Pharmacological Inhibition of CDK8 in Triple-Negative Breast Cancer Cell Line MDA-MB-468 Increases E2F1 Protein, Induces Phosphorylation of STAT3 and Apoptosis
Abstract
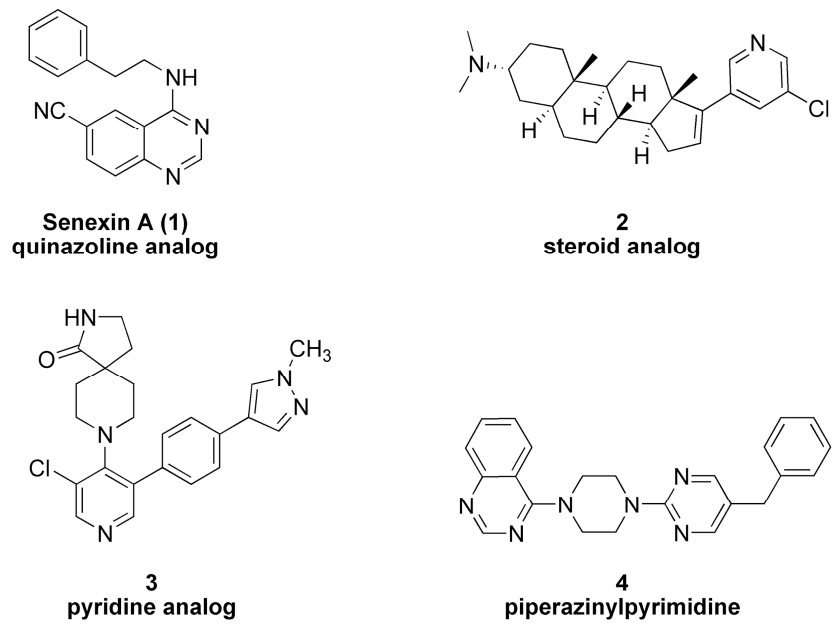
1. Introduction
2. Results
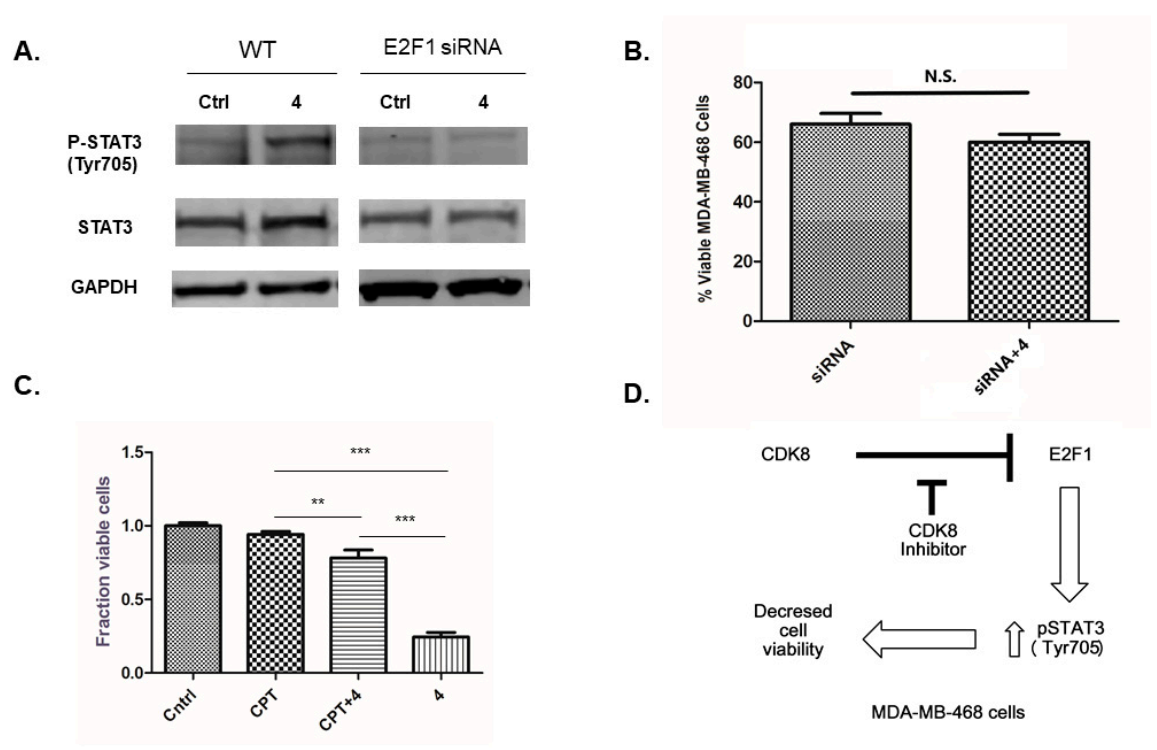
2.1. Effects of 4 on Cell Viability
2.2. Effects of Inhibitor 4 on β–Catenin, STAT1, STAT3 and E2F1 Proteins
2.3. Effects of E2F1 RNAi on STAT3 Protein
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Chemicals and Antibodies
4.2. Cell Viability
4.3. Apoptosis Assay
4.4. Immunoblotting
4.5. Statistics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Galbraith, M.D.; Donner, A.J.; Espinosa, J.M. CDK8: A positive regulator of transcription. Transcription 2010, 1, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Firestein, R.; Bass, A.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Isil, G.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates β–catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol. Res. 2019, 139, 471–488. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef]
- Knuesel, M.T.; Meyer, K.D.; Bernecky, C.; Taatjes, D.J. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivate function. Genes Dev. 2009, 23, 439–451. [Google Scholar] [CrossRef]
- Tsai, K.-L.; Sato, S.; Tomomori-Sato, C.; Conaway, R.C.; Conaway, J.W.; Asturias, F.J. A conserved mediator–CDK8 kinase module association regulates mediator–RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013, 20, 611–621. [Google Scholar] [CrossRef]
- Knuesel, M.T.; Meyer, K.D.; Donner, A.J.; Espinosa, J.M.; Taatjes, D.J. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 2009, 29, 650–661. [Google Scholar] [CrossRef]
- Zhao, J.; Ramos, R.; Demma, M. CDK8 regulates E2F1 transcriptional activity through S375 phosphorylation. Oncogene 2013, 32, 3520–3530. [Google Scholar] [CrossRef]
- Bancerek, J.; Poss, Z.C.; Steinparzer, I.; Sedlyarov, V.; Pfaffenwimmer, T.; Mikulic, I.; Dlöken, L.; Müller, M.; Taatjes, D.J.; Kovarik, P. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 2013, 38, 250–262. [Google Scholar] [CrossRef]
- Rzymski, T.; Mikula, M.; Zylkiewicz, E.; Dreas, A.; Wiklik, K.; Golas, A.; Wojcik, K.; Masiejczyk, M.; Wrobel, A.; Dolata, I.; et al. SEL120-34A is a novel CDK8 inhibitor active in AML cells with high levels of serine phosphorylation of STAT1 and STAT5 transactivation domains. Oncotarget 2017, 8, 33779–33795. [Google Scholar] [CrossRef]
- Kapoor, A.; Goldberg, M.S.; Cumberland, L.K.; Ratnakumar, K.; Segura, M.F.; Emanuel, P.O.; Menendez, S.; Vardabasso, C.; Leroy, G.; Vidal, C.I.; et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 2010, 468, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Bragelmann, J.; Klumper, N.; Offermann, A.; Von Massenhausen, A.; Bohm, D.; Deng, M.; Queisser, A.; Sanders, C.; Syring, I.; Merseburger, A.S.; et al. Pan-cancer analysis of the mediator complex transcriptome identifies CDK19 and CDK8 as therapeutic targets in advanced prostate cancer. Clin. Cancer Res. 2017, 23, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Kang, W.; Wang, L.; Sun, B.; Ma, J.; Zheng, C.; Sun, J.; Tian, Z.; Yang, X.; Xiao, W. E2F1 renders prostate cancer cell resistant to ICAM-1 mediated antitumor immunity by NF-κB modulation. Mol. Cancer 2014, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.C.; Farmaki, E.; Altilia, S.; Schools, G.P.; West, D.K.; Chen, M.; Chang, B.-D.; Puzyrev, A.T.; Lim, C.-U.; Rokow-Kittell, R.; et al. Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities. Proc. Natl. Acad. Sci. USA 2012, 109, 13799–13804. [Google Scholar] [CrossRef]
- Solum, E.; Hansen, T.V.; Aesoy, R.; Herfindal, L. New CDK8 inhibitors as anti-leukemic agents—Design, synthesis and biological evaluation. Bioorg. Med. Chem. 2020, 28, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Czodrowski, P.; Mallinger, A.; Wienke, D.; Esdar, C.; Pöschke, O.; Busch, M.; Rohdich, F.; Eccles, S.A.; Ortiz-Ruiz, M.-J.; Schneider, R.; et al. Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening. J. Med. Chem. 2016, 59, 9337–9349. [Google Scholar] [CrossRef] [PubMed]
- Shallal, H.M.; Russu, W.A. Discovery, Synthesis, and Investigation of the Antitumor Activity of Novel Piperazinylpyrimidine Derivatives. Euro. J. Med. Chem. 2011, 46, 2043–2057. [Google Scholar] [CrossRef]
- Mallinger, A.; Schiemann, K.; Rink, C.; Stieber, F.; Calderini, M.; Crumpler, S.; Stubbs, M.; Adeniji-Popoola, O.; Poesche, O.; Busch, M.; et al. Discovery of Potent, Selective, and Orally Bioavailable Small Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19. J. Med. Chem. 2016, 59, 1078–1101. [Google Scholar] [CrossRef]
- Koehler, M.F.T.; Bergeron, P.; Blackwood, E.M.; Bowman, K.; Clark, K.R.; Firestein, R.; Kiefer, J.R.; Maskos, K.; McCleland, M.L.; Orren, L.; et al. Development of a Potent, Specific CDK8 Kinase Inhibitor Which Phenocopies CDK8/19 Knockout Cells. ACS Med. Chem. Lett. 2016, 7, 223–228. [Google Scholar] [CrossRef]
- Dale, T.; Clarke, P.A.; Esdar, C.; Waalboer, D.; Adeniji-Popoola, O.; Ortiz-Ruiz, M.-J.; Mallinger, A.; Samant, R.S.; Czodrowski, P.; Musil, D.; et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nature Chem. Biol. 2015, 11, 973–980. [Google Scholar] [CrossRef]
- He, S.-B.; Yuan, Y.; Wang, L.; Yu, M.-J.; Zhu, Y.-B.; Zhu, X.-G. Effects of cyclin-dependent kinase 8 specific siRNA on the proliferation and apoptosis of colon cancer cells. J. Exp. Clin. Cancer Res. 2011, 30, 109. [Google Scholar] [CrossRef] [PubMed]
- Dolled-Filhart, M.; McCabe, A.; Giltnane, J.; Cregger, M.; Camp, R.L.; Rimm, D.L. Quantitative In situ Analysis of β-Catenin Expression in Breast Cancer Shows Decreased Expression Is Associated with Poor Outcome. Cancer Res. 2006, 66, 5487–5494. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.E.; Kitagawa, M.; Flavall, R.A.; Fu, X.Y. Activation of the STAT signalling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 1997, 17, 5328–5337. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.J.; Ji, J.-Y.; Yang, F.; Di Stefano, L.; Herr, A.; Moon, N.-S.; Kwon, E.-J.; Haigis, K.M.; Näär, A.M.; Dyson, N.J. E2F1 represses β-catenin transcription and is antagonized by both pRb and CDK8. Nature 2008, 455, 552–556. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, X.; Bao, Y.; Mo, J.; Du, H.; Hu, J.; Zhang, X. E2F1 silencing inhibits migration and invasion of osteosarcoma cells via regulating DDR1 expression. Int. J. Oncol. 2017, 51, 1639–1650. [Google Scholar] [CrossRef]
- Jackson, N.M.; Ceresa, B.P. EGFR-mediated apoptosis via STAT3. Exp. Cell Res. 2017, 356, 93–103. [Google Scholar] [CrossRef]
- Sun, B.; Wingate, H.; Swisher, S.G.; Keyomarsi, K.; Hunt, K.K. Absence of pRb facilitates E2F1-induced apoptosis in breast cancer cells. Cell Cycle 2010, 9, 1122–1130. [Google Scholar] [CrossRef]
- Xu, D.; Li, C.-F.; Zhang, X.; Gong, Z.; Chan, C.-H.; Lee, S.-W.; Jin, G.; Rezaeian, A.-H.; Han, F.; Wang, J.; et al. Skp2–macroH2A1–CDK8 axis orchestrates G2/M transition and tumorigenesis. Nat. Commun. 2015, 6, 6641–6655. [Google Scholar] [CrossRef]
- McDermott, M.S.J.; Chumanevich, A.A.; Lim, C.-U.; Liang, J.; Chen, M.; Altilia, S.; Oliver, D.; Rae, J.M.; Shtutman, M.; Kiaris, H.; et al. Inhibition of CDK8 mediator kinase suppresses estrogen dependent transcription and the growth of estrogen receptor positive breast cancer. Oncotarget 2017, 8, 12558–12575. [Google Scholar] [CrossRef]
- Li, X.-Y.; Luo, Q.-F.; Wei, C.-K.; Li, D.-F.; Li, J.; Fang, L. siRNA-mediated silencing of CDK8 inhibits proliferation and growth in breast cancer cells. Int. J. Clin. Exp. Pathol. 2014, 7, 92–100. [Google Scholar]
- Li, X.-Y.; Luo, Q.-F.; Wei, C.-K.; Li, D.-F.; Li, J.; Fang, L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int. J. Clin. Med. 2014, 7, 32–40. [Google Scholar]
- Tormo, E.; Bellester, S.; Adam-Artigues, A.; Burgues, O.; Alonso, E.; Bermejo, B.; Menendez, S.; Zazo, S.; Madoz-Gurpide, J.; Rovira, A.; et al. The miRNA-449 family mediates doxorubicin resistance in triple-negative breast cancer by regulating cell cycle factors. Sci. Rep. 2019, 9, 5316. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wen, P.; Hu, G.; Gao, Y.; Qi, X.; Zhu, K.; Chen, S.; Wu, L.; Xu, A.; Zhao, G. Antagonizing CDK8 Sensitizes Colorectal Cancer to Radiation Through Potentiating the Transcription of e2f1 Target Gene apaf1. Front. Cell Dev. Biol. 2020, 8, 408. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spear, J.M.; Lu, Z.; Russu, W.A. Pharmacological Inhibition of CDK8 in Triple-Negative Breast Cancer Cell Line MDA-MB-468 Increases E2F1 Protein, Induces Phosphorylation of STAT3 and Apoptosis. Molecules 2020, 25, 5728. https://doi.org/10.3390/molecules25235728
Spear JM, Lu Z, Russu WA. Pharmacological Inhibition of CDK8 in Triple-Negative Breast Cancer Cell Line MDA-MB-468 Increases E2F1 Protein, Induces Phosphorylation of STAT3 and Apoptosis. Molecules. 2020; 25(23):5728. https://doi.org/10.3390/molecules25235728
Chicago/Turabian StyleSpear, Jensen M., Zhixin Lu, and Wade A. Russu. 2020. "Pharmacological Inhibition of CDK8 in Triple-Negative Breast Cancer Cell Line MDA-MB-468 Increases E2F1 Protein, Induces Phosphorylation of STAT3 and Apoptosis" Molecules 25, no. 23: 5728. https://doi.org/10.3390/molecules25235728
APA StyleSpear, J. M., Lu, Z., & Russu, W. A. (2020). Pharmacological Inhibition of CDK8 in Triple-Negative Breast Cancer Cell Line MDA-MB-468 Increases E2F1 Protein, Induces Phosphorylation of STAT3 and Apoptosis. Molecules, 25(23), 5728. https://doi.org/10.3390/molecules25235728






