRef]
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov 2006, 5, 785–799. [Google Scholar] [CrossRef]
- Verma, S.; Dixit, R.; Pandey, K.C. Cysteine Proteases: Modes of Activation and Future Prospects as Pharmacological Targets. Front. Pharmacol. 2016, 7, 107. [Google Scholar] [CrossRef]
- Caglic, D.; Pungercar, J.R.; Pejler, G.; Turk, V.; Turk, B. Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. J. Biol. Chem. 2007, 282, 33076–33085. [Google Scholar] [CrossRef]
- Suay, I.; Arenal, F.; Asensio, F.J.; Basilio, A.; Cabello, M.A.; Díez, M.T.; García, J.B.; del Val, A.G.; Gorrochategui, J.; Hernández, P.; et al. Screening of basidiomycetes for antimicrobial activities. Antonie Van Leeuwenhoek 2000, 78, 129–139. [Google Scholar] [CrossRef]
- Gonzalez-Menendez, V.; Martin, J.; Siles, J.A.; Gonzalez-Tejero, M.R.; Reyes, F.; Platas, G.; Tormo, J.R.; Genilloud, O. Biodiversity and chemotaxonomy of Preussia isolates from the Iberian Peninsula. Mycol. Prog. 2017, 16, 713–728. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Trougakos, I.P.; Kletsas, D.; Chen, Q.M.; Gonos, E.S. Partial proteasome inhibition in human fibroblasts triggers accelerated M1 senescence or M2 crisis depending on p53 and Rb status. Aging Cell 2008, 7, 717–732. [Google Scholar] [CrossRef]
- Grune, T.; Merker, K.; Jung, T.; Sitte, N.; Davies, K.J.A. Protein oxidation and degradation during postmitotic senescence. Free Radic. Biol. Med. 2005, 39, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Sklirou, A.D.; Ralli, M.; Dominguez, M.; Papassideri, I.; Skaltsounis, A.-L.; Trougakos, I.P. Hexapeptide-11 is a novel modulator of the proteostasis network in human diploid fibroblasts. Redox Biol. 2015, 5, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Balantinou, E.; Trougakos, I.P.; Chondrogianni, N.; Margaritis, L.H.; Gonos, E.S. Transcriptional and posttranslational regulation of clusterin by the two main cellular proteolytic pathways. Free Radic. Biol. Med. 2009, 46, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.J.; Storrie, B. A simple DNA-dependent fluorescence enhancement assay for cell number. J. Histochem. Cytochem. 1981, 29, 326–328. [Google Scholar] [CrossRef]
- Portugal, J.; Waring, M.J. Assignment of DNA binding sites for 4′,6-diamidine-2-phenylindole and bisbenzimide (Hoechst 33258). A comparative footprinting study. Biochim. Biophys. Acta. 1988, 949, 158–168. [Google Scholar] [CrossRef]
- De Pedro, N.; Cautain, B.; Melguizo, A.; Vicente, F.; Genilloud, O.; Peláez, F.; Tormo, J.R. Mitochondrial complex I inhibitors, acetogenins, induce HepG2 cell death through the induction of the complete apoptotic mitochondrial pathway. J. Bioenerg. Biomembr. 2013, 45, 153–164. [Google Scholar] [CrossRef]
- Huang, M.; Liu, J.; Zhang, S.; Mei, X.; Yang, X. Effects of bioactive extracts from four edible mushrooms on the lifespan of Drosophila melanogaster. Mycology 2011, 2, 54–58. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–4 are available from the authors. |
| Compound | EtOAc LL Extract (μg mg−1) | EtOAc Extract (μg mg−1) |
|---|---|---|
| 1 | 17.57 ± 0.18 | 2.68 ± 0.17 |
| 2 * | 2.46 ± 0.13 | 1.03 ± 0.05 |
| 3 * | 7.99 ± 0.08 | 1.39 ± 0.12 |
| 4 * | 4.43 ± 0.04 | 0.89 ± 0.03 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgousaki, K.; Tsafantakis, N.; Gumeni, S.; Lambrinidis, G.; González-Menéndez, V.; Tormo, J.R.; Genilloud, O.; Trougakos, I.P.; Fokialakis, N. Biological Evaluation and In Silico Study of Benzoic Acid Derivatives from Bjerkandera adusta Targeting Proteostasis Network Modules. Molecules 2020, 25, 666. https://doi.org/10.3390/molecules25030666
Georgousaki K, Tsafantakis N, Gumeni S, Lambrinidis G, González-Menéndez V, Tormo JR, Genilloud O, Trougakos IP, Fokialakis N. Biological Evaluation and In Silico Study of Benzoic Acid Derivatives from Bjerkandera adusta Targeting Proteostasis Network Modules. Molecules. 2020; 25(3):666. https://doi.org/10.3390/molecules25030666
Chicago/Turabian StyleGeorgousaki, Katerina, Nikolaos Tsafantakis, Sentiljana Gumeni, George Lambrinidis, Victor González-Menéndez, Jose R. Tormo, Olga Genilloud, Ioannis P. Trougakos, and Nikolas Fokialakis. 2020. "Biological Evaluation and In Silico Study of Benzoic Acid Derivatives from Bjerkandera adusta Targeting Proteostasis Network Modules" Molecules 25, no. 3: 666. https://doi.org/10.3390/molecules25030666
APA StyleGeorgousaki, K., Tsafantakis, N., Gumeni, S., Lambrinidis, G., González-Menéndez, V., Tormo, J. R., Genilloud, O., Trougakos, I. P., & Fokialakis, N. (2020). Biological Evaluation and In Silico Study of Benzoic Acid Derivatives from Bjerkandera adusta Targeting Proteostasis Network Modules. Molecules, 25(3), 666. https://doi.org/10.3390/molecules25030666










