Abstract
Two new epimeric bibenzylated monoterpenes machaerifurogerol (1a) and 5-epi-machaerifurogerol (1b), and four known isoflavonoids (+)-vestitol (2), 7-O-methylvestitol (3), (+)-medicarpin (4), and 3,8-dihydroxy-9-methoxypterocarpan (5) were isolated from Machaerium Pers. This plant was previously assigned as Machaerium multiflorum Spruce, from which machaeriols A-D (6–9) and machaeridiols A-C (10–12) were reported, and all were then re-isolated, except the minor compound 9, for a comprehensive antimicrobial activity evaluation. Structures of the isolated compounds were determined by full NMR and mass spectroscopic data. Among the isolated compounds, the mixture 10 + 11 was the most active with an MIC value of 1.25 μg/mL against methicillin-resistant Staphylococcus aureus (MRSA) strains BAA 1696, −1708, −1717, −33591, and vancomycin-resistant Enterococcus faecium (VRE 700221) and E. faecalis (VRE 51299) and vancomycin-sensitive E. faecalis (VSE 29212). Compounds 6–8 and 10–12 were found to be more potent against MRSA 1708, and 6, 11, and 12 against VRE 700221, than the drug control ciprofloxacin and vancomycin. A combination study using an in vitro Checkerboard method was carried out for machaeriols (7 or 8) and machaeridiols (11 or 12), which exhibited a strong synergistic activity of 12 + 8 (MIC 0.156 and 0.625 µg/mL), with >32- and >8-fold reduction of MIC’s, compared to 12, against MRSA 1708 and −1717, respectively. In the presence of sub-inhibitory concentrations on polymyxin B nonapeptide (PMBN), compounds 10 + 11, 11, 12, and 8 showed activity in the range of 0.5–8 µg/mL for two strains of Acinetobacter baumannii, 2–16 µg/mL against Pseudomonas aeruginosa PAO1, and 2 µg/mL against Escherichia coli NCTC 12923, but were inactive (MIC > 64 µg/mL) against the two isolates of Klebsiella pneumoniae.
1. Introduction
The genus Machaerium Pers. (Fabaceae) consists of approximately 130 species, which are primarily distributed in the tropical Americas [1]. It is a genus of shrubs or lianas and small to medium-sized trees occurring throughout Southern Mexico to Brazil and Northern Argentina and Peru. These species are indigenous to all climatic regions ranging from equatorial rainforests to the verges of dry and cold deserts [2,3,4]. Several species of this genus are used in traditional medicines are considered to have multiple medicinal properties. Generally, various plant parts of Machaerium are used as an antitussive, and the sap is used to cure aphthous ulcers of the mouth [4]. M. floribundum is used to treat diarrhea and menstrual cramps [4]. The presence of a wide array of secondary metabolites from Machaerium, including flavonoids, terpenoids, and oxygenated phenolic compounds, together with their bioactivities, was recently reviewed by Amen et al. (2015) [2].
Earlier studies on one of the Machaerium species (Manuel Rimachi, Y-12161), named M. multiflorum Spruce, yielded four unique (+)-trans-hexahydrodibenzopyrans (HHDBP), machaeriols A-D, and three 5,6-seco-HHDBPs, machaeridiols A-C [5,6]. An unprecedented structural similarity for the HHDBP nucleus was observed in machaeriol and hexahydrocannabinol, and the 5,6-seco-HHDBP nucleus in machaeridiol and dihydrocannabidiol. Since these are the first reports of novel phytocannabinoids from a higher plant other than Cannabis, a recollection of the plant material was necessary from the original source. Unfortunately, there is an absence of documentary evidence for the existence of the species M. multiflorum. This species name was not included in the regional Floras as well as in major online databases (i.e., the International Plant Names Index (http://www.ipni.org/index.html) and The Plant List (http://www.theplantlist.org). Therefore, it was assumed that the plant sample was misidentified and was given the name combination M. multiflorum Spruce in error. An investigation was carried out on the identity of the plant, and a re-examination of the voucher specimen (Rimachi # 12161) at the Missouri Botanical Garden (MBG) concluded that this species should be treated only as an unidentified species of Machaerium Pers., as determined by the collection information (Manuel Rimachi, Y. 12161) [7].
The significance of the chemistry and biological activity of these aralkyl class of phytocannabinoid-type compounds led to the re-examination of the n-hexane and DCM fractions of the stem bark EtOH extract of the original plant material [5,6], as well as previously unexamined root and leaf extracts, which showed significant enhancement of antimicrobial activity against the various strains of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin resistant Enterococci (VRE). MRSA and VRE represent two potential threats to human health. According to the Centers for Disease Control and Prevention (CDC), MRSA can cause serious health problems, such as bloodstream infections and pneumonia. CA-MRSA occurs with a higher incidence rate in the United States and in particular amongst people who are in close physical contact, such as football athletes and childcare workers [8]. A recent national estimate for invasive MRSA incidence rates showed one in three people carry S. aureus in their nose and two in 100 people carry MRSA. Enterococci bacteria have the ability to survive for months in humans and animals. Similar to MRSA, VRE infections are commonly acquired by hospitalized patients. Enterococcal infections can be lethal, particularly those caused by VRE. According to the CDC, the number of nosocomial VRE isolates increased in the United States 20-fold, between 1989 and 1993. VRE is now the second to third most common cause of nosocomial infections in the USA [9].
In order to acquire substantial quantities of machaeriol A-D (6–9) and machaeridiol A-C (10–12) for comprehensive antimicrobial evaluations against MRSA and VRE, a reinvestigation was conducted on stem bark, leaves, and roots of the original plant material. During the course of this work, the novel epimeric mixture of bibenzylated furanoid monoterpenes, machaerifurogerol (1a), and 5-epi-machaerifurogerol (1b), together with the known isoflavons (+)-vestitol (2) and 7-O-methylvestitol (3), and pterocarpans (+)-medicarpin (4) and 3,8-dihydroxy-9-methoxypterocarpan (5), as well as previously isolated [5,6] machaeriol A-C (6–8) and machaeridiol A-C (10–12), were isolated. In this study, we report the correction of the previously reported botanical identity of the plant M. multiflorum, the structure elucidation of compounds 1–5, and comprehensive antimicrobial activities of compounds 1–8 and 10–12.
2. Results and Discussion
2.1. Botanical Identity of Machaerium sp. (Rimachi 12161)
The regional floras and other relevant publications were consulted for possible botanical identification of the species [7,10]. The voucher specimen (Rimachi 12161) was confirmed to belong to the genus Machaerium, based on the morphological features and available field information such as habitat, leaf, inflorescence, and fruit characters. The leaves showed partial similarities with those of M. leiophyllum var. leiophyllum and M. glabrum. However, complete identification of the specimen was not possible due to a lack of information on necessary diagnostic features, such as presence or absence of spines, features of stipules, and floral characters. It is possible that the specimen could represent an un-described taxon. The description (vide infra) is based on a single herbarium specimen and the associated collection information available from the original collection.
The Machaerium sp. (12161) plant was found to grow on sandy soils in open forests in Maynas, near Loreto, Peru, at an altitude of about 140–160 m. It is a woody liana with cylindrical stems, imparipinnate leaves with 17–21 leaflets, green flowers in axillary panicles, and 1-seeded samaroid fruits with a terminal wing showing reticulate venations.
2.2. Phytochemical Constituents
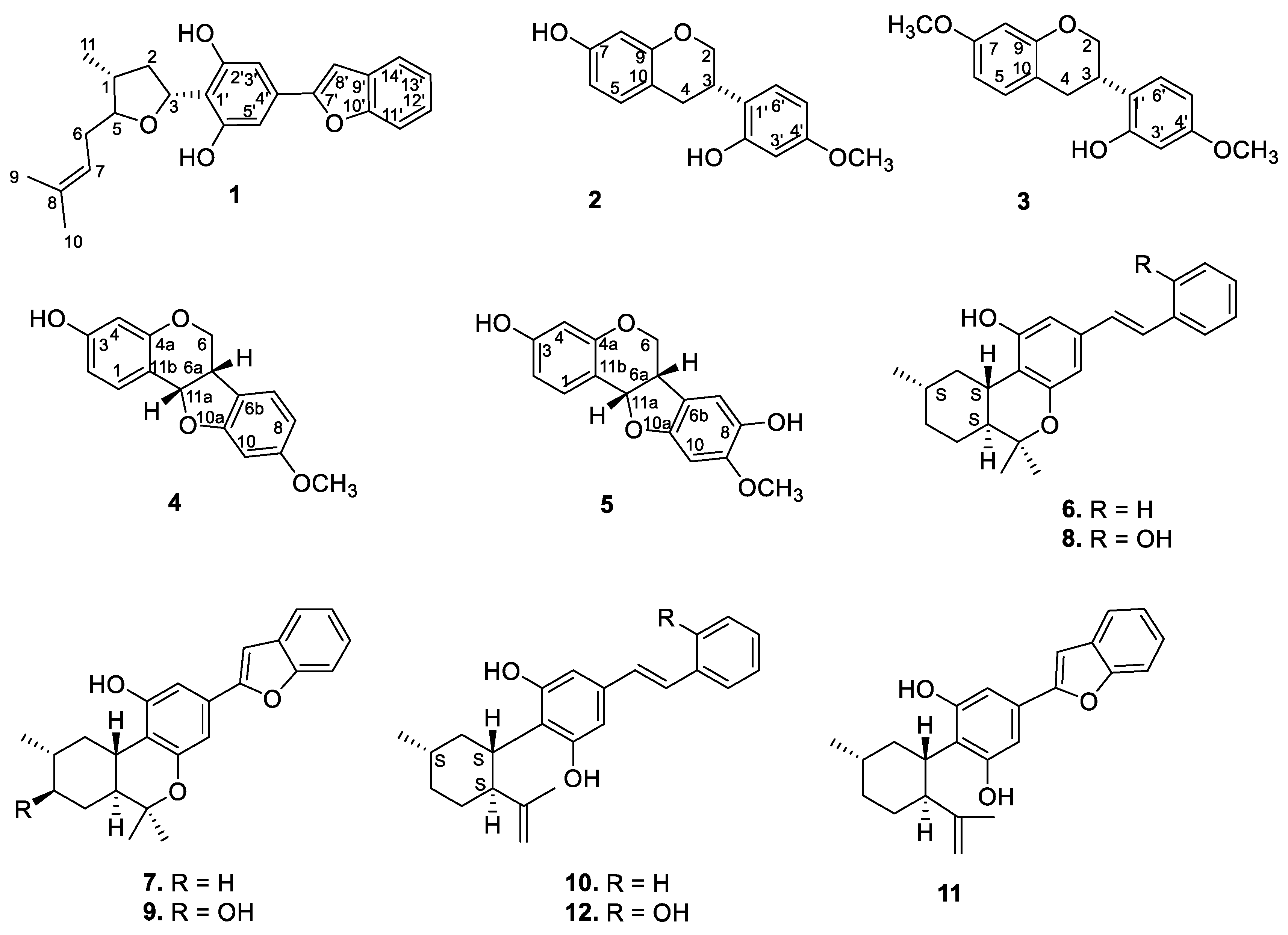
The dried EtOH extract of the stem bark was fractionated with n-hexane, followed by dichloromethane (DCM), and resulted in the isolation of compounds 1–5 (Figure 1 and Figure S1) (see the Experimental Section).
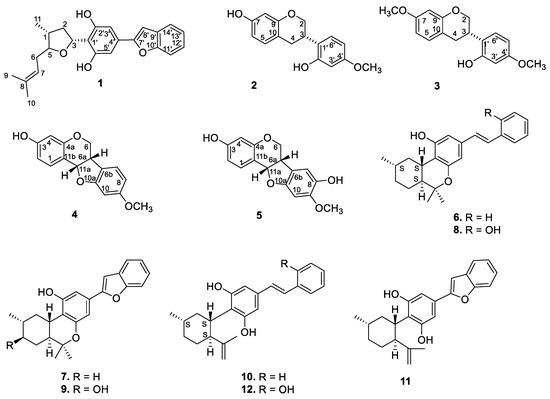
Figure 1.
Structures of the isolated compounds.
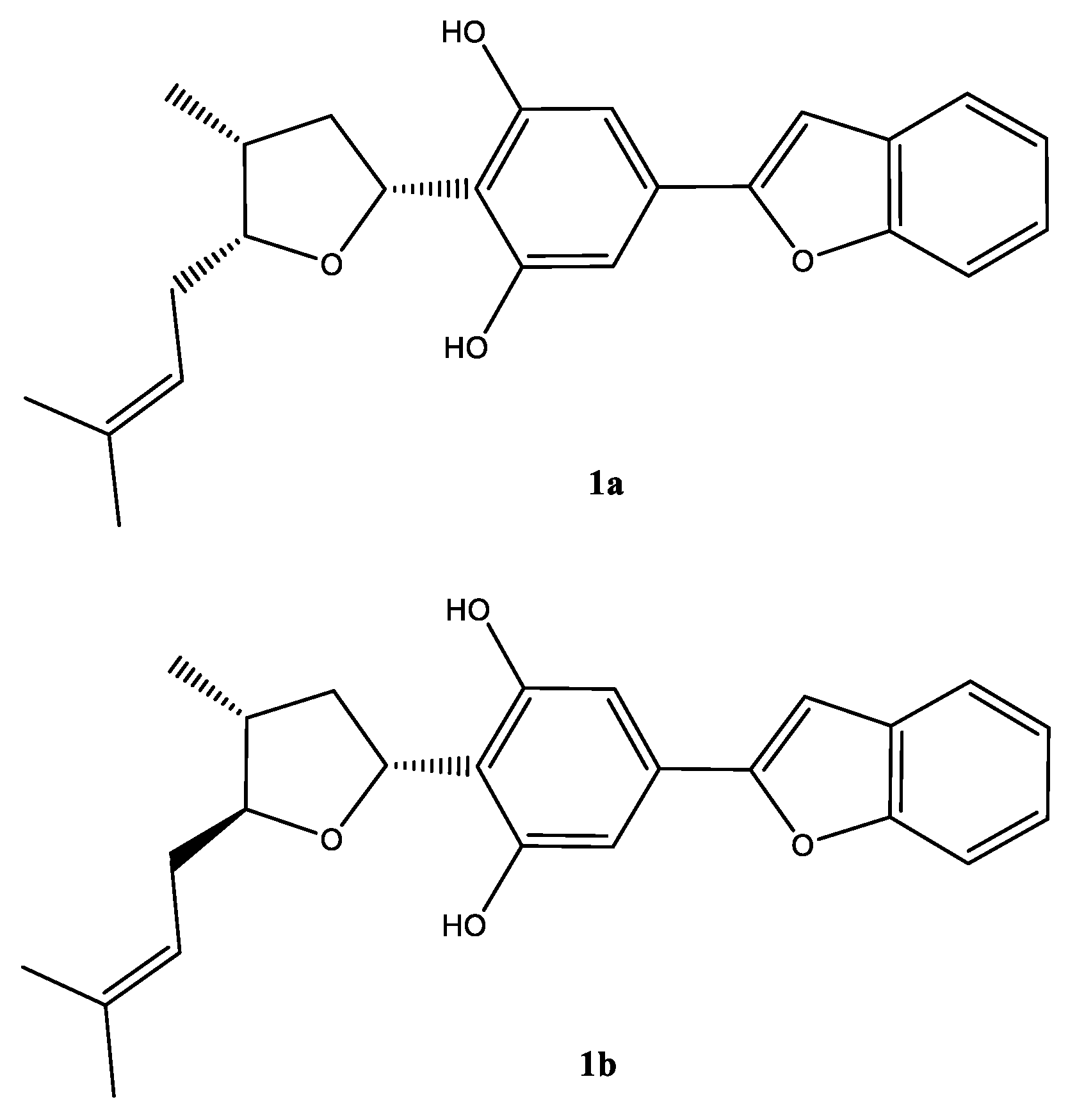
Chromatographic separation of the DCM fraction 20–24 that led to the isolation of compound 1, showed a single peak upon LC–MS analysis, which showed a protonated molecular ion peak at m/z 379.1906 [M + H]+ in its ESI–HRMS, suggesting the molecular formula C28H29O3. A careful analysis of the 1H and 13C NMR spectra (Table 1), and 2D NMR COSY, HMQC, HMBC, and NOESY spectra (Figures S2–S11) suggested that the compound was a mixture of C-5 epimers 1a and 1b (Figure 2).

Table 1.
1H and 13C NMR (in CDCl3) data for epimeric compounds 1a and 1b.
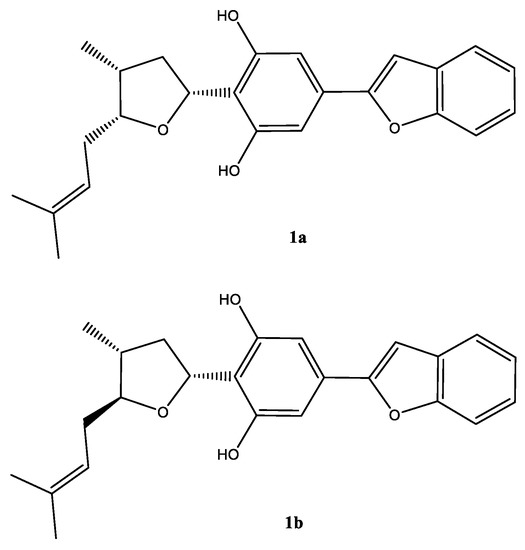
Figure 2.
Structure of the epimeric compound (1a) (major) and (1b) (minor) in mixture.
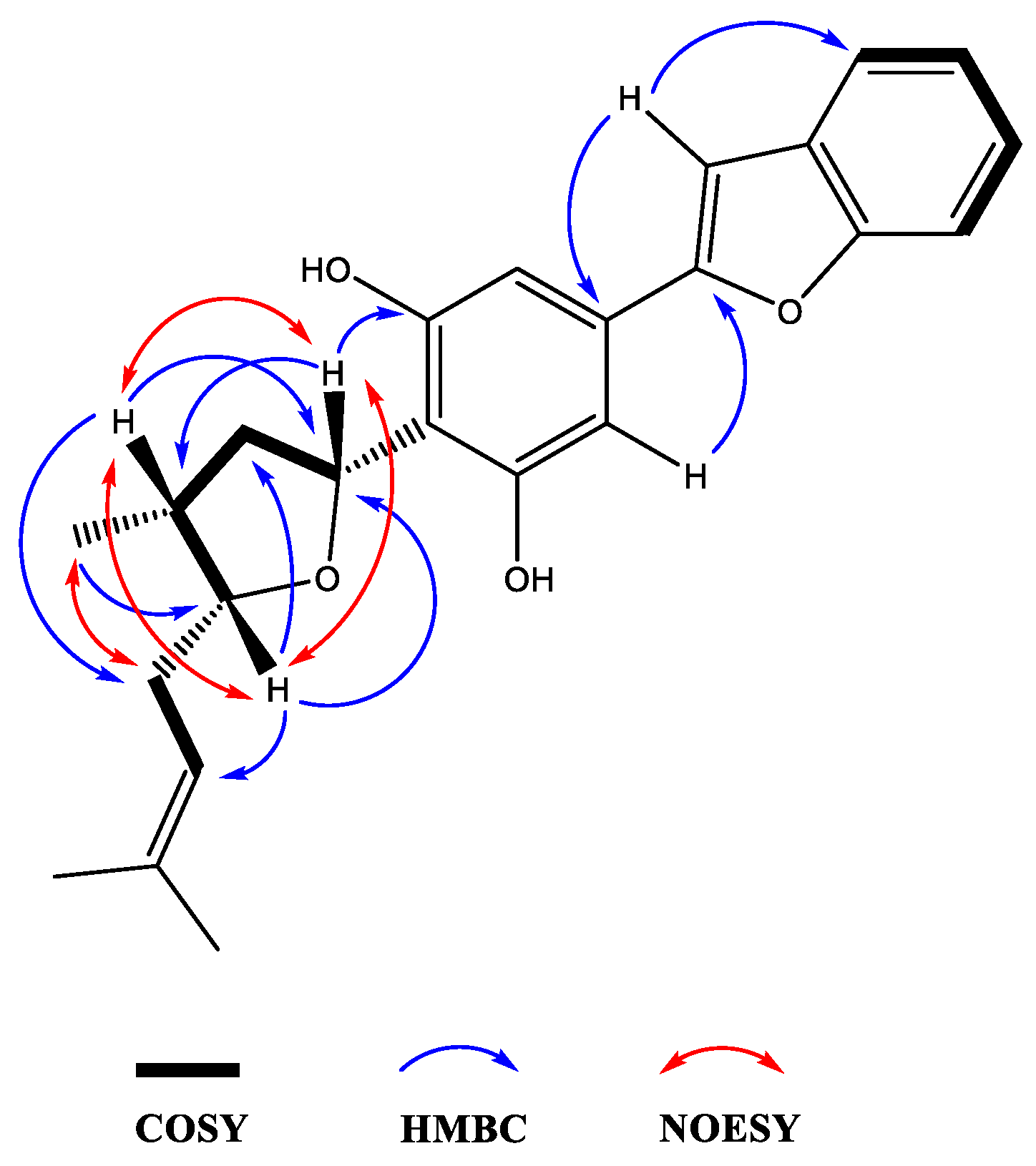
Moreover, the NMR data were found to be partially comparable with the machaeridiol C (11) [6], suggesting the presence of a benzofuran side chain (δH 7.18–7.52) (4H) attached to the substituted resorcinol moiety. The HMBC spectrum (1a; Figure 2) showed 2J- and 3J correlations between the H-8′ at δH 6.89 (brs) and the two sp2-hybridized carbons at C-4′ and C-7′ (δC 130.6 and 154.9, respectively), supporting the attachment of C-4′ of the resorcinol unit to the C-7′ of benzofuran ring. The 1H NMR spectrum also showed signals at δH 6.83 (2H) for two identical protons (H-3′ and H-5′), suggesting the presence of C-1′,2′,4′,6′-tetra substituted resorcinol ring with two oxygenated carbons at C-2′ and C-6′ (δC 155.3 and 155.4). In addition, the 1H NMR spectrum (1a) showed signals at δH 4.0 (1H, m, H-5) and 5.32 (1H, dd, J = 1.2, 7.6 Hz, H-3) for the tetrahydrofuran ring, and at δH 1.02 (3H, d, J = 7.2 Hz, H-11) for a Me-group. The HMBC (Figure 3) spectrum showed correlations between the H-3 (δH 5.32) and the three sp2 hybridized carbons at C-1′, C-2′, and C-6′ (δC 113.3, 155.3, and 155.4, respectively), supporting the attachment of the tetrahydrofuran ring at C-3 (δC 77.7) to C-1′ position. The HMBC spectrum also showed cross peaks between the methyl protons H-11 (δH 1.02) and C-1, C-2, and C-5 (δC 35.2, 41.8, and 83.1), supporting the attachment of the methyl (C-11, δC 15.5) to the tetrahydrofuran at C-1 (δC 35.2). The 1H, 13C, and 2D NMR spectra supported the presence of the 2-methylbut-2-ene unit. This was confirmed by the HMBC spectrum, which showed correlations between the H-6 at (δH 2.4) and the three carbons at C-5, C-7, and C-8 (δC 83.1, 120.0, and 134.5, respectively), confirming the attachment of the methylbut-2-ene unit to the tetrahydrofuran moiety at C-5. The relative configurations at C-1, C-3, and C-5 of the tetrahydrofuran were assigned via careful analysis of NOESY correlations for 1a. In the NOESY spectrum (assigned for 1a), H-5 (δH 4.0) showed correlation with H-1 (δH 2.4) and H-3 (δH 5.32), indicating the cofacial (β)-orientation of the three groups. Additionally, NOESY showed cross peaks between H-1 (δH 2.4.) and H-3 (δH 5.32), which supported their presence in the same plane of the molecule like H-5. On the other hand, such nOe signals were not evident in the NOESY of the minor compound 1b. Moreover, in its 1H NMR spectrum, H-5 and H-3 were deshielded at δH 4.21 (1H, m) and 5.59 (1H, dd, J = 12.0, 8.0 Hz), respectively, for the tetrahydrofuran ring, and at δH 1.02 (3H, d, J = 7.2 Hz, H-11) for an Me-group. Moreover, a CD analysis for this compound revealed a weak spectrum in the range of 250–500 nm, which is reflective of the epimeric nature of the compound. Based on the foregoing discussion and comparing the NMR data with compound 11 [6], the structure 1a and 1b were determined for machaerifurogerol and 5-epi-machaerifurogerol, respectively.
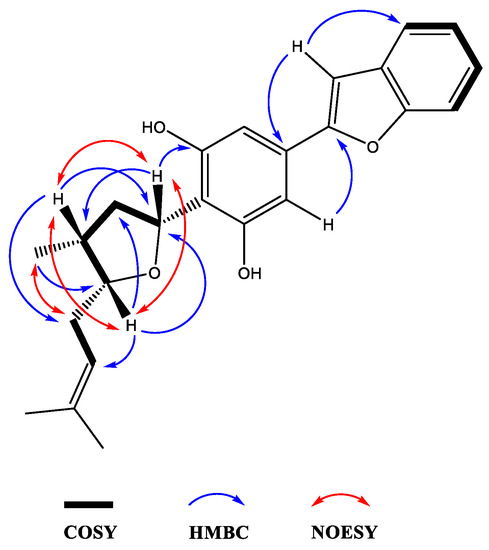
Figure 3.
Key 2D NMR COSY, HMBC, and NOESY correlations of compound 1a.
During the course of isolation, four isoflavonoid derivatives (2–5) and previously reported machaeriols (6–8) and machaeridiols (10–12) were isolated from the DCM partition of the stem bark and leaves extracts. However, the minor compound 9 could not be isolated due to a paucity of material. Compounds 2 and 3 were identified as known isoflavons (+)-vestitol and 7-O-methylvestitol, while 4 and 5 were identified as known peterocarpans. (+)-medicarpin and 3,8-dihydroxy-9-methoxy-pterocarpan, respectively, previously reported from Machaerium vestitum and Cuban propolis [10,11]. Compounds 4 and 5 were also reported from Cuban propolis [11]. The 1H and 13C NMR spectroscopic data (see Table S1) of compounds 2–5 were in agreement with those reported [11,12]. In addition, examination of the leaves of Machaerium sp. also yielded compounds 6–8 and 10–12, as well as their presence in the root extract. The identities of compounds 6–12 were established by NMR spectra and by direct comparison with authentic samples (TLC, HPLC/ LC–MS).
2.3. Antimicrobial Activity against Gram-Positive Species and Fungi
The availability of machaeriols A-C (6–8) and machaeridiols (10–12) [5,6] offered the opportunity to carry out a comprehensive investigation of antimicrobial activity. Among the tested fractions, DCM-25-32 (enriched with compounds 8 and 10–12) and DCM-10-15 (enriched with compound 12) were the most active against bacteria S. aureus, and methicillin-resistant S. aureus (MRSA), and the fungi Candida glabrata, C. krusei, and Cryptococcus neoformans, with IC50 values of <0.8, <0.8, <0.8, 5.35, <0.8 μg/mL, and <0.8, 1.95, 3.0, 6.07, 12.58 μg/mL, respectively (Table 2). Antibacterial activities of 6–8, 10–12, and a mixture 10 + 11 (1:1) were evaluated against methicillin-resistant S. aureus (ATCC 1708, 1696, and 1717), the ex vivo MRSA XEN31 strain, and vancomycin-resistant Enterococci (VRE; Enterococcus faecium ATCC 700221), low-level VRE (E. faecalis ATCC 51299), and the vancomycin-sensitive strain (VSA; E. faecalis ATCC 29212) (Table 3 and Table 4). Compound 11 and mixture 10 + 11 (1:1) showed the most potent activity against MRSA BAA 1696, BAA 1708, BAA 1717, and BBA 33591 with IC50/MIC/MBC values of 0.43/1.25/5 μg/mL, 0.38/1.25/1.25 μg/mL, 0.38/1.25/2.5 μg/mL, 0.71/1.25/1.25 μg/mL; and 0.41/1.25/10 μg/mL, 0.34/1.25/1.25 μg/mL, 0.39/1.25/1.25 μg/mL and 0.61/1.25/10 μg/mL, respectively. On the other hand, compound 8 and mixture 10 + 11 were found to be the most potent against E. faecium ATCC 700221 and E. faecalis ATCC 51299, (VRE) and E. faecalis ATCC 29212 (VSE) with IC50/MIC/MBC of 0.48/1.25/2.5 μg/mL, 1.02/1.25/5 μg/mL and 1.16/2.5/2.5 μg/mL; and 0.49/1.25/2.5 μg/mL, 0.70/1.25/5 μg/mL, and 0.72/1.25/5 μg/mL, respectively (Table 3 and Table 4). The activities of compounds 6–8 and 10–12 were found to be more potent than ciprofloxacin and vancomycin against the MRSA BBA 1708 strain, while 6, 8, and 10 + 11 were more active against VRE 700221 than the positive controls.

Table 2.
Antimicrobial activity (IC50 in μg/mL) of Machaerium DCM fractions a.

Table 3.
Anti-MRSA activities (in μg/mL) of compounds 6–8 and 10–12.

Table 4.
Anti-VRE activities (in μg/mL) of compounds 6–8 and 10–12.
2.4. Antimicrobial Combination Studies
In light of the strong antimicrobial activity of the DCM fraction 25–32, which is enriched with compounds 8, 10–12 (Table 3), a combination study using an in vitro Checkerboard method [13,14] was carried out for machaeriol (7 or 8) and machaeridiol (11 or 12), to evaluate the synergy of combination treatment against the strains of MRSA and Enterococcus (VRE) (Table 4). Among these compounds, a combination of machaeridiol B (12; at MIC 5 μg/mL) and machaeriol C (8; at ½ MIC 1.25 μg/mL) exhibited a potent activity, with the MIC values of 0.156 and 0.625 μg/mL exhibiting a >32- and >8-fold reduction of MICs, compared to those observed for 12, against MRSA 1708 and MRSA 1717 strains. When these two compounds were tested with an inverse concentration (i.e., MIC of 8; 2.5 μg/mL + ½ MIC of 12; 2.5 μg/mL), a strong synergism was also observed, but to a lesser extent. When tested against VRE (E. faecium 700221), this combination showed synergism with the MIC values of 1.25 μg/mL, a >4-fold reduction of MIC compared to 12 (Table 5).

Table 5.
Combination study (MIC in μg/mL) a of compounds 7, 8, 11, and 12 by Checkerboard assay against MRSA and VRE.
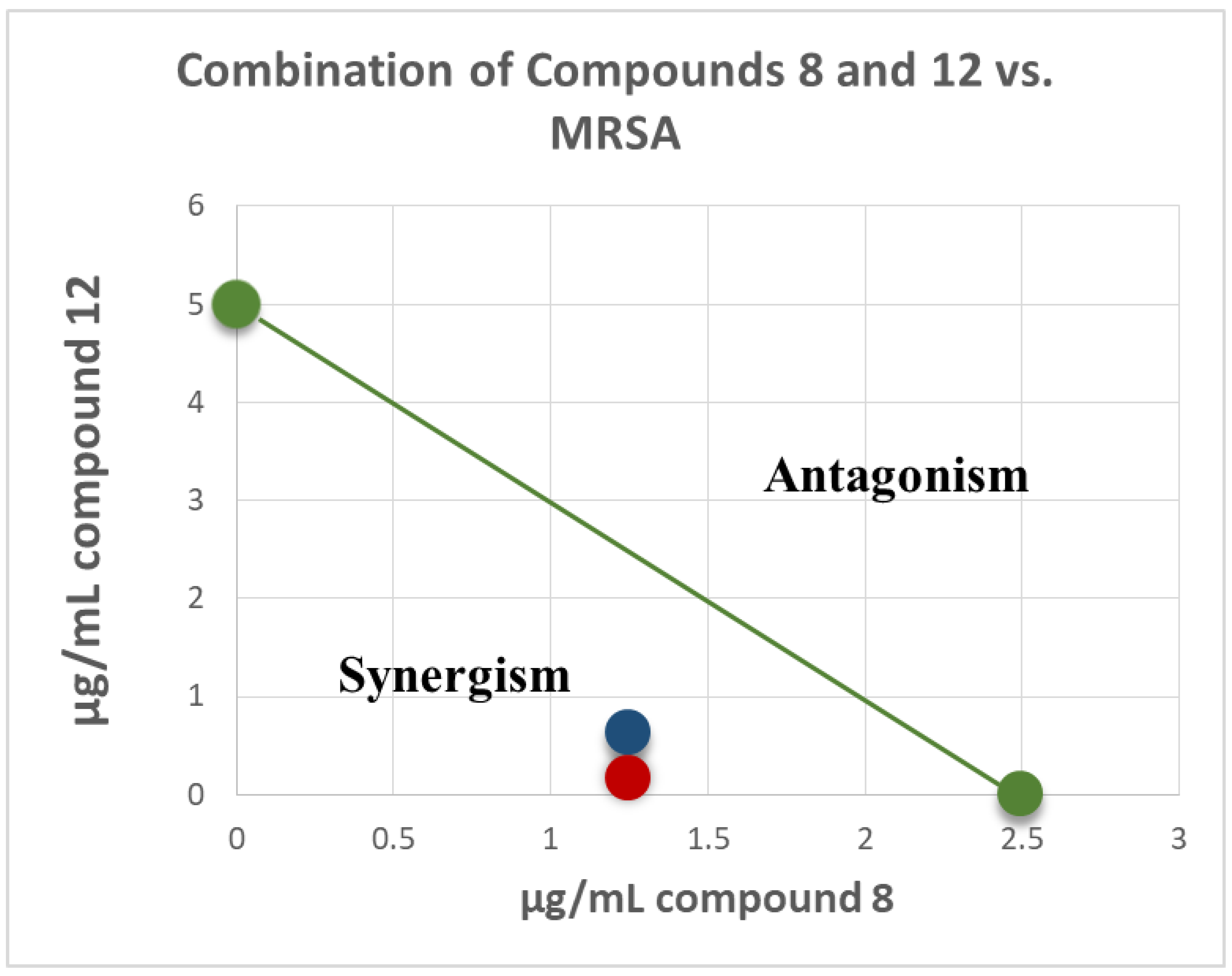
Isobologram showing synergistic activity of the combination of compounds 8 and 12 in MRSA 1708 (red) and 1717 (blue) are presented in Figure 4. The green series represents the additivity line of compounds 8 and 12 (green dots represent the MIC of each compound alone; the green line represents all possible additive combinations). The red (MRSA 1708) and blue (1717) dots represent the combination of compounds 8 and 12, and show that they fell below the additivity line (the combination of the compounds produces a synergistic effect beyond additivity). This synergism between machaeriol (HHDBP) and machaeridiol (seco-HHDBP) could be due to different molecular targets affected by these two molecules. A combination study of compounds 8 and 12 with antibiotics, either methicillin or ciprofloxacin, did not show any additive or synergistic effects.
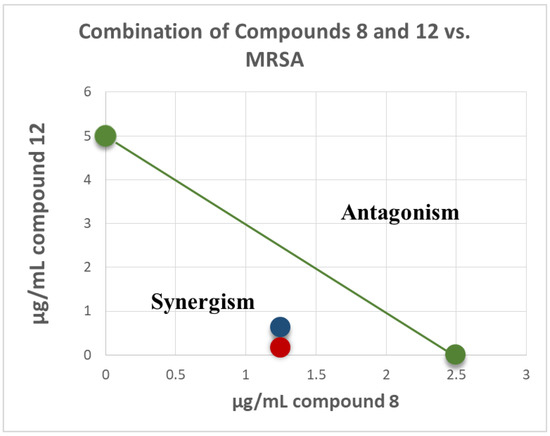
Figure 4.
Isobologram of combination of compounds 8 and 12 in MRSA 1708 (red) and 1717 (blue). The green series represents the additivity line of compounds 8 and 12 green dots represent the MIC of each compound alone; the green line represents all possible additive combinations). The red (MRSA 1708) and blue (1717) dots represent the combination of compounds 8 and 12, and show that they fell below the additivity line (the combination of compounds produces a synergistic effect beyond additivity).
2.5. Antimicrobial Activity against Gram-Negative Species
The activity of the compounds was determined in the Gram-negative species of the ESKAPEE panel, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli (Table 6). None of the tested compounds displayed any antimicrobial activity in the Gram-negative species up to a concentration of 128 µg/mL. However, when the outer membrane was permeabilized using the membrane permeabilizer polymyxin-B-nonapeptide (PMBN), MICs as low as 0.5 µg/mL were observed, suggesting an intracellular target. With the exception of 6 + 7, all compounds tested displayed a good activity in A. baumannii and E. coli strains and in P. aeruginosa strain PAO1. The P. aeruginosa strain NCTC 13437 is a near-pan drug resistant strain, and this was reflected in the MICs to compounds 6 + 7, 10 + 11, 11, and 12, although 8 had an MIC of 8 µg/mL in the presence of PMBN. None of the compounds displayed activity in the two K. pneumoniae strains, even in the presence of PMBN, suggesting the target/pathway might be missing or modified in this species.

Table 6.
Gram-negative MIC Values (in µg/mL) of compounds 6–8 and 10–12 in the presence of the membrane permeabilizer polymyxin-B-nonapeptide at 30 µg/mL.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were recorded at ambient temperature using a Rudolph Research Analytical Autopol IV automatic polarimeter. IR spectra were obtained using a Bruker Tensor 27 instrument. NMR spectra were acquired on a Varian Mercury 400 MHz NMR spectrometer at 400 (1H) and 100 MHz (13C) in CDCl3, using the residual solvent as an internal standard. Multiplicity determinations (DEPT) and 2D NMR spectra (HMQC, HMBC, NOESY) were obtained using the standard Bruker pulse programs. ESI-HRMS were acquired by direct injection using a Water Xevo G2-S TOF with electrospray ionization (ESI). TLC was carried out on pre-coated silica gel 60 F254 (EMD Chemicals Inc, Darmstadt, Germany) using toluene-EtOAc (9:1) and n-hexane-EtOAc (7.5:2.5) as solvents. Centrifugal preparative TLC (CPTLC, using a Chromatotron, Harrison Research Inc. model 8924, tagged with a fraction collector) was carried out on 6 mm custom-made RP C18 silica gel [15], and silica gel P254 (Analtech) 1, 2, and 4 mm rotors, using H2O-MeOH, EtOAc-:n-hexane, and CHCl3 as eluents. SPE cartridges C18 (Supelco Inc., Bellefonte, PA, USA) were used in the fractionation work. Purifications were performed on prep-HPLC (silica gel-100 A 250 × 15.00, 5 µM; Phenomenex Luna, Torrance, CA, USA) using an HPLC Delta Prep 4000 equipped with a dual wavelength detector Model 2487 adjusted at 210 and 254 nm (Waters Corporation, Milford, MA, USA), Preparative HPLC was carried out on Waters LC module I plus, using Phenomenex C18, 22 mm, λ 254, flow 15 mL/min, 0–2 min [90% H2O; 10% MeCN], 2–45 min, 10% MeCN→ 100% MeN, 45–50 min 100% MeCN]. Samples were dried using a Savant Speed Vac Plus SC210A concentrator. The compounds were visualized by spraying the TLC plates with 1% vanillin-H2SO4 spray reagent.
3.2. Plant Material
The stem bark, leaves, and roots of Machaerium Pers. (Manuel Rimachi, Y.-12161), previously identified as M. multiflorum Spruce [5] by (Late) Professor Sydney T. McDaniel, was collected in November, 1997, from open sandy forest near Loreto (Maynas), Peru. The voucher specimen (Manuel Rimachi Y. 12161) is deposited at the Missouri Botanical Garden (http://www.tropicos.org/Specimen/100326687).
3.3. Extraction and Isolation of Compounds from Stem Bark and Leaves
The powdered stem bark (0.5 kg) was extracted by percolation with 95% EtOH (3 × 2 L) and the combined extracts were evaporated under reduced pressure (yield 17.7 g). A portion of the dried EtOH extract (15 g) was percolated with n-hexane, followed by DCM, and finally the residual extract was washed with MeOH (each 200 mL × 3). The n-hexane, DCM, and MeOH fractions were separately filtered and dried, which afforded 3.8, 8.9, and 4.5 g, respectively. The antimicrobial activity was detected in the DCM fraction (IC50 < 20 μg/mL against S. aureus and MRSA). A portion of the dried DCM fraction (1.65 g) was fractionated by CPTLC with a Chromatotron® instrument, using a 4 mm custom-made C18 RP silica gel ChromatoRotorTM [15], eluting with a gradient of 60% to 100% MeCN-H2O to afford 30 fractions. The fractions were pooled by TLC analyses.
Fractions 1–8 (475 mg) were combined and further subjected to CPTLC, using a 4 mm silica gel P254 disc, and gradient elution with MeCN:DCM. Elution with 2% MeCN:DCM afforded medicarpin (4; 4.5 mg), followed by elution with 4% MeCN:DCM, which gave 3,8-dihydroxy-9-methoxy-pterocarpan (5; 7.5 mg), and finally elution with 5% MeCN:DCM yielded vestitol (2; 9.8 mg). The combined fractions 10–15 (70 mg) was subjected to preparative C18 RP-HPLC, using 90% MeCN:H2O as solvent, which afforded machaeridiol B (12), followed by machaeridiol A (10) and machaeridiol C (11). Similarly, combined fractions 25–32 (100 mg) was also separated by preparative C18 RP-HPLC, which afforded additional quantities of 10–12 [total yields: 10 (10 mg), 11; (18 mg), 12 (21 mg)] and machaeriol C (8; 34.6), however, the minor compound machaeriol D (9) could not be re-isolated due to a paucity of material. Further elution with 75% MeCN:H2O afforded 13 fractions, which contained the mixture of two compounds 6 + 7, (50 mg). The mixture was then separated by preparative C18 RP-HPLC (column: ODS prodigy 10µ, 250 × 10 mm; detector: UV-254 nm), using 95% MeCN:H2O as solvent, which afforded 6 (16 mg), followed by 7 (16 mg). Finally, the dried n-hexane fraction (77 mg) was subjected to CPTLC, using a 2 mm C18 RP rotor, and eluted with 65% MeCN:H2O, which afforded 7-O-methylvestitol (3; 8 mg). A sub-fraction of DCM (15 mg) was subjected to prep-HPLC (Waters LC module I plus, using Phenomenex C18, 2 mm), which afforded compound 1a+1b (5 mg). The structures of (+)-vestitol (2), 7-O-methylvestitol (3), (+)-medicarpin (4) and 3,8-dihydroxy-9-methoxypterocarpan (5) were determined by physical and spectroscopic data (1H and 13C NMR, see SI 1), and also by comparison with those reported [11,12]. The structures of the re-isolated compounds 6–8 and 10–12 were identified by NMR data [5,6] and by direct comparison (TLC, HPLC/LC–MS) with their respective authentic samples available in our laboratories. Finally, powdered leaves (560 g) and root bark (50 g) of Machaerium sp. were extracted using the method described previously [5,6], and compounds 6–8 and 10–12 were isolated from leaves as describe below.
The powdered leaf was percolated with n-hexane, followed by DCM and EtOH (each 3 × 2 L) to yield 5, 14, and 9 g of extracts, respectively. A portion of the DCM extract (10 g) was subjected to reversed phase (RP) cartridge (10 G, 60 mL Giga tube), and eluted with MeCN-H2O to afford 30 fractions. The combined fractions 20–21 (102 mg; eluted by 60–65% MeCN-H2O) were subjected to centrifugal preparative thin layer chromatograph (CPTLC, 1 mm Si gel P254 disc), eluting with 0.5–1% MeCN-DCM to yield 12 (11.4 mg). Fraction 22 (60 mg; eluted with 75% MeCN-H2O) was further subjected to CPTLC (1 mm RP-C18 ChromatoRotor), eluted with 50–100% H2O-CH3CN to afford 115 fractions, of which fractions 42–45 and 90–115 eluting with 80% and 90% MeCN-H2O yielded 10 (2.7 mg) and 11 (4.5 mg), respectively. Combined fractions 46–89 (28.4 mg) were enriched with 12 (+ traces of 10 + 11). Similarly, RP cartridge purified fractions 23 and 24 (60 and 70 mg; eluted with 70 and 80% MeCN-H2O, respectively) were further purified (1 mm RP-C18 ChromatoRotor) by eluting separately with 50–100% H2O-MeCN to yield a mixture of 8 + 10 + 11 (32 mg and 31 mg respectively). The above enriched mixtures were further purified preparative RP-HPLC, using 90% MeCN-H2O as solvent to afford compounds 12, 11 + 12, 11, and 8 (5, 32, 4 and 31.7 mg, respectively).
A portion of n-hexane extract (2.5 g) was fractionated with CPTLC (6 mm, Si gel P254 disc) eluting with 5% DCM in hexane to yield 10 fractions. The fractions 3–7 (840 mg) that enriched with compounds 6 and 7 were combined and further attempted to purify with an additional CPTLC (4 mm, Si gel P254 disc) eluting with 5% DCM in hexane to yield semi-pure 6 (40 mg), 6+7 (50 mg), and semi-pure 7 (6 mg). In addition, the presence of these compounds in leaves, stem bark, and root extracts were confirmed by HPLC and LC–MS (vide infra).
3.4. Machaerifurogerol (1a) and 5-epi-Machaerifurogerol (1b)
Amorphous solid; [α]26D +5.8 (c 0.05, MeOH); IR (KBr) υ max 3341 (OH), 2924, 1631, 1574, 1452, 1248, 961, 801, 613, 591 cm−1; 1H and 13C NMR, see Table 1; HRESIMS m/z 379.1906 [M + H]+ (calcd. for C24H27O4, 379.1865).
3.5. Identification of Compounds 6–8 and 10–12 by LC–MS
LC–MS analysis was carried out on an Agilent system using Luna 5 μ C18 (2), 150 × 4.6 mm, λ 254, flow 1 mL/min, gradient 0–2 min [95% H2O; 5% MeCN], 2–30 min, 5% MeCN→ 100% MeCN, 30–35 min 100% MeCN, 35–45 min [95% H2O; 5% MeCN]. The retention times (Rt) of the compounds 6 (m/z 349.2 [M + H]+; C24H29O2), 8 (365.2 [M + H]+; C24H29O3), 7 (363.2 [M + H]+; C24H27O3), 10 (349.2 [M + H]+; C24H29O2); 11 (363.2 [M + H]+; C24H27O3), and 12 (365.2 [M + H]+; C24H29O2) were found to be 4.4, 4.5, 9.9, 10.0, 9.4, and 9.7 min−1, respectively. Compounds 6–9 and 10–12 were identified from leaves, stem bark, and root extracts through HPLC and LC–MS.
3.6. Antimicrobial Assays
All organisms were obtained from the American Type Culture Collection (Manassas, VA, USA) or the National Collection of Type Cultures (Colindale, UK), unless specified otherwise. These included the yeasts Candida albicans ATCC 90028, C. glabrata ATCC 90030, and C. krusei ATCC 6258; the fungi Cryptococcus neoformans ATCC 90113 and Aspergillus fumigatus ATCC 204305; and the bacteria Escherichia coli ATCC 35218, NCTC 12923, Klebsiella pneumoniae NCTC 13368, M6 (Colindale, UK), Acinetobacter baumannii AYE (ATCC BAA-1710), ATCC 17978, Pseudomonas aeruginosa ATCC 27853, PAO1 (Manoil collection, University of Washington, Washington, DC, USA), NCTC 13437, Mycobacterium intracellulare ATCC 23068, methicillin-resistant Staphylococcus aureus ATCC 33591 (MRSa), USA-300 MRSa (ATCC BAA-1717), USA-400 MRSa (ATCC BAA-1696), Mupirocin-resistant S. aureus (ATCC BAA-1708), Enterococcus faecium ATCC 700221 (VRE), E. faecalis ATCC 29212 (Vancomycin-sensitive) and Enterococcus faecium ATCC 51299 (Vancomycin-intermediate). Drug controls ciprofloxacin, methicillin and vancomycin (ICN Biomedicals, Aurora, OH, USA) for bacteria and amphotericin B (ICN Biomedicals) for yeasts and fungi were included in each assay. Susceptibility testing was performed using a modified version of the CLSI (formerly NCCLS) method [16,17,18]. M. intracellulare was tested using a modified Franzblau method [18]. Samples were serially diluted in 20% DMSO/saline and transferred in duplicates to 96-well flat-bottomed microplates. Microbial inocula were prepared by correcting the OD630 of microbe suspensions in incubation broth to give final target inocula. All organisms were read at either 530 nm, using the Biotek Powerwave XS plate reader (Bio-Tek Instruments, Winooski, VT, USA) or 544ex/590em, (M. intracellulare, A. fumigatus) using the Polarstar Galaxy Plate Reader (BMG Lab Technologies, Ortenburg, Germany), prior to and after incubation. Minimum fungicidal or bactericidal concentrations were determined by removing 5 μL from each clear well, followed by transferring to agar, and incubating. The MFC/MBC was defined as the lowest test concentration that kills the organism (allows no growth on agar).
Gram-negative MICs were determined using the CLSI microbroth dilution method, modified as described previously [19]. Bacteria were added at a starting concentration of 5 × 105 cfu/mL and incubated for 20 h at 37 °C in the dark. Absorbance at OD600 was then read using the CLARIOstar plate reader (BMG Lab Technologies, Germany). The MIC was defined as the lowest concentration where visible growth could not be detected, equivalent to an OD600 of 0.1. MICs were also determined in the presence of the membrane permeabilizer, polymyxin-B-nonapeptide (PMBN) following the same method, with an additional step; after the 2-fold dilution of compound was prepared and before the bacteria were added, PMBN was added to all wells at a final concentration of 30 µg/mL. This concentration was shown to not significantly inhibit growth of the test panel.
3.7. Antimicrobial Combination Study by Checkerboard Method
The combination study of the compounds was carried out using a standard Checkerboard method [13,14]. Strains were grown on Eugon agar at 35 °C, prior to assays. Test samples were dissolved in DMSO (2 mg/mL) to the desired concentrations, and serially-diluted with 20% DMSO/saline. Samples were transferred to 96 well assay plates (10 µL) in a checkerboard layout. Inocula were prepared by suspending growth from agar in 0.9% saline, determining the OD630, and correcting in incubation broth (cation-adjusted Mueller-Hinton, Difco) to afford 5 × 105 colony forming units per mL, after addition to samples (180 µL) using standard inocula calculations. Final sample test concentrations were 1/100th the DMSO stock concentrations. The assay plates were read at 530 nm prior to and after incubation at 35 °C for 18–20 h. IC50s of each test compound were calculated using the XLfit 4.2 software (IDBS, Alameda, CA, USA) using the fit model 201. After incubation, all 96 wells were also pinned to Eugon Agar and incubated at 35 °C overnight to determine bactericidal activity. Fractional inhibitory concentrations (FICs) were calculated to evaluate possible synergy with FICS < 0.5 synergistic.
4. Conclusions
Based on our investigation carried out on the identity of the plant, and a re-examination of the voucher specimen (Rimachi # 12161) at the MOBOT, it can now be concluded that this species should be treated only as an unidentified species of Machaerium Pers., as determined by the collection information (Manuel Rimachi, Y. 12161). This appears to be the first report of macharifurogerol (1a) and its epimer 1b from a natural source. In addition, the isolation of isoflavons (2 and 3) and pterocarpans (4 and 5) from this Machaerium species (Rimachi 12161) illustrated that these isoflavonoids are typical chemotaxonomic markers of the genus Machaerium [11,12]. Machaeriols and its biogenetic precursor machaeridiols are only isolated from this species (#12161) from the genus Machaerium, which are analogous to hexahydrocannabinol (HHC) and dihydrocannabidiol base skeletons of Cannabis and its variants, in higher plants [6]. The only other bibenzyl analogue of Δ9-THC, perrottetinen, was previously reported from the liverwort Radula perrottetii [20]. It is intriguing to note that the strong MRSA and VRE inhibitory activities, together with their antiparasitic activities [5,6] of the isolated compounds of Machaerium (12161) is contributed by HHDBP machaeriols and their 5,6-seco analogs machaeridiols. The stereo-specific total synthesis of machaeriol A-D (6–9) and mechaeridiol B (12) was reported [21,22,23,24]. In addition, analogs of machaeriols and related HHC were recently synthesized, which showed anticancer activity [25]. It was anticipated that these phytocannabinoids could serve as potential template for anti-MRSA and anti-VRE lead candidates, because of their inherent inhibitory activities alone, as well as strong synergistic activity when tested in combination with machaeriol and machaeridiol. The observation of significant activity in permeabilized multidrug resistant Gram-negative pathogens, also offers the potential for optimization of the chemical scaffold to generate analogues with better cell permeability. These compounds might provide important new leads for WHO priority Gram-negative bacterial pathogens.
Supplementary Materials
The following are available online, NMR and HRMS spectra (Figures S1–S11) of compound 1, and Table for NMR data (Table S1) of compounds 2–5 are provided in supporting information.
Author Contributions
I.M., J.M.S., and K.M.R., conceptualized the study; I.M., M.A.I., M.K., and J.Z., planned the experiments and spectral analysis for chemistry work; M.W., execute LCMS work; M.R.J., planned antimicrobial and Checkerboard assay at UM; J.M.S., C.H., M.C., B.M., and T.A.-A., executed antimicrobial work at KCL and PHE; V.R., and I.M., authenticated and provided critical information on plant authentication; M.A.I., M.R.J., M.A.I., V.R., M.W., J.Z., J.M.S., and K.M.R., prepared the original draft of the manuscript. All the authors contributed to the writing and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the USDA Agricultural Research Service Specific Cooperative Agreement No. 58-6060-6-015, and the NIH, NIAID, Division of AIDS, grant no. AI 27094. Work at PHE was supported by Grant in aid funding through an Open Innovation programme (Project 111742).
Acknowledgments
The authors sincerely thank Manuel Rimachi Y. and (Late) Sydney T. McDaniel (Mississippi State University) for the collection of plant material; Andrew Townesmith, Missouri Botanical Garden, St. Louis, MO, USA; Andrew Sanders, UCR Herbarium, California, USA; and Fabiana Filardi, Instituto de Pesquisas Jardim Botânico do Rio de Janeiro, Brazil, for their expert opinion on the voucher specimen. Marwa Hasan, Y. Wang, F. Wiggers, and M. Wright, NCNPR, UM, for assisting and conducting chemistry work, HRMS, NMR experiments and biological assays, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mabberley, D.J. Mabberley’s Plant-Book: A Portable Dictionary of Plants, Their Classification and Uses; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Amen, Y.M.; Marzouk, A.M.; Zaghloul, M.G.; Afifi, M.S. The genus Machaerium (Fabaceae): Taxonomy, phytochemistry, traditional uses and biological activities. Natl. Prod. Res. 2015, 29, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Joly, L.G.; Guerra, S.; Septimo, R.; Solis, P.N.; Correa, M.; Gupta, M.; Levy, S.; Sandberg, F. Ethnobotanical inventory of medicinal plants used by the Guaymi Indians in Western Panama. Part I. J. Ethnopharmacol. 1987, 20, 145–171. [Google Scholar] [CrossRef]
- Polhill, R.M.; Raven, P.H. Advances in Legume Systematics, Part 1; Royal Botanic Gardens: Kew, UK, 1981. [Google Scholar]
- Muhammad, I.; Li, X.-C.; Dunbar, D.C.; ElSohly, M.A.; Khan, I.A. Antimalarial (+)-trans-hexahydrodibenzopyran derivatives from Machaerium multiflorum. J. Nat. Prod. 2001, 64, 1322–1325. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, I.; Li, X.-C.; Jacob, M.R.; Tekwani, B.L.; Dunbar, D.C.; Ferreira, D. Antimicrobial and Antiparasitic (+)-trans-Hexahydrodibenzopyrans and Analogues from Machaerium multiflorum. J. Nat. Prod. 2003, 66, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Filardi, F.L.R.; Lima, H.C.D. The diversity of Machaerium (Leguminosae: Papilionoideae) in the Atlantic Forest: Three new species, nomenclatural updates, and a revised key. Syst. Bot. 2014, 39, 145–159. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef]
- Salgado, C.D. The risk of developing a vancomycin-resistant Enterococcus bloodstream infection for colonized patients. Am. J. Infect. Control 2008, 36, S175. [Google Scholar] [CrossRef]
- Lozano, P.; Klitgaard, B.B. The genus Machaerium (Leguminosae: Papilionoideae: Dalbergieae) in Ecuador. Brittonia 2006, 58, 124–150. [Google Scholar] [CrossRef]
- Piccinelli, A.L.; Campo Fernandez, M.; Cuesta-Rubio, O.; Márquez Hernández, I.; De Simone, F.; Rastrelli, L. Isoflavonoids isolated from Cuban propolis. J. Agric. Food Chem. 2005, 53, 9010–9016. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, K.; Ollis, W.D.; Redman, B.T.; Sutherland, I.O.; Gottlieb, O.R. Vestitol and vesticarpan, isoflavonoids from Machaerium vestitum. Phytochemistry 1978, 17, 1413–1415. [Google Scholar] [CrossRef]
- Norden, C.W.; Wentzel, H.; Keleti, E. Comparison of techniques for measurement of in vitro antibiotic synergism. J. Infect. Dis. 1979, 140, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Renneberg, J. Definitions of antibacterial interactions in animal infection models. J. Antimicrob. Chemother. 1993, 31 (Suppl. D), 167–175. [Google Scholar] [CrossRef] [PubMed]
- Ilias, M.; Samoylenko, V.; Gillium, V.D. Preparation of Pre-Coated Rp-Rotors and Universal Chromatorotors, Chromatographic Separation Devices and Methods for Centrifugal Preparative Chromatography. Google Patents US20140224740A1, 14 August 2014. [Google Scholar]
- Wikler, M.A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard. CLSI (NCCLS) 2006, 26, M7–A7. [Google Scholar]
- Michael, A.; Vishnu, C.; Ana, E.; Mahmoud, A.; Linda, L.; Frank, C. NCCLS, Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard, M27-A2; CLSI: Wayne, PA, USA, 2002. [Google Scholar]
- NCCLS. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes; Approved Standard; NCCLS: Wayne, PA, USA, 2003. [Google Scholar]
- Bock, L.; Hind, C.; Sutton, J.; Wand, M. Growth media and assay plate material can impact on the effectiveness of cationic biocides and antibiotics against different bacterial species. Lett. Appl. Microbiol. 2018, 66, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Shimamura, T.; Ishii, H.; Renner, M.; Braggins, J.; Asakawa, Y. New bibenzyl cannabinoid from the New Zealand liverwort Radula marginata. Chem. Pharm. Bull. 2002, 50, 1390–1392. [Google Scholar] [CrossRef] [PubMed]
- Chittiboyina, A.G.; Reddy, C.R.; Watkins, E.B.; Avery, M.A. First synthesis of antimalarial Machaeriols A and B. Tetrahedron Lett. 2004, 45, 1689–1691. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Q.; Zheng, J.; Zhang, J.; Pan, X.; She, X. A general route to 5, 6-seco-hexahydrodibenzopyrans and analogues: First total synthesis of (+)-Machaeridiol B and (+)-Machaeriol B. Tetrahedron 2007, 63, 1014–1021. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Q.; Chen, B.; Lu, J.; Wang, H.; She, X.; Pan, X. Total Synthesis of (+)-Machaeriol D with a Key Regio-and Stereoselective SN2′ Reaction. Angew. Chem. Int. Ed. 2006, 45, 3651–3653. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Lee, Y.R. A short total synthesis for biologically interesting (+)-and (−)-machaeriol A. Synlett 2008, 2008, 1643–1646. [Google Scholar]
- Thapa, D.; Lee, J.S.; Heo, S.W.; Lee, Y.R.; Kang, K.W.; Kwak, M.K.; Choi, H.G.; Kim, J.A. Novel hexahydrocannabinol analogs as potential anti-cancer agents inhibit cell proliferation and tumor angiogenesis. Eur. J. Pharmacol. 2011, 650, 64–71. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–8 and 10–12 are available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).