New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies
Abstract
:1. Introduction
2. Experimental
2.1. Synthesis
2.1.1. General Procedure for the Synthesis of Carboxylic Acid Hydrazides (Series A)
2.1.2. General Procedure for the Synthesis of Thiosemicarbazides (Series B)
B13. 4-(2,4-Dichlorophenyl)-1-(pyridin-3-yl)acetylthiosemicarbazide
B14. 4-(3,4-Dichlorophenyl)-1-(pyridin-4-yl)acetylothiosemicarbazide
2.1.3. General Procedure for the Synthesis of 1,2,4-triazole Derivatives (Series C)
C1. 4-(2-Fluorphfenyl)-3-(pyridin-2-yl)-1,2,4-triazoline-5-thione
C2. 4-(2-Chlorophenyl)-3-(pyridin-2-yl)-1,2,4-triazoline-5-thione
C3. 4-(2,4-Dichlorofenylo)-3-(pyridin-2-yl)-1,2,4-triazoline-5-thione
C4. 4-(3,4-Dichlorofenylo)-3-(pyridin-2-yl)-1,2,4-triazoline-5-thione
C5. 4-(2-Chlorofenylo)-3-(pyridin-3-yl)-1,2,4-triazoline-5-thione
C6. 4-(4-Nitrofenylo)-3-(pyridin-3-yl)-1,2,4-triazoline-5-thione
C7. 4-(2,4-Dichlorofenylo)-3-(pyridin-3-yl)-1,2,4-triazoline-5-thione
C8. 4-(3,4-Dichlorofenylo)-3-(pyridin-3-yl)-1,2,4-triazoline-5-htione
C9. 4-(2-Fluorofenylo)-3-(pyridin-4-yl)-1,2,4-triazoline-5-thione
C10. 4-(2,4-Dichlorophenyl)-3-(pyridin-4-yl)-1,2,4-triazoline-5-thione
C11. 4-Phenyl-3-(pyridin-2-yl)-1,2,4-triazoline-5-thione
C12. 4-(4-Methoxyphenyl)-3-(pyridin-3-yl)-1,2,4-triazoline-5-thione
C13. 4-(2,4-Dichlorophenyl)-3-(pyridin-3-ylmethyl)-1,2,4-triazoline-5-thione
C14. 4-(3,4-Dichlorophenyl)-3-(pyridin-4-ylmethyl)-1,2,4-triazoline-5-thione
2.2. X-ray Structure Determination
2.3. Theoretical Calculations
2.4. Antimycobacterial Assay
2.5. Molecular Docking
3. Results and Discussion
3.1. Chemistry
3.2. X-ray Analysis
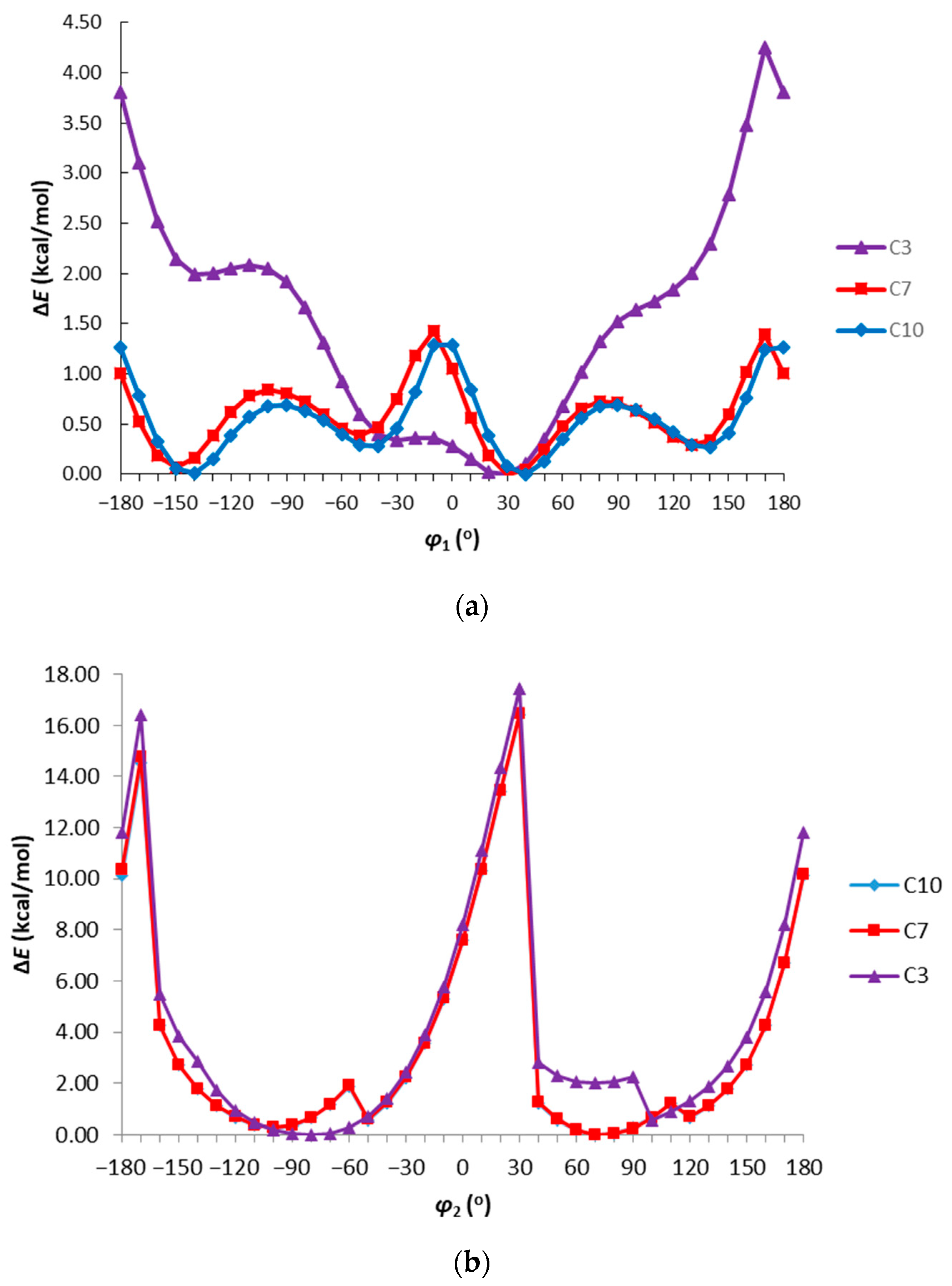
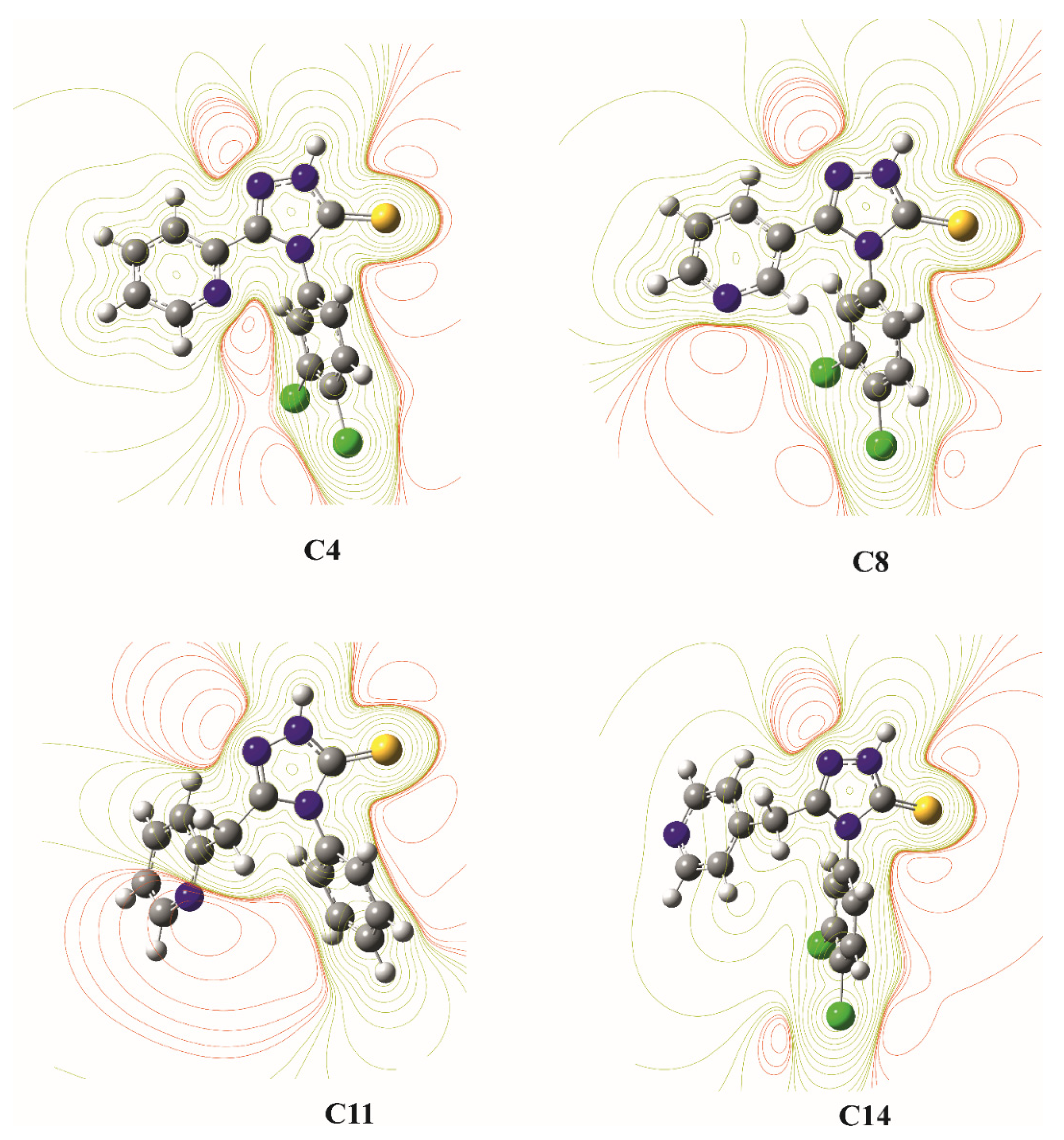
3.3. Theoretical Calculations
3.4. Antimycobacterial Activity
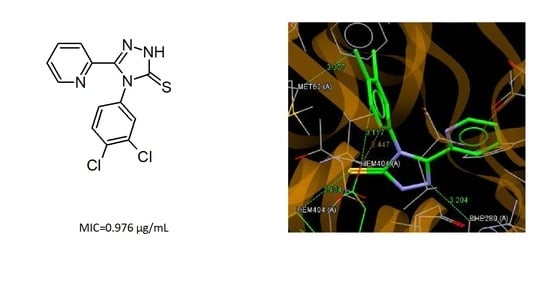
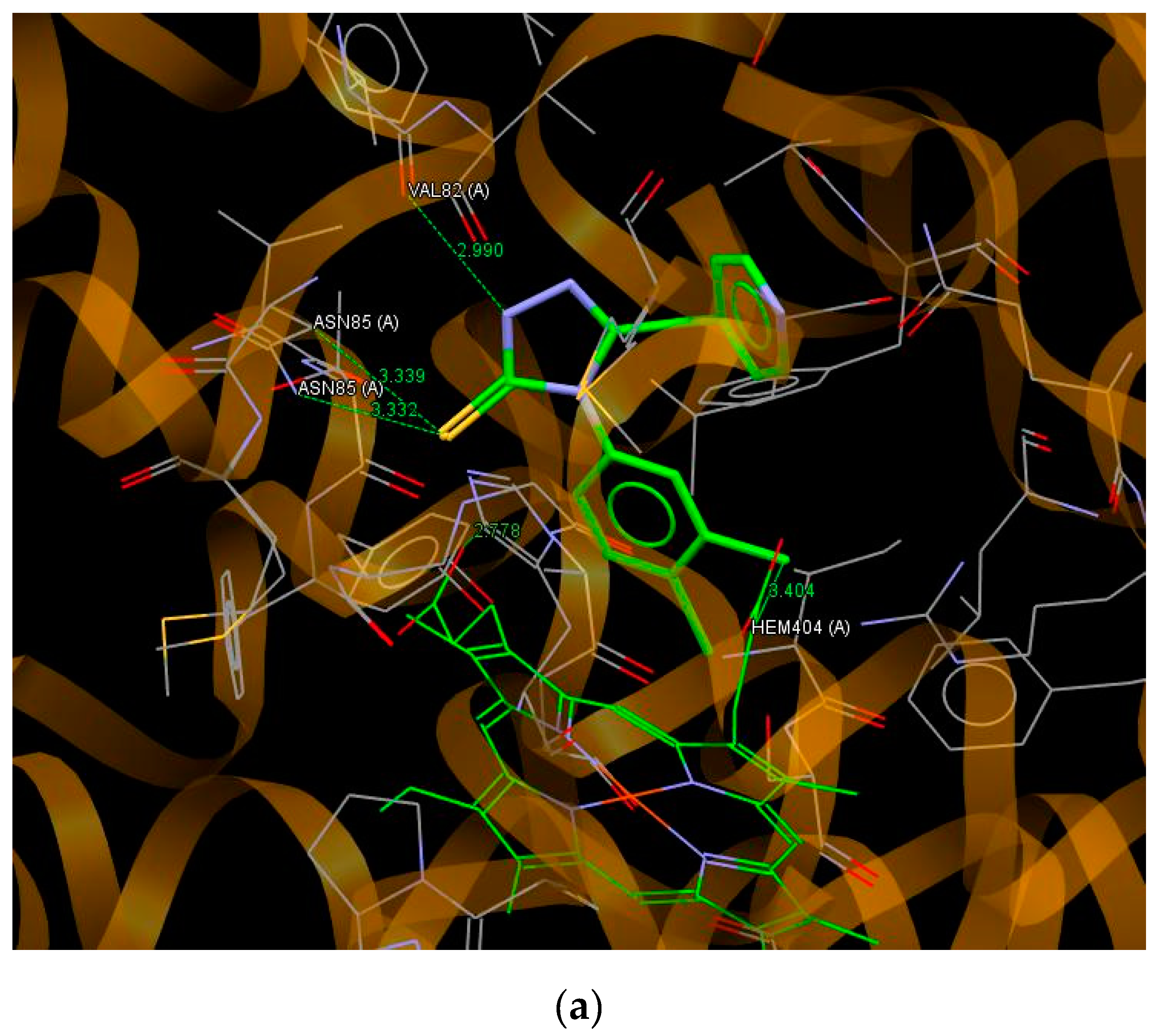
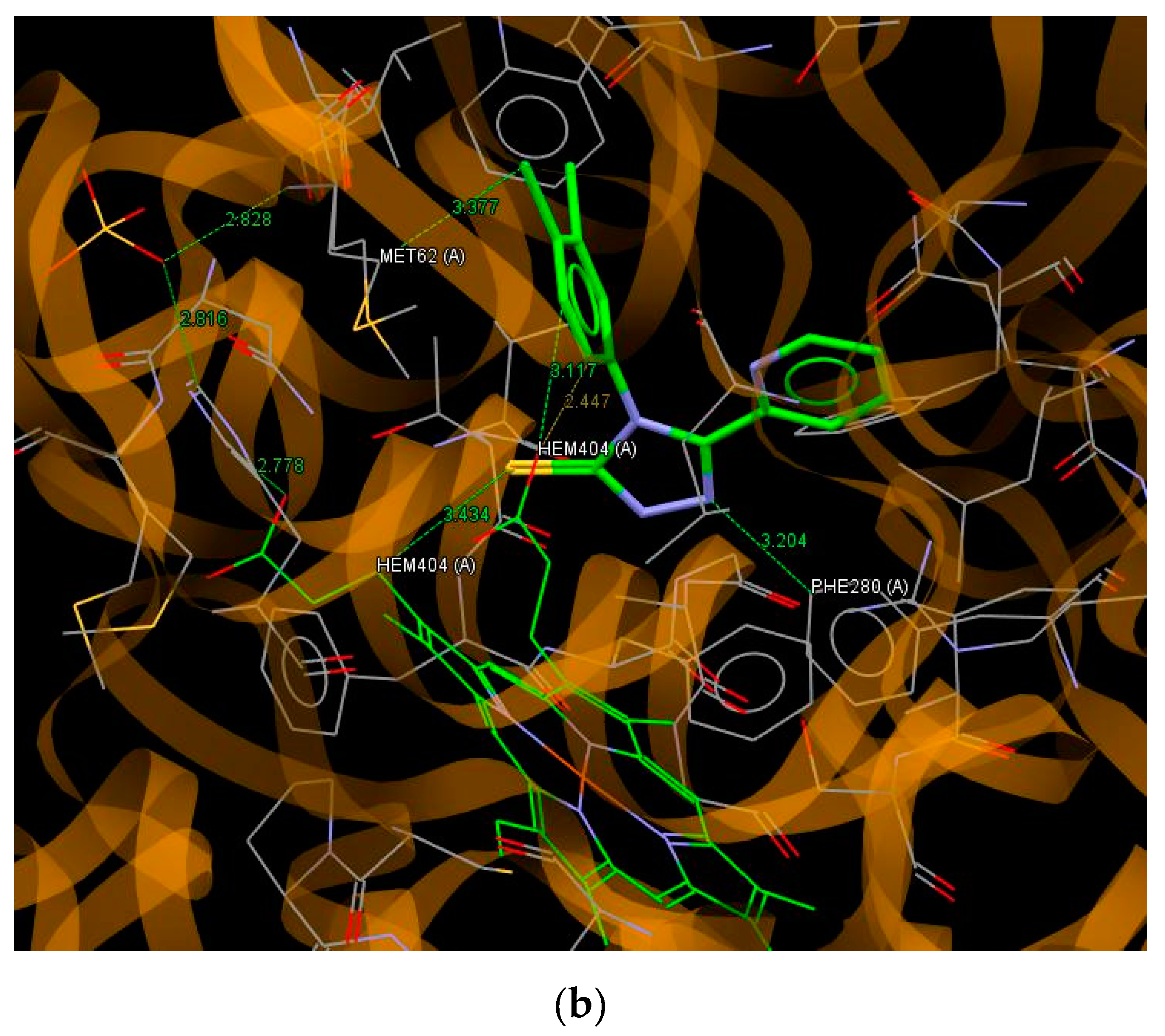
3.5. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Beard, C.; Loughman, R.; Smith, A.; Speijers, J. Baseline sensitivity to Tyree triazole fungicides in Pyrenophora tritici-repentis. Aust. Plant Pathol. 2009, 28, 168–172. [Google Scholar] [CrossRef]
- Wu, C.; Sun, J.; Zhang, A.; Liu, W. Dissipation and Enantioselective Degradation of Plant Growth Retardants Paclobutrazol and Uniconazole in Open Field, Greenhouse, and Laboratory Soils. Environ. Sci. Technol. 2013, 47, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Kokalj, A.; Kovacevic, N.; Peljhan, S.; Finšgar, M.; Lesar, A.; Milošev, I. Triazole, Benzotriazole, and Naphthotriazole as Copper Corrosion Inhibitors: I. Molecular Electronic and Adsorption Properties. ChemPhysChem 2011, 12, 3547–3555. [Google Scholar] [CrossRef] [PubMed]
- Asami, T.; Min, Y.K.; Nagata, N.; Yamagishi, K.; Takatsuto, S.; Fujioka, S.; Murofushi, N.; Yamaguchi, I.; Yoshida, S. Characterization of Brassinazole, a Triazole-Type Brassinosteroid Biosynthesis Inhibitor. Plant Physiol. 2000, 123, 93–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekhar, M.M.; Nagarjuna, U.; Padmavathi, V.; Padmaja, A.; Padmaja, A.; Vijaya, T. Synthesis and antimicrobial activity of pyrimidinyl 1,3,4-oxadiazoles, 1,3,4-thiadiazoles and 1,2,4-triazoles. Eur. J. Med. Chem. 2018, 145, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dubovis, M.; Rudakov, G.; Kulagin, A.; Tsarkova, K.; Popkov, S.; Goloveshkin, A.; Charkaer, G. A new metod of synthesis of substituted 1-(1H-imidazole-4-yl)-1H-1,2,3-triazoles and their fungicidal actvity. Tetrahedron 2018, 78, 672–683. [Google Scholar] [CrossRef]
- Wu, J.; Ni, T.; Chai, X.; Wang, T.; Wang, H.; Chen, J.; Jin, Y.; Zhang, D.-Z.; Yu, S.; Jiang, Y. Molecular docking, design, synthesis and antifungal activity study of novel triazole derivatives. Eur. J. Med. Chem. 2018, 143, 1840–1846. [Google Scholar] [CrossRef]
- Cao, X.; Wang, W.; Wang, S.; Bao, L. Asymmetric synthesis of novel triazole derivatives and their in vitro antiviral activity and mechanism of action. Eur. J. Med. Chem. 2017, 139, 718–725. [Google Scholar] [CrossRef]
- Wu, M.-J.; Wu, D.-M.; Chen, J.-B.; Zhao, J.-F.; Gong, L.; Gong, Y.-X.; Li, Y.; Yang, X.-D.; Zhang, H. Synthesis and anti-proliferative activity of allogibberic acid derivatives containing 1,2,3-triazole pharmacophore. Bioorganic Med. Chem. Lett. 2018, 28, 2543–2549. [Google Scholar] [CrossRef]
- Mustafa, M.; Abdelhamid, D.; Abdelhafez, E.M.N.; Ibrahim, M.A.; Gamal-Eldeen, A.M.; Aly, O.M. Synthesis, antiproliferative, anti-tubulin activity, and docking study of new 1,2,4-triazoles as potential combretastatin analogues. Eur. J. Med. Chem. 2017, 141, 293–305. [Google Scholar] [CrossRef]
- Yamada, M.; Takahashi, T.; Hasegawa, M.; Matsumura, M.; Ono, K.; Fujimoto, R.; Kitamura, Y.; Murata, Y.; Kakusawa, N.; Tanaka, M.; et al. Synthesis, antitumor activity, and cytotoxicity of 4-substituted 1-benzyl-5-diphenylstibano-1H-1,2,3-triazoles. Bioorganic Med. Chem. Lett. 2018, 28, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Saadaoui, I.; Krichen, F.; Ben-Salah, B.; Ben-Mansour, R.; Miled, N.; Bougatef, A.; Kossentini, M. Design, synthesis and biological evaluation of Schiff bases of 4-amino-1,2,4-triazole derivativates as potent angiotensin converting enzyme inhibitors and antioxidant activities. J. Mol. Str. 2019, 1180, 344–354. [Google Scholar] [CrossRef]
- Pawar, S.; Upadhyay, P.; Burade, S.; Kumbhar, N.; Patil, R.; Dhavale, D. Synthesis and anti-leishmanial activity of TRIS-glycine-B-alanine dipeptidic triazole dendron coated with nonameric mannoside glycocluster. Carbohydr. Res. 2019, 485, 1–7. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Z.; Gao, C.; Ren, Q.-C.; Chang, L.; Lv, Z.-S.; Feng, L.-S. Triazole derivatives and their anti-tubercular activity. Eur. J. Med. Chem. 2017, 138, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, W.; Jones, S.; Zubieta, J. Solid state coordination chemistry of metal-1,2,4 triazolates and the related metal-5-(pyrid-4-yl)tetrazolates. CrystEngComm 2011, 13, 4457–4485. [Google Scholar] [CrossRef]
- Kamboj, V.K.; Verma, P.K.; Dhanda, A.; Ranjan, S. 1,2,4-Triazole Derivatives as Potential Scaffold for Anticonvulsant Activity. Central Nerv. Syst. Agents Med. Chem. 2015, 15, 17–22. [Google Scholar] [CrossRef]
- Zhou, C.-H.; Gan, L.; Zhang, Y.; Zhang, F.; Wang, G.; Jin, L.; Geng, R. Review on supermolecules as chemical drugs. Sci. China Ser. B Chem. 2009, 52, 415–458. [Google Scholar] [CrossRef]
- Chu, X.-M.; Wang, C.; Wang, W.-L.; Liang, L.-L.; Liu, W.; Gong, K.-K.; Sun, K.-L. Triazole derivatives and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 2019, 166, 206–223. [Google Scholar] [CrossRef]
- Xu, M.; Peng, Y.; Zhu, L.; Wang, S.; Ji, J.; Rakesh, K. Triazole derivatives as inhibitors of Alzheimer’s disease: Current developments and structure-activity relationships. Eur. J. Med. Chem. 2019, 180, 656–672. [Google Scholar] [CrossRef]
- Ceesay, M.M.; Couchman, L.; Smith, M.; Wade, J.; Flanagan, R.J.; Pagliuca, A. Triazole antifungals used for prophylaxis and treatment of invasive fungal disease in adult haematology patients: Trough serum concentrations in relation to outcome. Med. Mycol. 2016, 54, 691–698. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.; Bimal, D.; Bohra, K.; Singh, B.; Ponnan, P.; Jain, R.; Varma-Basil, M.; Muty, J.; Thirumal, M.; Prasad, A. Synthesis and antimycobacterial activity of 1-(B-D-Ribofuranosyl)-4-coumarinyloxymethyl-/-coumarinyl-1,2,3-triazole. Eur. J. Med. Chem. 2018, 150, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Sajja, Y.; Vanguru, S.; Vulupala, H.R.; Bantu, R.; Yogeswari, P.; Sriram, D.; Nagarapu, L. Design, synthesis and in vitro anti-tuberculosis activity of benzo[6,7]cyclohepta[1,2-b]pyridine-1,2,3-triazole derivatives. Bioorganic Med. Chem. Lett. 2017, 27, 5119–5121. [Google Scholar] [CrossRef] [PubMed]
- Wilde, F.; Lemmerhirt, H.; Emmrich, T.; Bednarski, P.J.; Link, A. 2-Pyridineacetic acid hydrazide Microwave assisted synthesis and evaluation of acylhydrazones as potential inhibitors of bovine glutathione peroxidise. Mol. Divers. 2014, 18, 307.e322. [Google Scholar] [CrossRef] [PubMed]
- Monjas, L.; Swier, L.J.Y.M.; Setyawati, I.; Slotboom, D.J.; Hirsch, A.K.H. Dynamic Combinatorial Chemistry to Identify Binders of ThiT, an S-Component of the Energy-Coupling Factor Transporter for Thiamine. ChemMedChem 2017, 12, 1693–1696. [Google Scholar] [CrossRef] [Green Version]
- Tran, S.B.; Maxwell, B.D.; Burrell, R.; Bonacorsi, S.J. The syntheses of isotopically labelled CB-1 antagonists for the treatment of obesity. J. Label. Compd. Radiopharm. 2016, 59, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Pitucha, M.; Woś, M.; Miazga-Karska, M.; Klimek, K.; Mirosław, B.; Pachuta-Stec, A.; Gładysz, A.; Ginalska, G. Synthesis, antibacterial and antiproliferative potential of some new 1-pyridinecarbonyl-4-substituted thiosemicarbazide derivatives. Med. Chem. Res. 2016, 25, 1666–1677. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Yun, W.; Lee, J.; Jang, S.; Won Park, S.; Kim, D.H.; Seon, K.P.; Hyun, J.; Jeong, K.; Ku, J.; et al. Synthesis and anti-endoplasmic reticulum stress activity of n-substituted-2-arylcarbonylhydrazine carbothioamides. Med. Chem. Res. 2019, 28, 2142–2152. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Zeng, F. Studies on acylthiosemicarbazides and related heterocyclic derivatives. (XII). Synthetic and spectroscopic studies on 1-nicotinoyl-4-aryl thiosemicarbazides and substituted 1,2,4-triazoles. Gaodeng Xuexiao Huaxue Xuebao 1990, 11, 148–153. [Google Scholar]
- Pitucha, M.; Karczmarzyk, Z.; Swatko-Ossor, M.; Wysocki, W.; Wos, M.; Chudzik, K.; Ginalska, G.; Fruziński, A. Synthesis, In Vitro Screening and Docking Studies of New Thiosemicarbazide Derivatives as Antitubercular Agents. Molecules 2019, 24, 251. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, M.E.; Rakha, T.H.; Bekheit, M.M. Ligational Behaviour of 1-Picolinoyl–4-phenyl–3-thiosemicarbazide (H2PTS) Towards Some Transition Metal Ions. Synth. React. Inorg. Met. Chem. 1996, 26, 1149–1161. [Google Scholar] [CrossRef]
- Dobosz, M.; Pitucha, M.; Wujec, M. The reactions of cyclization of thiosemicarbazide derivatives to 1,2,4-triazole or 1,3,4-thiadiazole system. Acta Polon. Pharm. 1996, 53, 31–38. [Google Scholar]
- Pitucha, M.; Janeczko, M.; Klimek, K.; Fornal, E.; Wos, M.; Pachuta-Stec, A.; Ginalska, G.; Kaczor, A.A. 1,2,4-Triazolin-5-thione derivatives with anticancer activity as CK1γ kinase inhibitors. Bioorganic Chem. 2020, 99, 103806. [Google Scholar] [CrossRef] [PubMed]
- Modzelewska-Banachiewicz, B.; Jagiełło-Wójtowicz, E.; Tokarzewska-Wielosz, E. Synthesis and biological activity of bis-1,2,4-triazole and bis-1,3,4-thiadiazole derivatives. Acta Polon. Pharm. 2000, 57, 199–204. [Google Scholar]
- CrysAlisPro, Agilent Technologies, Version 1.171.37.35h, Release 09-02-2015 CrysAlis171.NET. Available online: https://www.selectscience.net/products/crysalispro/?prodID=197116 (accessed on 19 December 2020).
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Cryst. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 03, Revision E.01; Gaussian Inc.: Wallingford, CT, USA, 2004. [Google Scholar]
- Dennington, R.; Keith, T.; Millam, J. GaussView 4.1; Semichem, Inc.: Shawnee Mission, KS, USA, 2007. [Google Scholar]
- Ghose, A.K.; Pritchett, A.; Crippen, G.M. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships III: Modeling hydrophobic interactions. J. Comput. Chem. 1988, 9, 80–90. [Google Scholar] [CrossRef]
- Hyper-Chem, Release 8.0.10 for Windows Molecular Modeling System; Hypercube Inc.: Gainesville, FL, USA, 2006; p. 32601.
- Turnidge, J.D.; Jorgensen, J.H. Antimicrobial Susceptibility Testing: General Cosiderations. In Manual of Clinical Microbiology, 7th ed.; Murray, P.R., Baron, E.J., Pfaller, M.A., Tenover, F.C., Yolken, R., Eds.; American Society for Microbiology: Washington, DC, USA, 1999; pp. 1469–1473. [Google Scholar]
- Taban, I.M.; Elshihawy, H.E.A.E.; Torun, B.; Zucchini, B.; Williamson, C.J.; Altuwairigi, D.; Ngu, A.S.T.; McLean, K.J.; Levy, C.W.; Sood, S.; et al. Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation. J. Med. Chem. 2017, 60, 10257–10267. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical Scoring Functions for Advanced Protein−Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of bond lengths determined by X-ray and neutron diffraction. Part 1. Bond lengths in organic compounds. J. Chem. Soc. Perkin Trans. 1987, S1–S19. [Google Scholar] [CrossRef]
- Pennington, L.D.; Moustakas, D.T. The Necessary Nitrogen Atom: A Versatile High-Impact Design Element for Multiparameter Optimization. J. Med. Chem. 2017, 60, 3552–3579. [Google Scholar] [CrossRef] [PubMed]
- Rode, N.D.; Sonawane, A.D.; Nawale, L.; Khedkar, V.M.; Joshi, R.A.; Likhite, A.P.; Sarkar, D.; Joshi, R.R. Synthesis, biological evaluation, and molecular docking studies of novel 3-aryl-5-(alkyl-thio)-1H-1,2,4-triazoles derivatives targeting Mycobacterium tuberculosis. Chem. Biol. Drug Des. 2017, 90, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Hudson, S.A.; McLean, K.J.; Surade, S.; Yang, Y.-Q.; Leys, D.; Ciulli, A.; Munro, A.W.; Abell, C. Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121. Angew. Chem. Int. Ed. 2012, 51, 9311–9316. [Google Scholar] [CrossRef] [PubMed]


| D–H…A | D–H | H…A | D…A | D–H…A |
|---|---|---|---|---|
| C1 | ||||
| N1–H1…S5i | 0.89(3) | 2.37(4) | 3.261(2) | 175(3) |
| i = −x, 2 − y, −z | ||||
| C12 | ||||
| N1–H1…N63i | 0.92(3) | 1.88(3) | 2.789(3) | 171(2) |
| i = x, 3/2 − y, −1/2 + z | ||||
| C13 | ||||
| N1–H1…N63i | 0.99(3) | 1.83(3) | 2.815(4) | 170(3) |
| C43–H43…N2ii | 0.93 | 2.53 | 3.435(4) | 164 |
| C46–H46…S5iii | 0.93 | 2.84 | 3.675(4) | 150 |
| C65–H65…S5iv | 0.93 | 2.87 | 3.714(4) | 152 |
| C31–H312…Sv | 0.97 | 2.87 | 3.552(4) | 129 |
| i = −1 + x, 1 + y, z; ii = x, −1+y, z; iii = −x, 1 − y, 1 − z; iv = −x, 1 − y, −z; v = 1 + x, y, z | ||||
| Compound | Dm | NBO Charge | EHOMO | ELUMO | ΔE | logP | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N4 | Npyridil | S | ||||||
| C1 | 5.631 | −0.381 | −0.274 | −0.455 | −0.455 | −0.207 | −134.17 | −45.45 | 88.68 | 4.95 |
| C2 | 5.736 | −0.381 | −0.276 | −0.462 | −0.451 | −0.205 | −133.80 | −45.42 | 88.38 | 5.33 |
| C3 | 6.001 | −0.389 | −0.273 | −0.460 | −0.451 | −0.200 | −136.58 | −48.19 | 88.39 | 5.85 |
| C4 | 6.262 | −0.382 | −0.274 | −0.462 | −0.451 | −0.202 | −137.66 | −49.22 | 88.44 | 5.85 |
| C5 | 3.265 | −0.384 | −0.277 | −0.471 | −0.444 | −0.206 | −136.88 | −43.70 | 93.18 | 5.40 |
| C6 | 6.515 | −0.382 | −0.270 | −0.471 | −0.440 | −0.193 | −144.86 | −73.52 | 77.33 | 0.97 |
| C7 | 3.502 | −0.384 | −0.276 | −0.474 | −0.443 | −0.202 | −139.61 | −46.19 | 93.42 | 5.81 |
| C8 | 3.901 | −0.384 | −0.275 | −0.465 | −0.441 | −0.206 | −140.92 | −47.72 | 93.20 | 5.91 |
| C9 | 2.870 | −0.382 | −0.258 | −0.455 | −0.442 | −0.196 | −138.88 | −49.32 | 89.55 | 5.02 |
| C10 | 1.587 | −0.381 | −0.265 | −0.472 | −0.439 | −0.206 | −141.92 | −51.26 | 90.65 | 5.91 |
| C11 | 4.538 | −0.394 | −0.308 | −0.476 | −0.469 | −0.222 | −132.84 | −32.45 | 100.39 | 4.75 |
| C12 | 3.208 | −0.384 | −0.281 | −0.461 | −0.444 | −0.217 | −133.86 | −42.17 | 91.68 | 4.63 |
| C13 | 3.694 | −0.393 | −0.294 | −0.482 | −0.444 | −0.205 | −138.47 | −38.39 | 99.94 | 5.85 |
| C14 | 4.688 | −0.392 | −0.291 | −0.480 | −0.448 | −0.205 | −140.64 | −39.24 | 101.40 | 5.85 |
| Inhibition Zone (mm) 250 μg of Compound PerWell | ||||||
|---|---|---|---|---|---|---|
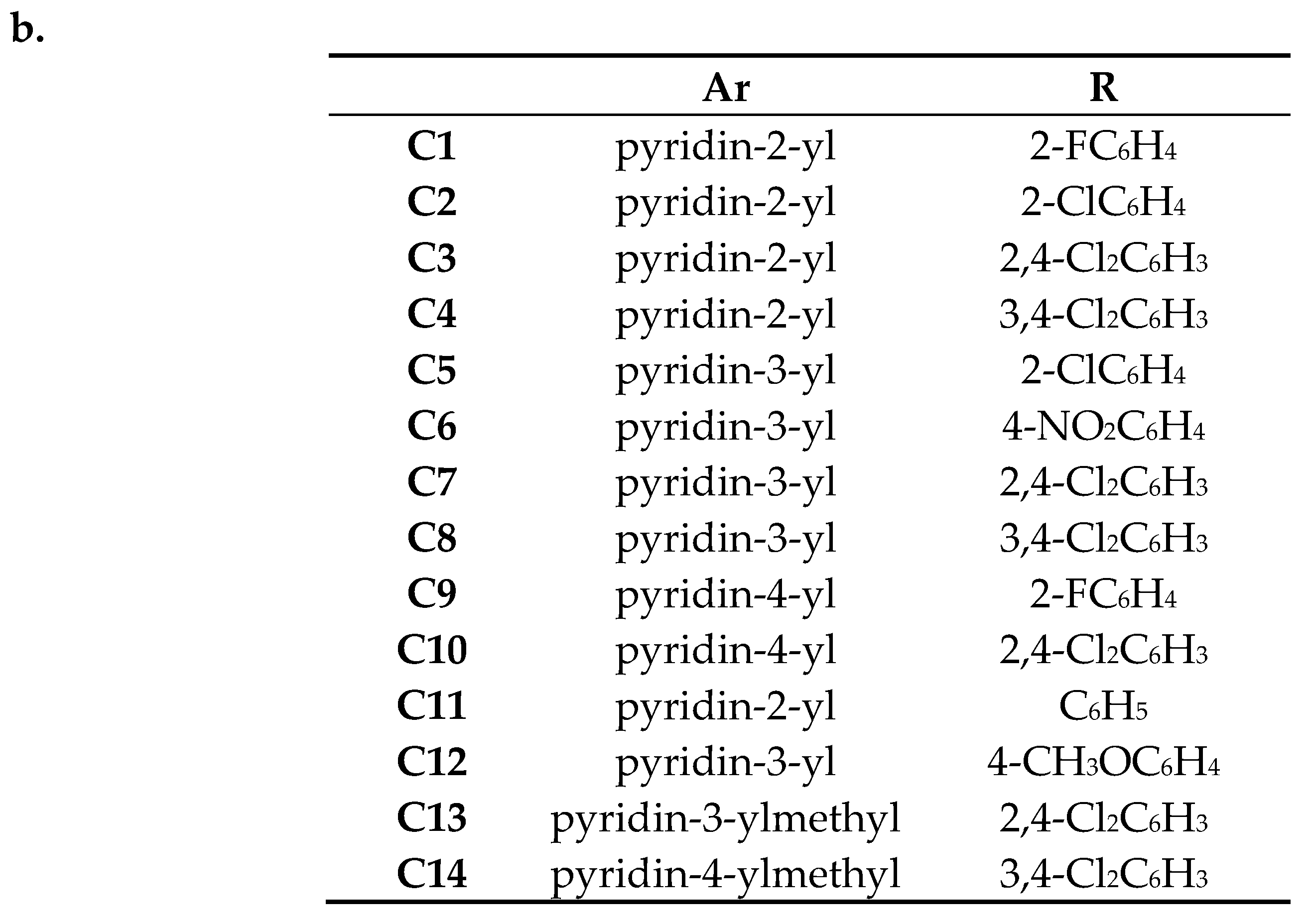
| Ar | R | Mycobacterium H37Ra | Mycobacterium Phlei | Mycobacterium Smegmatis | Mycobacterium Timereck | |
| C1 | pyridin-2-yl | 2-FC6H4 | 15.1 ± 0.61 | 15.1 ± 0.53 | 16.0 ± 0.81 | 15.1 ± 0.67 |
| C2 | pyridin-2-yl | 2-ClC6H4 | 19.6 ± 0.43 | 21.5 ± 1.12 | 17.2 ± 0.98 | 15.3 ± 0.84 |
| C3 | pyridin-2-yl | 2,4-Cl2C6H3 | 10.2 ± 0.8 | 17.0 ± 0.78 | 17.9 ± 0.77 | 20.1 ± 1.15 |
| C4 | pyridin-2-yl | 3,4-Cl2C6H3 | 26.9 ± 0.78 | 22.7 ± 0.91 | 22.3 ± 1.05 | 21.5 ± 0.9 |
| C5 | pyridin-3-yl | 2-ClC6H4 | 12.3 ± 0.78 | 0.0 | 10.2 ± 0.86 | 16.5 ± 0.41 |
| C6 | pyridin-3-yl | 4-NO2C6H4 | 0.0 | 0.0 | 0.0 | 10.6 ± 0.53 |
| C7 | pyridin-3-yl | 2,4-Cl2C6H3 | 0.0 | 8.9 ± 0.66 | 0.0 | 0.0 |
| C8 | pyridin-3-yl | 3,4-Cl2C6H3 | 22.2 ± 0.72 | 21.9 ± 0.29 | 23.1 ± 0.6 | 24.7 ± 0.87 |
| C9 | pyridin-4-yl | 2-FC6H4 | 10.6 ± 0.53 | 10.9 ± 0.66 | 12.3 ± 0.92 | 19.6 ± 0.53 |
| C10 | pyridin-4-yl | 2,4-Cl2C6H3 | 0.0 | 0.0 | 0.0 | 0.0 |
| C11 | pyridin-2-yl | C6H5 | 35.9 ± 0.49 | 34.0 ± 0.93 | 35.3 ± 0.7 | 35.1 ± 0.58 |
| C12 | pyridin-3-yl | 4-CH3OC6H4 | 10.6 ± 0.85 | 0.0 | 0.0 | 0.0 |
| C13 | pyridin-3-ylmethyl | 2,4-Cl2C6H3 | 12.0 ± 0.67 | 0.0 | 0.0 | 0.0 |
| C14 | pyridin-4-ylmethyl | 3,4-Cl2C6H3 | 20.7 ± 0.29 | 0.0 | 18.2 ± 0.53 | 18.6 ± 0.98 |
| MIC (µg/mL) | ||||||
|---|---|---|---|---|---|---|
| Ar | R | Mycobacterium H37Ra | Mycobacterium Phlei | Mycobacterium Smegmatis | Mycobacterium Timereck | |
| C1 | pyridin-2-yl | 2-FC6H4 | 62.5 | 500 | >500 | >500 |
| C4 | pyridin-2-yl | 3,4-Cl2C6H3 | 0.976 | 7.81 | 125 | 62.5 |
| C8 | pyridin-3-yl | 3,4-Cl2C6H3 | 62.5 | 31.25 | 125 | 62.5 |
| C11 | pyridin-2-yl | C6H5 | 62.5 | 31.25 | 250 | 62.5 |
| C14 | pyridin-2-ylmethyl | 3,4-Cl2C6H3 | 62.5 | 125 | >500 | 125 |
| Compound | fCHEMPLP | Ligand—Amino Acids Interactions |
|---|---|---|
| C1 | 56.67 | F42…C(HEME); S5…C(MET62); N1…O(HEME); N2…C(HEME) |
| C2 | 57.79 | N1…O(HEME); N2…C(HEME); S5…C(MET62) |
| C3 | 54.59 | S5…C(HEME); N1…O(VAL82) |
| C4 | 57.68 | N2…C(PHE280); S5…C(HEME); C45…O(HEME); C46…O(HEME); Cl44…(MET62) |
| C5 | 57.04 | S5…C(MET62); N2…C(PHE280) |
| C6 | 67.83 | S5…C(MET62) × 2; N63…N(ARG386); O…O(SER237) |
| C7 | 55.14 | S5…C(MET62); N63…C(PHE280); Cl44…(C(PHE280); Cl42…O(HEME) |
| C8 | 53.67 | N63…N(ARG386); Cl43…C(VAL78) |
| C9 | 58.71 | S5…C((MET62); N64…O(SER237) |
| C10 | 60.20 | N2…C(HEME); N64…N(ARG386); C63…O(SER237); C43…O(HEME) |
| C11 | 65.77 | S5…C(MET62) × 2; S5…C(HEME) |
| C12 | 55.89 | N1…O(VAL82) |
| C13 | 66.94 | S5…N(ASN85); S5…C(ASN85); N64…C(MET62); N1…O(VAL82); N2…C(MET62) |
| C14 | 68.29 | N1…O(VAL82); S5…N(ASN85); S5…C(ASN85); Cl43…C(HEME) |
Sample Availability: Samples of the compounds B13, B14, C1–C14 are available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karczmarzyk, Z.; Swatko-Ossor, M.; Wysocki, W.; Drozd, M.; Ginalska, G.; Pachuta-Stec, A.; Pitucha, M. New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies. Molecules 2020, 25, 6033. https://doi.org/10.3390/molecules25246033
Karczmarzyk Z, Swatko-Ossor M, Wysocki W, Drozd M, Ginalska G, Pachuta-Stec A, Pitucha M. New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies. Molecules. 2020; 25(24):6033. https://doi.org/10.3390/molecules25246033
Chicago/Turabian StyleKarczmarzyk, Zbigniew, Marta Swatko-Ossor, Waldemar Wysocki, Monika Drozd, Grazyna Ginalska, Anna Pachuta-Stec, and Monika Pitucha. 2020. "New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies" Molecules 25, no. 24: 6033. https://doi.org/10.3390/molecules25246033
APA StyleKarczmarzyk, Z., Swatko-Ossor, M., Wysocki, W., Drozd, M., Ginalska, G., Pachuta-Stec, A., & Pitucha, M. (2020). New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies. Molecules, 25(24), 6033. https://doi.org/10.3390/molecules25246033