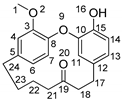
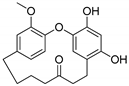
Galeon: A Biologically Active Molecule with In Silico Metabolite Prediction, In Vitro Metabolic Profiling in Rat Liver Microsomes, and In Silico Binding Mechanisms with CYP450 Isoforms
Abstract
1. Introduction
2. Results and Discussion
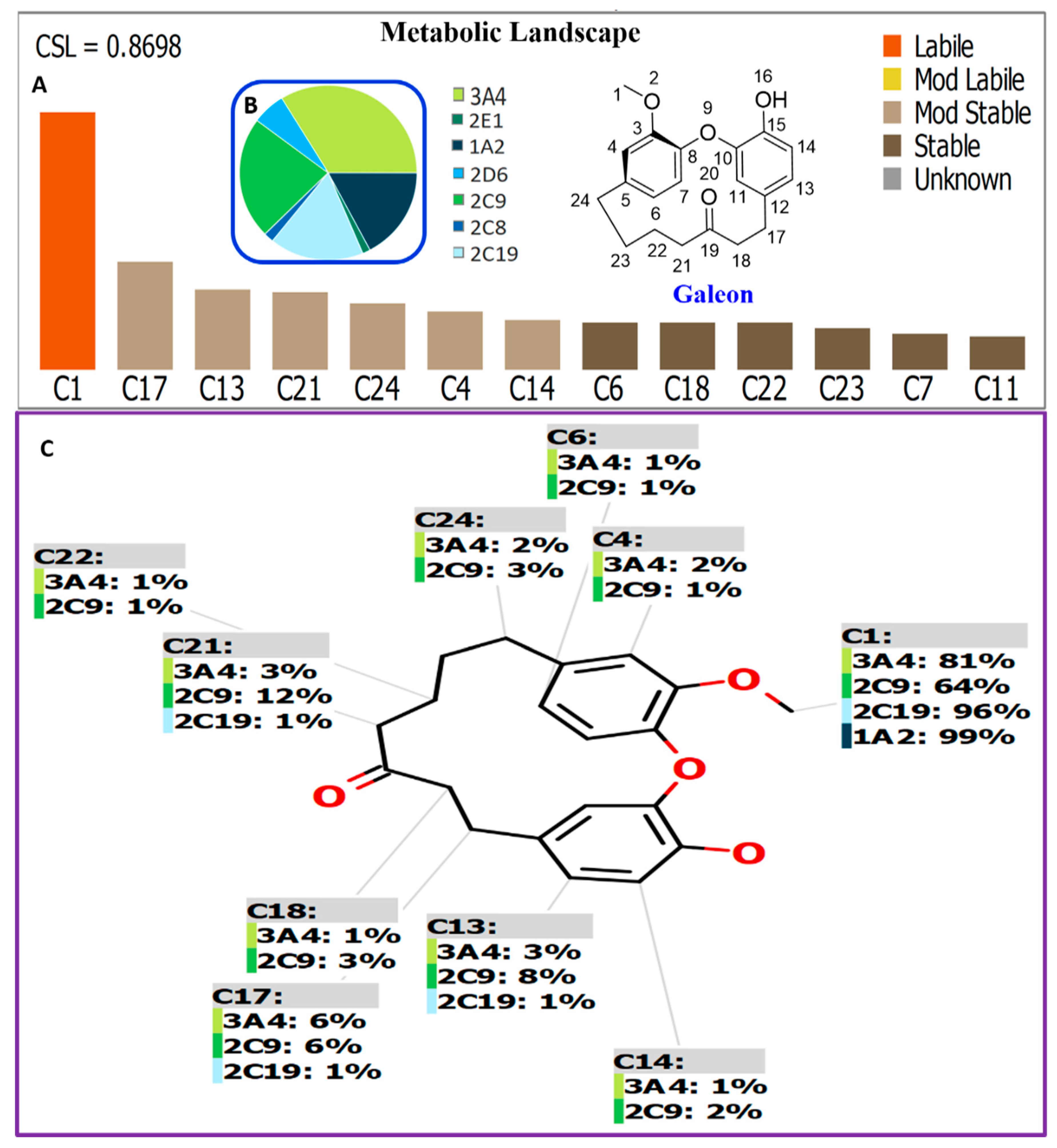
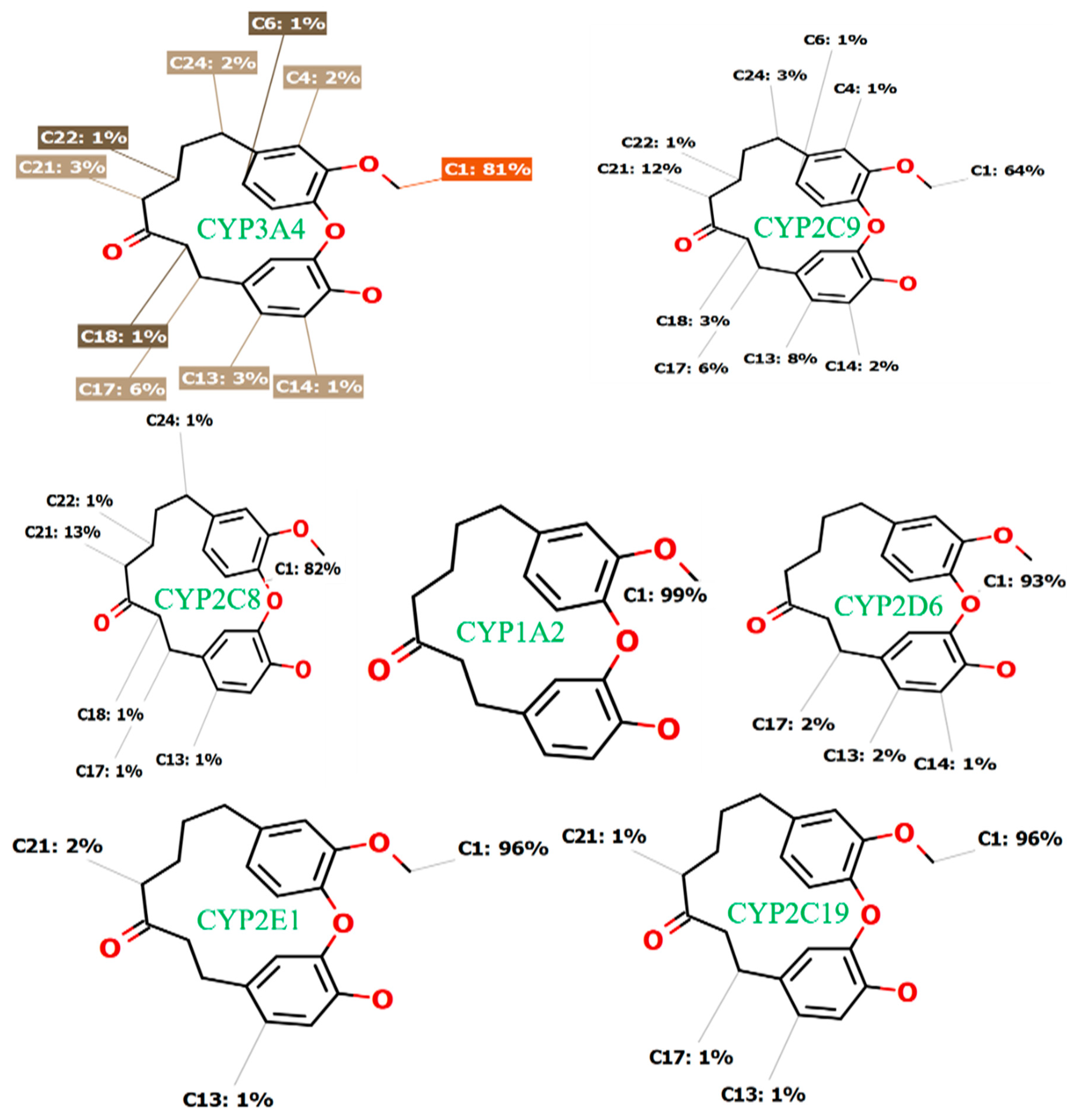
2.1. In Silico Prediction of Galeon Metabolites Using the CYP450 Metabolism Module of the StarDrop® Software
2.2. In Silico Galeon Structural Alert Sites and Toxicity Prediction
2.3. In Vitro Metabolic Profiling of Galeon
2.3.1. Mass Spectrometry of Galeon
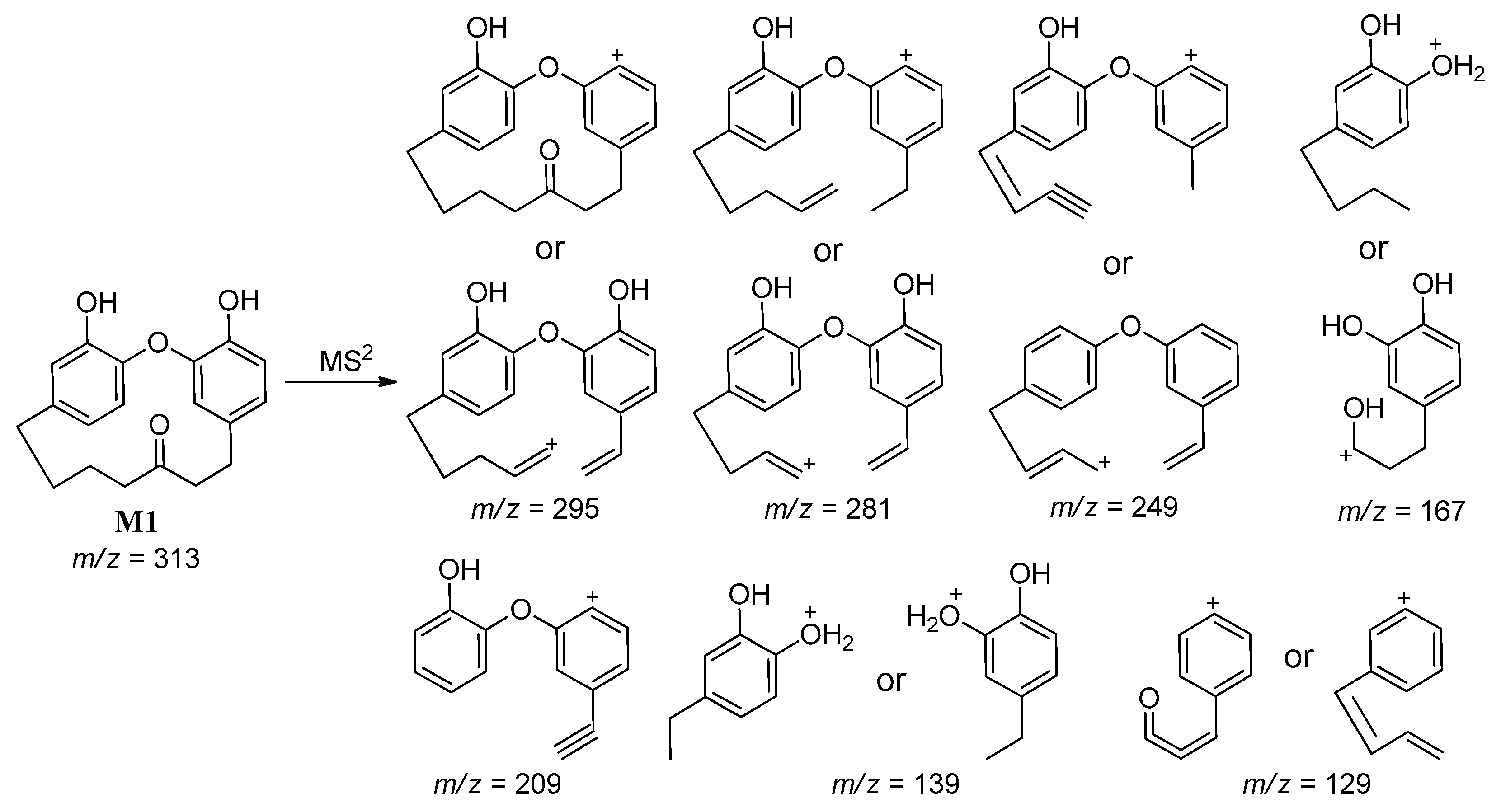
2.3.2. Identification of Phase-I Metabolites
2.3.3. Identification of Reactive Metabolites
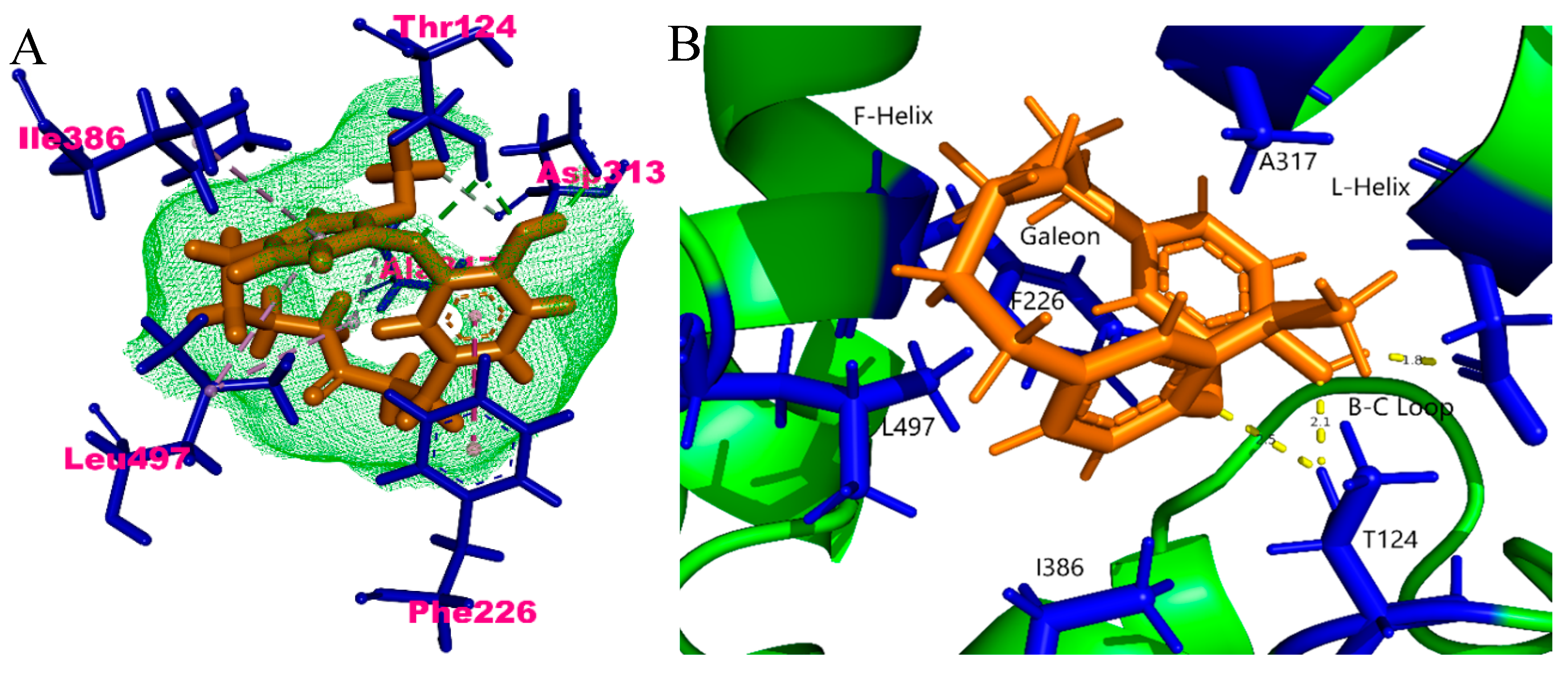
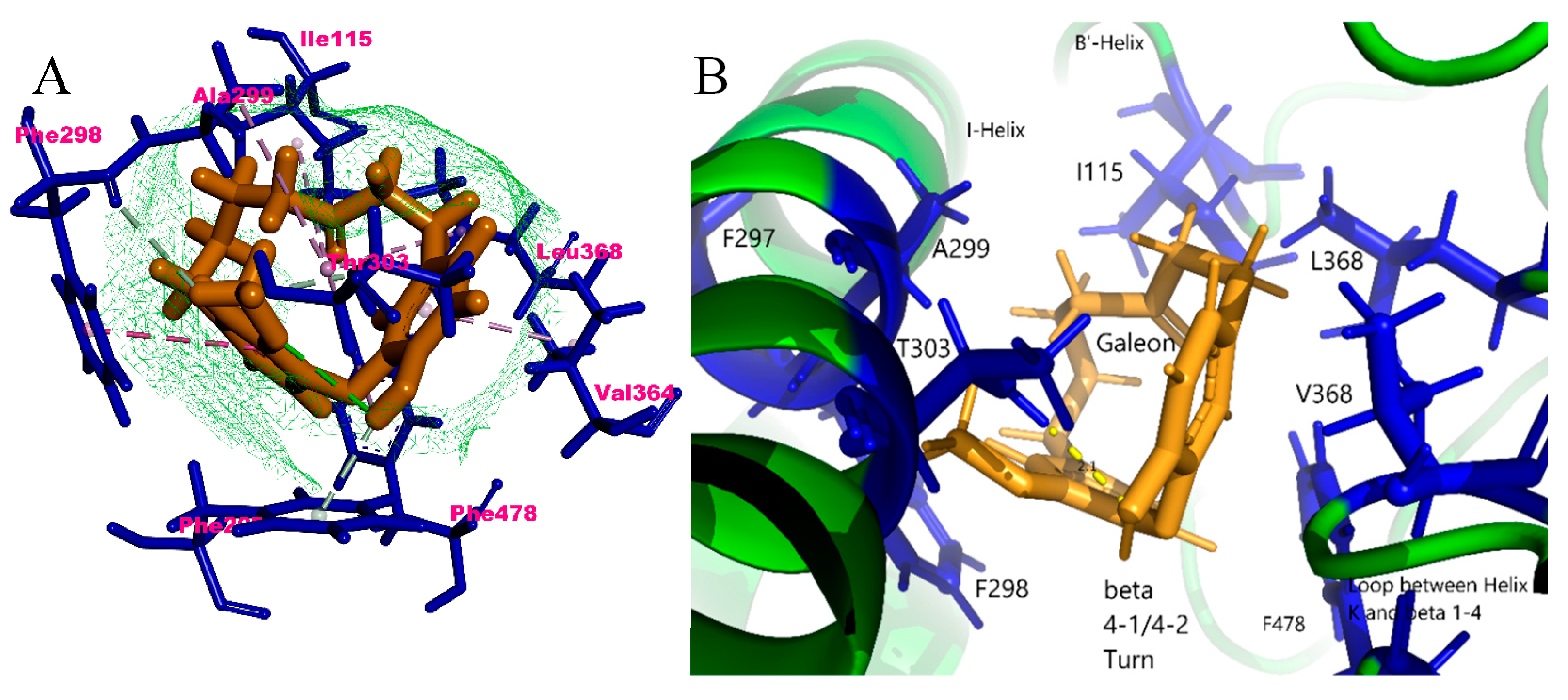
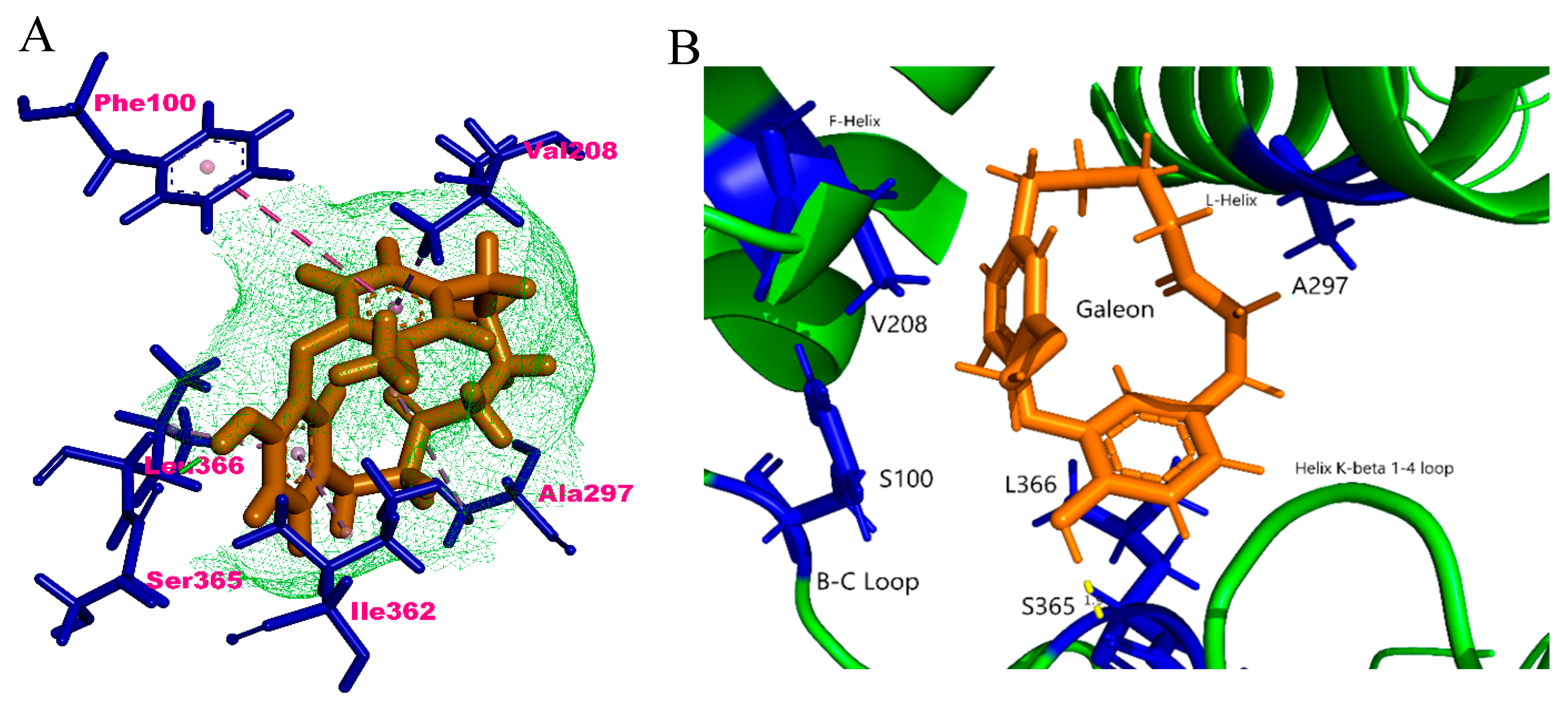
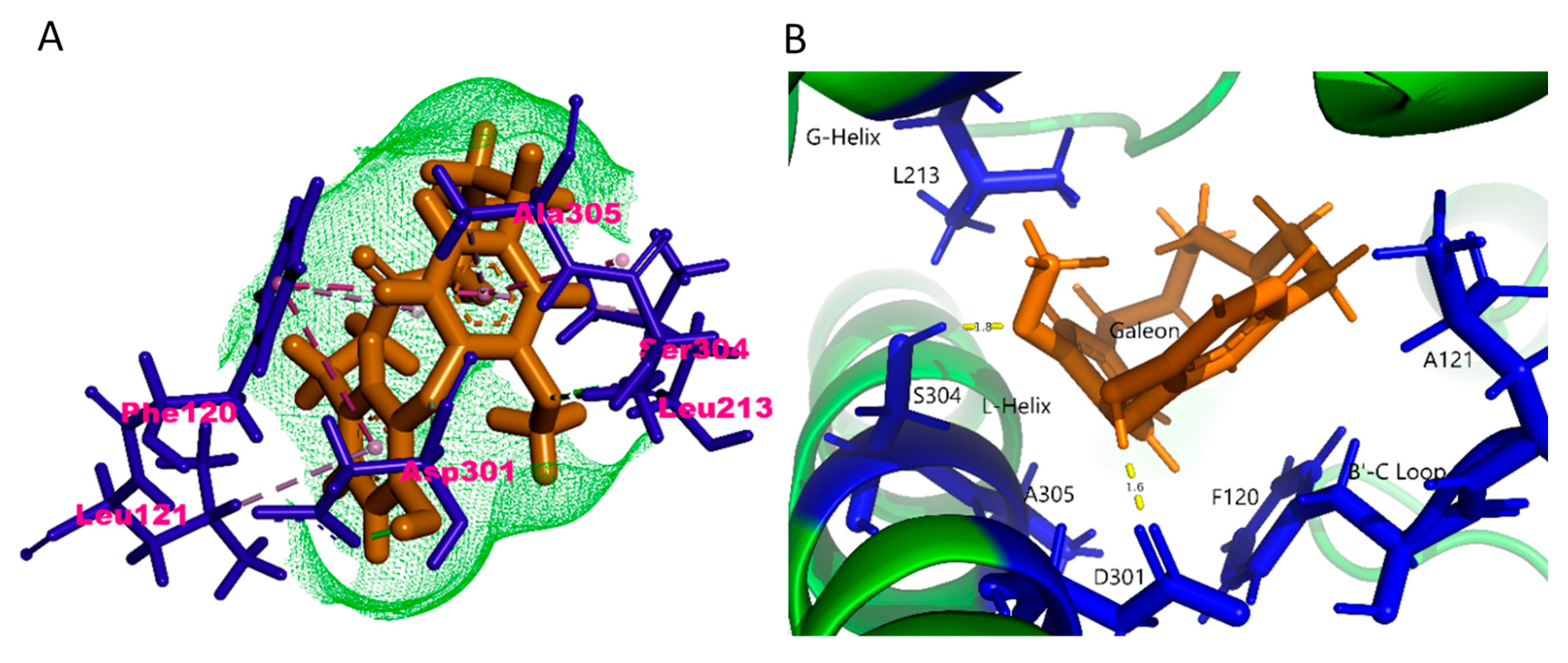
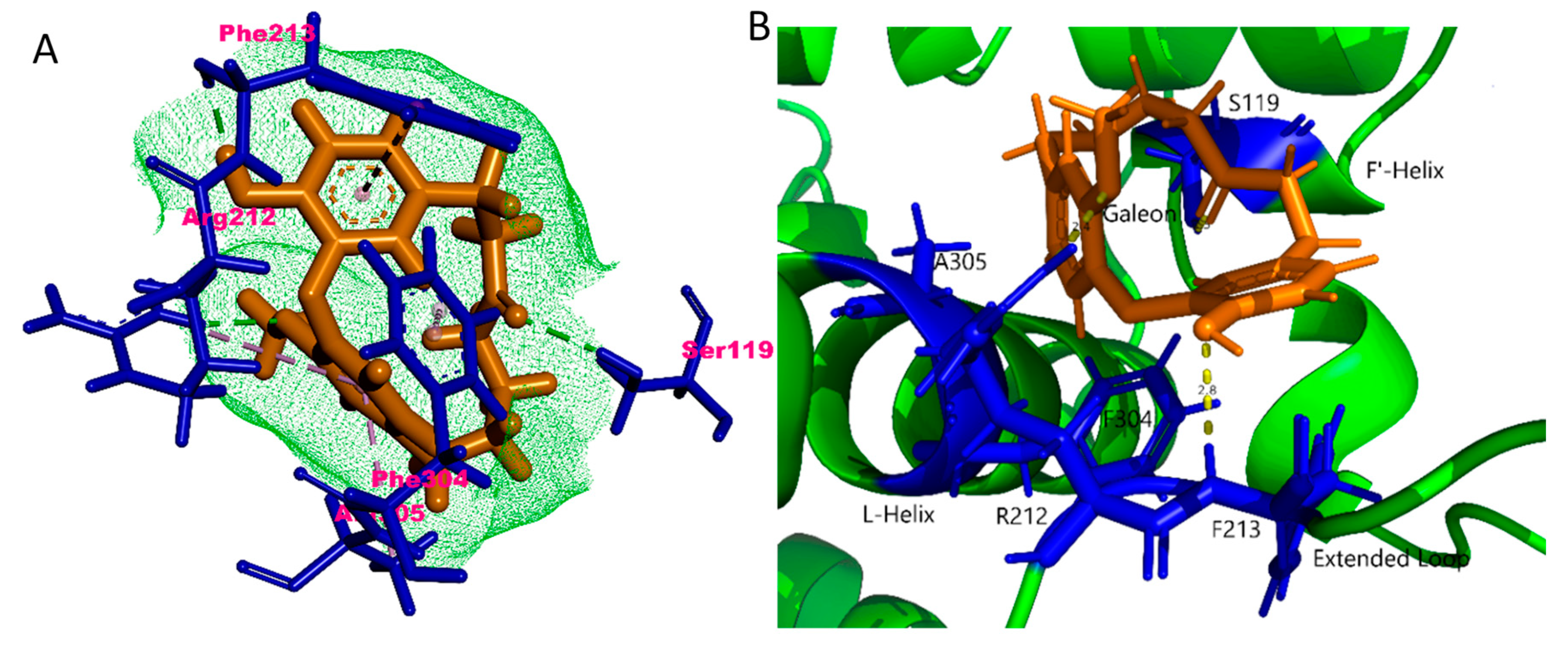
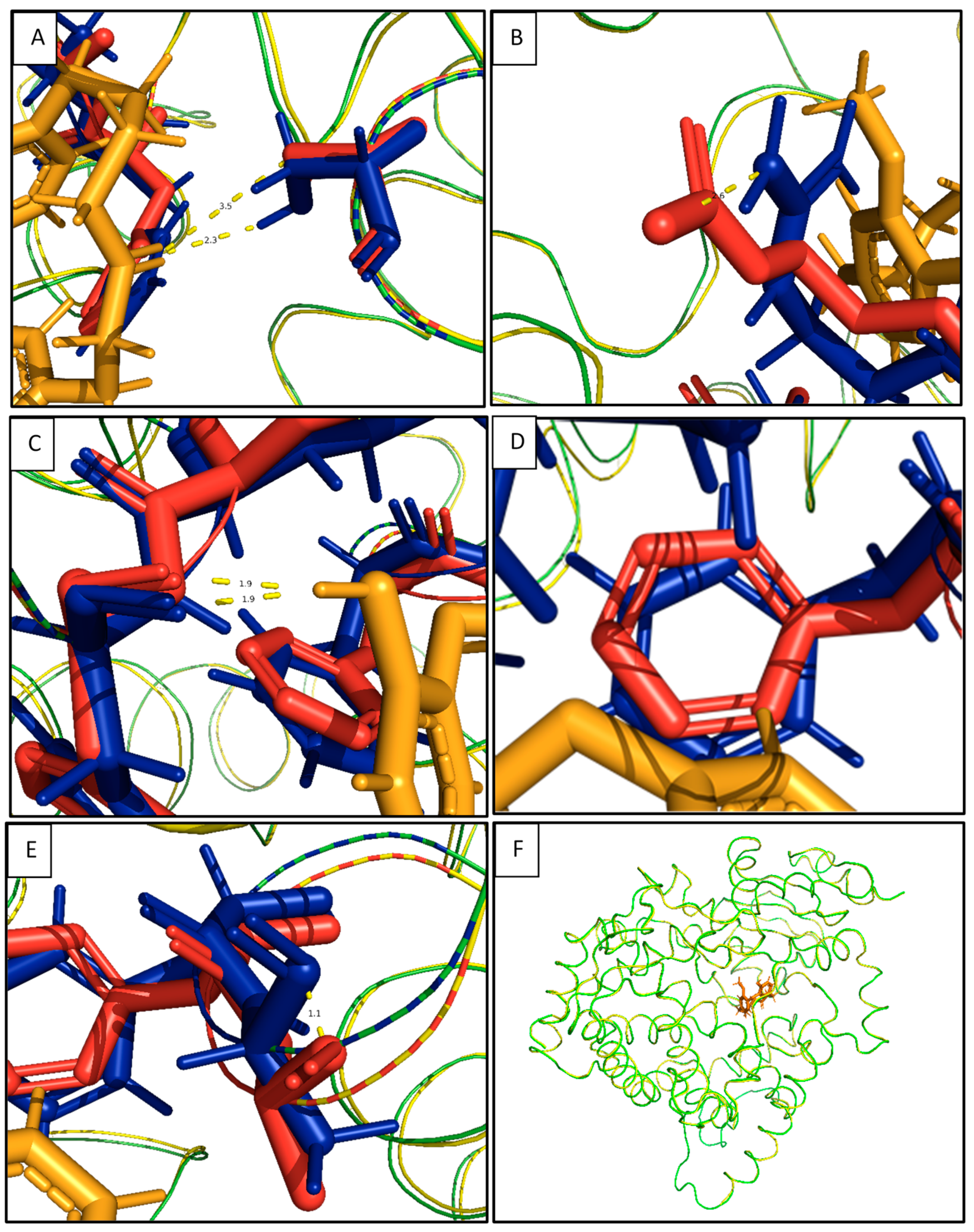
2.4. In Silico Binding Mechanism of Galeon with Cytochrome P450 Isoforms
3. Materials and Methods
3.1. In Silico Study
3.2. Chemicals, Reagents, and Instrumentation
3.3. RLMs Incubations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Attwa, M.W.; Darwish, H.W.; Alhazmi, H.A.; Kadi, A.A. Investigation of metabolic degradation of new ALK inhibitor: Entrectinib by LC-MS/MS. Clin. Chim. Acta 2018, 485, 298–304. [Google Scholar] [CrossRef]
- Attwa, M.W.; Kadi, A.A.; Darwish, H.W.; Abdelhameed, A.S. Investigation of the metabolic stability of olmutinib by validated LC-MS/MS: Quantification in human plasma. RSC Adv. 2018, 8, 40387–40394. [Google Scholar] [CrossRef]
- Attia, S.M. Deleterious effects of reactive metabolites. Oxid. Med. Cell Longev. 2010, 3, 238–253. [Google Scholar] [CrossRef]
- Yang, L.; Hyunji, J.; Md, A.; Hwa-Jong, L.; Wencui, Y.; Mohammad, S.A.; Moinul, K.; Adnan, A.K.; Yurngdong, J.; Youngjoo, K.; et al. Synthesis, Biological Evaluation and Molecular Docking Study of Cyclic Diarylheptanoids as Potential Anticancer Therapeutics. Anti-Cancer Agents Med. Chem. 2020, 20, 464–475. [Google Scholar] [CrossRef]
- Kadi, A.A.; Yin, W.; Rahman, A.M. In-vitro metabolic profiling study of potential topoisomerase inhibitors ‘pyrazolines’ in RLMs by mass spectrometry. J. Chromatogr. B 2019, 1114, 125–133. [Google Scholar] [CrossRef]
- Rahman, A.M.; Lu, Y.; Lee, H.-J.; Jo, H.; Yin, W.; Alam, M.S.; Cha, H.; Kadi, A.A.; Kwon, Y.; Jahng, Y. Linear diarylheptanoids as potential anticancer therapeutics: Synthesis, biological evaluation, and structure–activity relationship studies. Arch. Pharmacal Res. 2018, 41, 1131–1148. [Google Scholar] [CrossRef]
- Islam, M.S.; Park, S.; Song, C.; Kadi, A.A.; Kwon, Y.; Rahman, A.M. Fluorescein hydrazones: A series of novel non-intercalative topoisomerase IIα catalytic inhibitors induce G1 arrest and apoptosis in breast and colon cancer cells. Eur. J. Med. Chem. 2017, 125, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Kadi, A.A.; Al-Shakliah, N.S.; Yin, W.; Rahman, A.M. In vitro investigation of metabolic profiling of newly developed topoisomerase inhibitors (ethyl fluorescein hydrazones, EtFLHs) in RLMs by LC–MS/MS. J. Chromatogr. B 2017, 1054, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Woo, H.; Jun, K.-Y.; Kadi, A.A.; Abdel-Aziz, H.A.; Kwon, Y.; Rahman, A.M. Design, synthesis, topoisomerase I & II inhibitory activity, antiproliferative activity, and structure–activity relationship study of pyrazoline derivatives: An ATP-competitive human topoisomerase IIα catalytic inhibitor. Bioorg. Med. Chem. 2016, 24, 1898–1908. [Google Scholar] [PubMed]
- Rahman, A.F.M.M.; Park, S.-E.; Kadi, A.A.; Kwon, Y. Fluorescein hydrazones as novel nonintercalative topoisomerase catalytic inhibitors with low DNA toxicity. J. Med. Chem. 2014, 57, 9139–9151. [Google Scholar] [CrossRef]
- Lee, K.-S.; Li, G.; Kim, S.H.; Lee, C.-S.; Woo, M.-H.; Lee, S.-H.; Jhang, Y.-D.; Son, J.-K. Cytotoxic diarylheptanoids from the roots of Juglans mandshurica. J. Nat. Prod. 2002, 65, 1707–1708. [Google Scholar] [CrossRef] [PubMed]
- Morihara, M.; Sakurai, N.; Inoue, T.; Kawai, K.-I.; Nagai, M. Two novel diarylheptanoid glucosides from Myrica gale var. tomentosa and absolute structure of plane-chiral galeon. Chem. Pharm. Bull. 1997, 45, 820–823. [Google Scholar] [CrossRef]
- Malterud, K.E.; Anthonsen, T.; Hjortas, J. 14-Oxa-[7.1]-metapara-cyclophanes from Myrica gale L., a new class of natural products. Tetrahedron Lett 1976, 3069–3072. [Google Scholar] [CrossRef]
- Rahman, A.M.; Al-Shakliah, N.S.; Yin, W.; Kadi, A.A. In vitro Investigation of Metabolic Profiling of a Potent Topoisomerase Inhibitors Fluorescein Hydrazones (FLHs) in RLMs by LC-MS/MS. J. Chromatogr. B 2017, 1054, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Peng, X.; Ruan, H. Diarylheptanoids from the fresh pericarps of Juglans hopeiensis. Fitoterapia 2019, 136, 104165. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Peng, X.; Zhou, J.; Zhou, M.; Ruan, H. Diarylheptanoids from the fresh pericarps of Juglans sigillata. Nat. Product Res. 2018, 32, 2457–2463. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Choi, S.Y.; Jeong, B.; Min, H.J.; Yang, H.; Bae, Y.S. Chemical constituents of branches and barks of Juglans mandshurica. Chem. Nat. Compd. 2018, 54, 342–343. [Google Scholar] [CrossRef]
- Ting, Y.-C.; Wang, H.-C.; Ko, H.-H.; Peng, C.-F.; Chang, H.-S.; Hsieh, P.-C.; Chen, I.-S. Biological evaluation of secondary metabolites from the roots of Myrica adenophora. Phytochemistry 2014, 103, 89–98. [Google Scholar] [CrossRef]
- Li, J.; Sun, J.-X.; Yu, H.-Y.; Chen, Z.-Y.; Zhao, X.-Y.; Ruan, H.-L. Diarylheptanoids from the root bark of Juglans cathayensis. Chin. Chem. Lett. 2013, 24, 521–523. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Wang, D.; Niu, F. Studies on constituents from pericarps of Juglans mandshurica with anti-tumor activity. Chin. Tradit. Herbal Drugs 2010, 41, 11–14. [Google Scholar]
- Li, Y.-X.; Ruan, H.-L.; Zhou, X.-F.; Zhang, Y.-H.; Pi, H.-F.; Wu, J.-Z. Cytotoxic diarylheptanoids from pericarps of Juglans cathayensis Dode. Chem. Res. Chin. Univ. 2008, 24, 427–429. [Google Scholar] [CrossRef]
- Inoue, T. Constituents of Acer nikoense and Myrica rubra. On Diarylheptanoids. Yakugaku Zasshi 1993, 113, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Ishida, J.; Kozuka, M.; Tokuda, H.; Nishino, H.; Nagumo, S.; Lee, K.-H.; Nagai, M. Chemopreventive potential of cyclic diarylheptanoids. Bioorg. Med. Chem. 2002, 10, 3361–3365. [Google Scholar] [CrossRef]
- Ding, Q.; Wang, Q.; He, H.; Cai, Q.; Wang, Q.; He, H. Asymmetric Synthesis of (-)-Pterocarine and (-)-Galeon via Chiral Phase Transfer-Catalyzed Atropselective Formation of Diarylether Cyclophane Skeleton. Org. Lett. 2017, 19, 1804–1807. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Zhang, C.-j.; Zhang, L.; Jiang, H. Total synthesis of macrocyclic diarylheptanoids galeon from natural products. Zhongyiyao Xuebao 2015, 43, 12–14. [Google Scholar]
- Salih, M.Q.; Beaudry, C.M. Chirality in diarylether heptanoids: Synthesis of myricatomentogenin, jugcathanin, and congeners. Org. Lett. 2012, 14, 4026–4029. [Google Scholar] [CrossRef]
- Pitsinos, E.N.; Vidali, V.P.; Couladouros, E.A. Diaryl Ether Formation in the Synthesis of Natural Products. Eur. J. Org. Chem. 2011, 1207–1222. [Google Scholar] [CrossRef]
- Jeong, B.-S.; Wang, Q.; Son, J.-K.; Jahng, Y. A versatile synthesis of cyclic diphenyl ether-type diarylheptanoids: Acerogenins, (±)-galeon, and (±)-pterocarine. Eur. J. Med. Chem. 2007, 1338–1344. [Google Scholar] [CrossRef]
- Al-Shakliah, N.S.; Attwa, M.W.; Kadi, A.A.; AlRabiah, H. Identification and characterization of in silico, in vivo, in vitro, and reactive metabolites of infigratinib using LC-ITMS: Bioactivation pathway elucidation and in silico toxicity studies of its metabolites. RSC Adv. 2020, 10, 16231–16244. [Google Scholar] [CrossRef]
- Attwa, M.W.; Kadi, A.A.; Abdelhameed, A.S. Phase I metabolic profiling and unexpected reactive metabolites in human liver microsome incubations of X-376 using LC-MS/MS: Bioactivation pathway elucidation and in silico toxicity studies of its metabolites. RSC Adv. 2020, 10, 5412–5427. [Google Scholar] [CrossRef]
- Attwa, M.W.; Kadi, A.A.; Abdelhameed, A.S.; Alhazmi, H.A. Metabolic Stability Assessment of New PARP Inhibitor Talazoparib Using Validated LC-MS/MS Methodology: In silico Metabolic Vulnerability and Toxicity Studies. Drug Des. Devel. Ther. 2020, 14, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.selectscience.net/products/stardrop-5---drug-discovery-software/?prodID=105769#tab-1 (accessed on 31 October 2020).
- Available online: https://www.optibrium.com/stardrop/stardrop6.5.php (accessed on 31 October 2020).
- Shin, Y.G.; Le, H.; Khojasteh, C.; Hop, C.E. Comparison of metabolic soft spot predictions of CYP3A4, CYP2C9 and CYP2D6 substrates using MetaSite and StarDrop. Comb. Chem. High Throughput Screen 2011, 14, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Segall, M.; Champness, E.; Obrezanova, O.; Leeding, C. Beyond profiling: Using ADMET models to guide decisions. Chem. Biodivers 2009, 6, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Marchant, C.A.; Briggs, K.A.; Long, A. In Silico Tools for Sharing Data and Knowledge on Toxicity and Metabolism: Derek for Windows, Meteor, and Vitic. Toxicol. Mech. Methods 2008, 18, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Hamza, A.; Zannat, S.S.; Fahad, R.; Rahman, A.; Hosen, S.M.Z.; Dash, R.; Hossain, M.K. Targeting galectin-3 by natural glycosides: A computational approach. Netw. Modeling Anal. Health Inform. Bioinform. 2020, 9, 14. [Google Scholar] [CrossRef]
- Sansen, S.; Yano, J.K.; Reynald, R.L.; Schoch, G.A.; Griffin, K.J.; Stout, C.D.; Johnson, E.F. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J. Biol. Chem. 2007, 282, 14348–14355. [Google Scholar] [CrossRef]
- Porubsky, P.R.; Meneely, K.M.; Scott, E.E. Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J. Biol. Chem. 2008, 283, 33698–33707. [Google Scholar] [CrossRef]
- Vaz, A.D.; McGinnity, D.F.; Coon, M.J. Epoxidation of olefins by cytochrome P450: Evidence from site-specific mutagenesis for hydroperoxo-iron as an electrophilic oxidant. Proc. Natl. Acad. Sci. USA 1998, 95, 3555–3560. [Google Scholar] [CrossRef]
- Vaz, A.D.; Pernecky, S.J.; Raner, G.M.; Coon, M.J. Peroxo-iron and oxenoid-iron species as alternative oxygenating agents in cytochrome P450-catalyzed reactions: Switching by threonine-302 to alanine mutagenesis of cytochrome P450 2B4. Proc. Natl. Acad. Sci. USA 1996, 93, 4644–4648. [Google Scholar] [CrossRef]
- Blobaum, A.L.; Kent, U.M.; Alworth, W.L.; Hollenberg, P.F. Novel reversible inactivation of cytochrome P450 2E1 T303A by tert-butyl acetylene: The role of threonine 303 in proton delivery to the active site of cytochrome P450 2E1. J. Pharmacol. Exp. Ther. 2004, 310, 281–290. [Google Scholar] [CrossRef]
- Rowland, P.; Blaney, F.E.; Smyth, M.G.; Jones, J.J.; Leydon, V.R.; Oxbrow, A.K.; Lewis, C.J.; Tennant, M.G.; Modi, S.; Eggleston, D.S.; et al. Crystal structure of human cytochrome P450 2D6. J. Biol. Chem. 2006, 281, 7614–7622. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; Cosme, J.; Vinković, D.M.; Ward, A.; Angove, H.C.; Day, P.J.; Vonrhein, C.; Tickle, I.J.; Jhoti, H. Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone. Science 2004. [Google Scholar] [CrossRef] [PubMed]
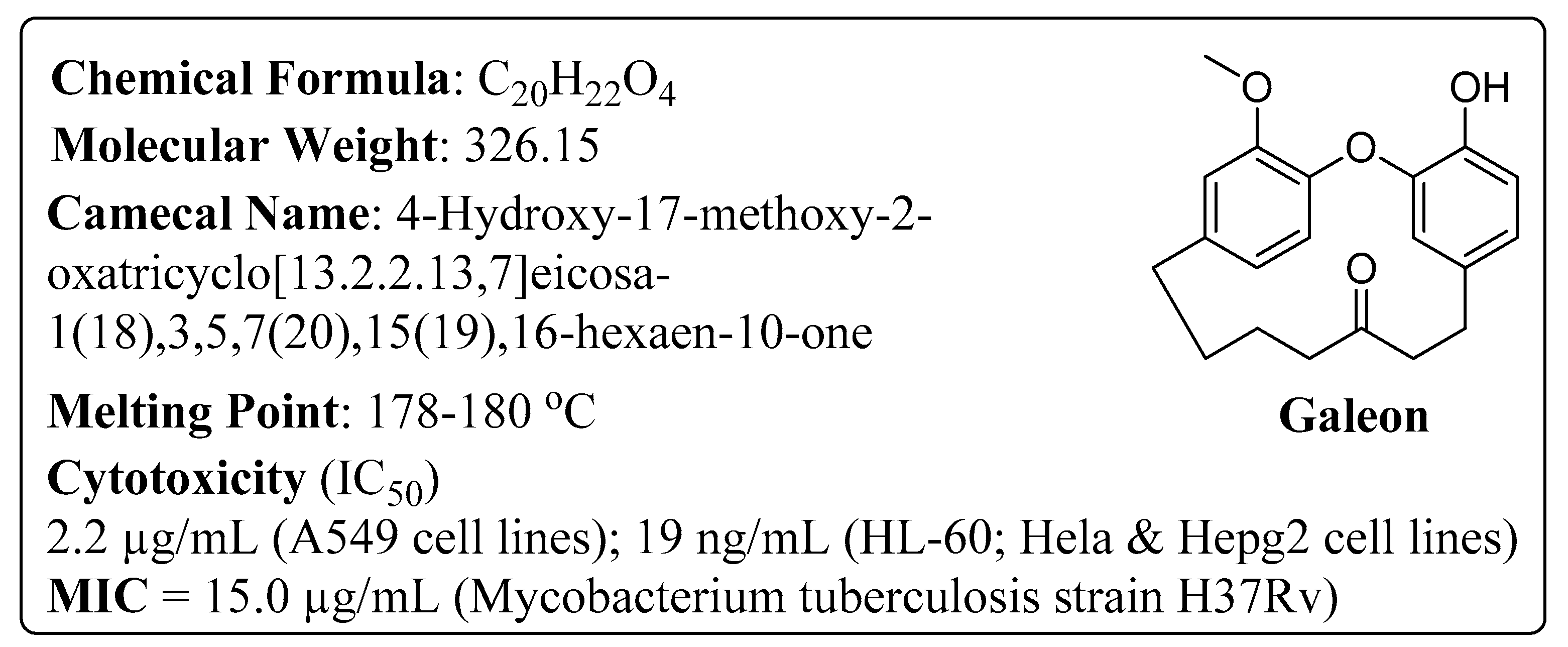





| Galeon and Its Metabolites | Skin Sensitization | Thyroid Toxicity | Chromosome Damage | Carcinogenicity | Teratogenicity, Genotoxicity, and Mutagenicity |
|---|---|---|---|---|---|
 Galeon | NA | NA | NA | NA | NA |
 Hydroxy(C11)-galeon | Plausible | NA | NA | NA | NA |
 Hydroxy(C13)-galeon | Plausible | Plausible | NA | Plausible | NA |
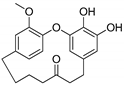 Hydroxy(C14)-galeon | Plausible | NA | Plausible | NA | NA |
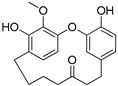 Hydroxy(C4)-galeon | Plausible | NA | NA | NA | NA |
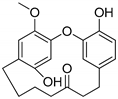 Hydroxy(C6)-galeon | Plausible | NA | NA | Plausible | NA |
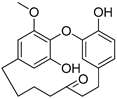 Hydroxy(C7)-galeon | Plausible | NA | NA | NA | NA |
| Metabolites | Nominal Observed Mass (m/z) | Retention Time (min) | Fragments (MS2 in m/z) | Possible Metabolic Reactions |
|---|---|---|---|---|
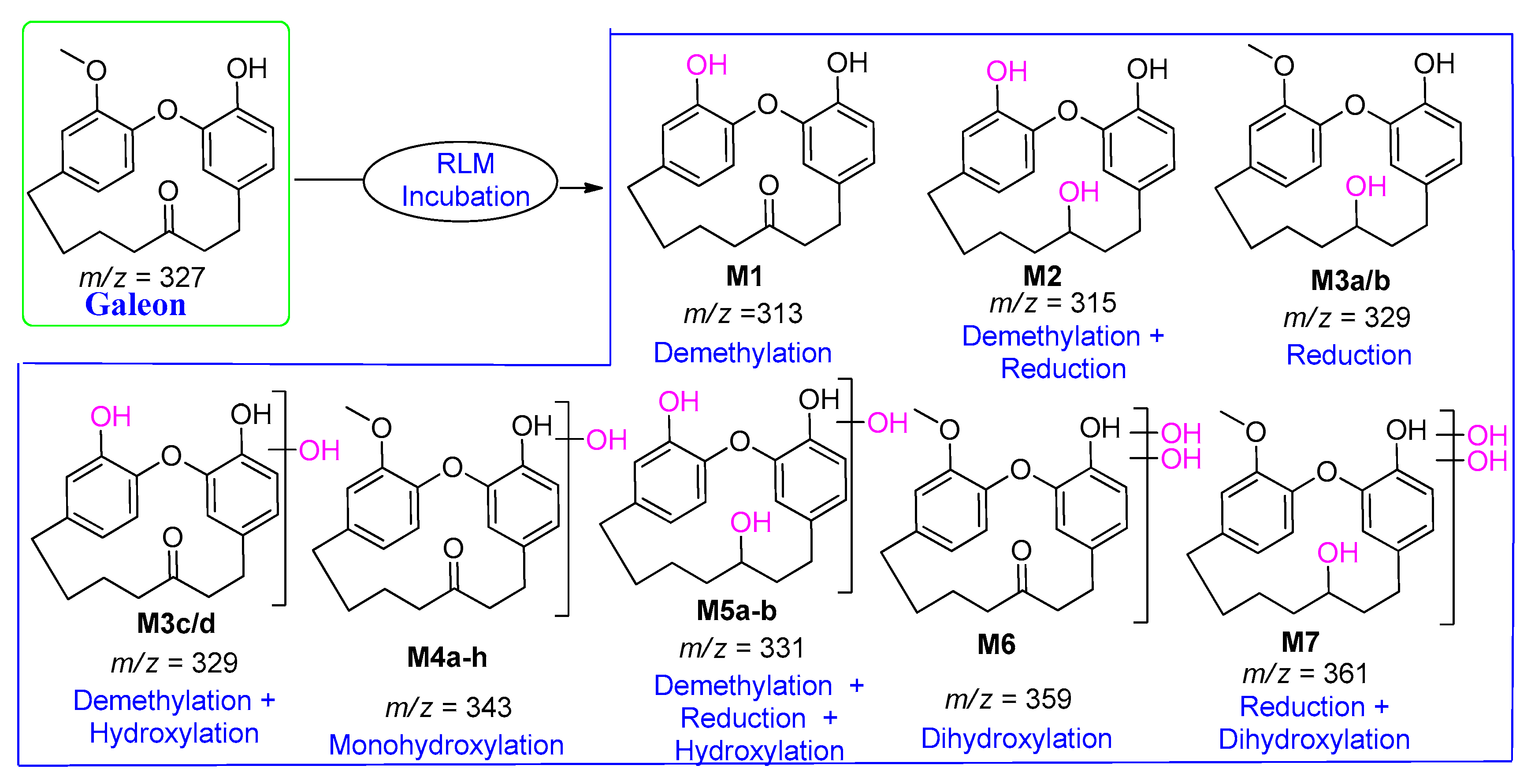
| M1 | 313 | 25.0 | 295, 281, 249, 209, 167, 139, 129 | Demethylation |
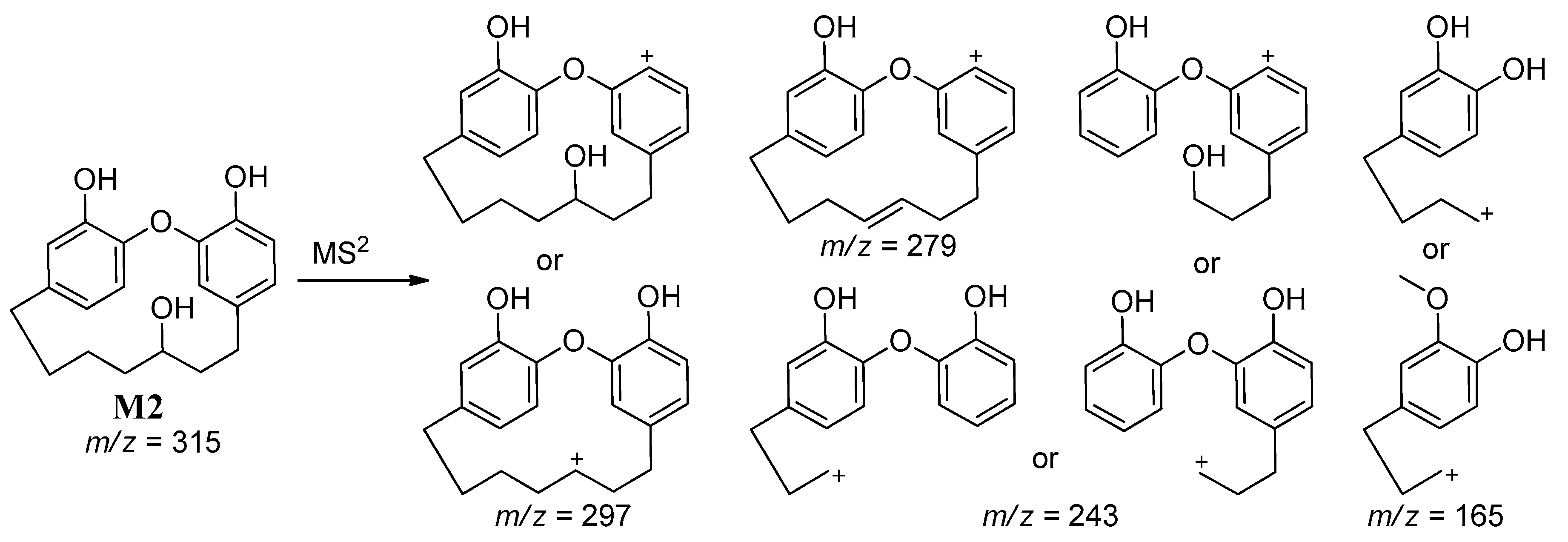
| M2 | 315 | 28.9 | 297, 279, 243, 165 | Demethylation with Reduction |
| M3a/b or M3c/d | 329 | 40.0 or 40.6 | 311, 257, 121 133, 124, 110 | Reduction or Demethylation with Hydroxylation |
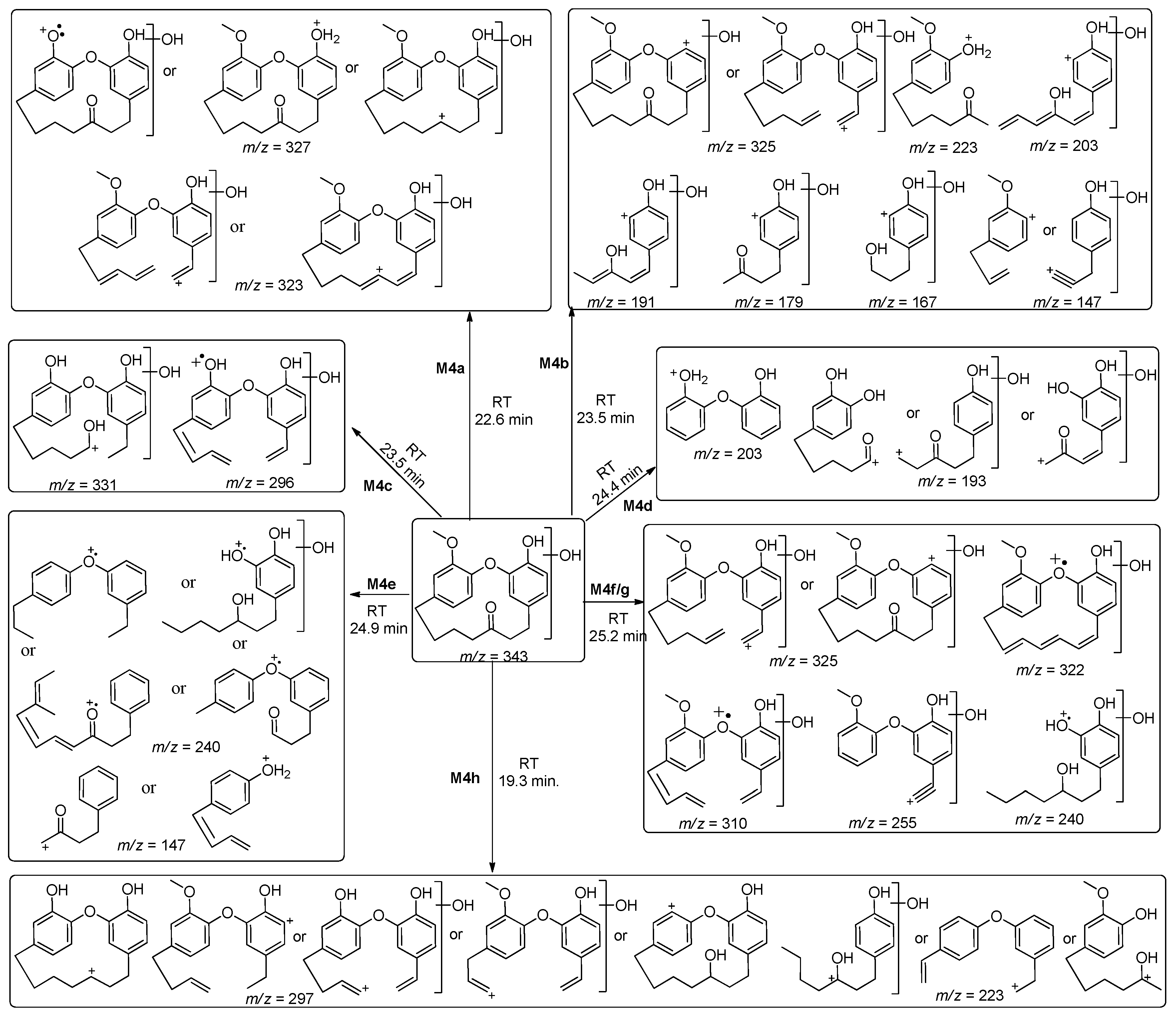
| M4a | 343 | 22.6 | 327, 323 | Hydroxylation |
| M4b | 23.5 | 325, 223, 203, 191, 179, 167, 147 | ||
| M4c | 24.0 | 331, 296 | ||
| M4d | 24.4 | 203, 193 | ||
| M4e | 24.9 | 240, 147 | ||
| M4f/g | 25.2 | 322, 310, 255, 240/325, 322, 310, 240 | ||
| M4h | 19.3 | 297, 223 | ||
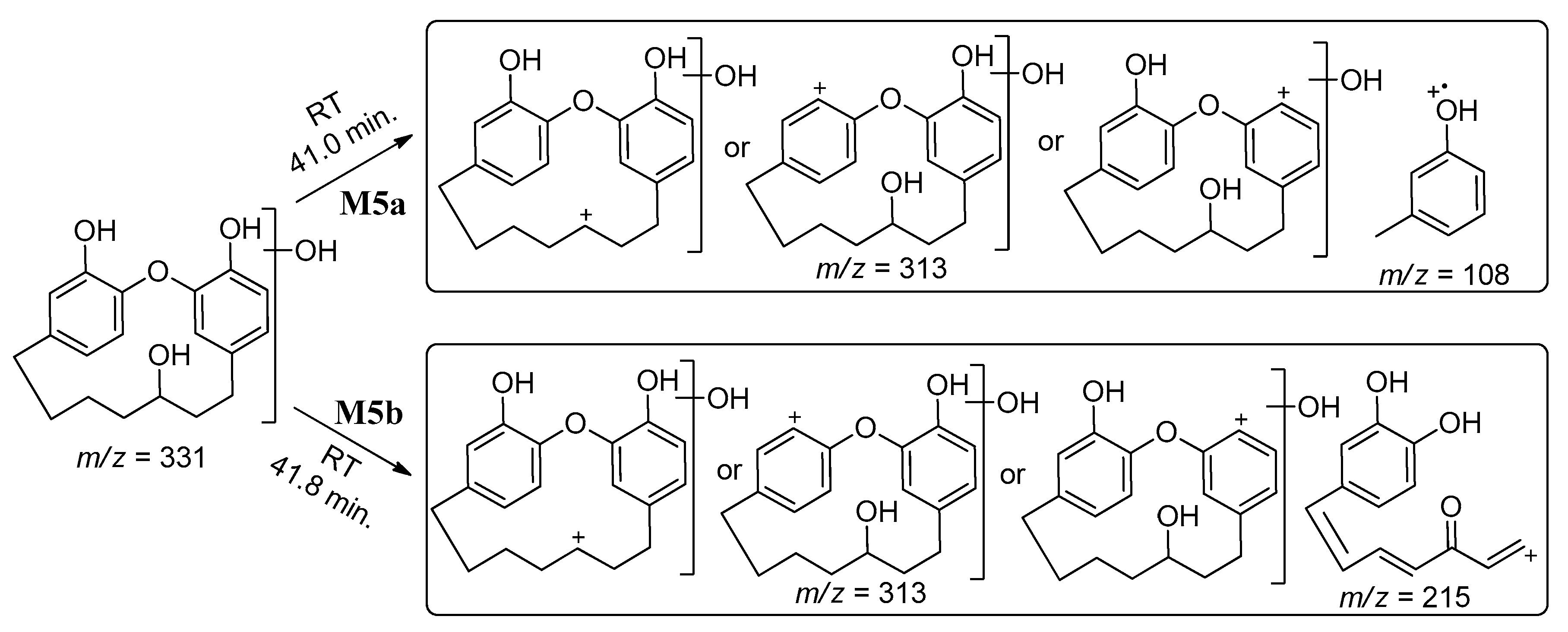
| M5a | 331 | 41.0 | 313, 108 | Demethylation with Hydroxylation and Reduction |
| M5b | 41.8 | 313, 215 | ||
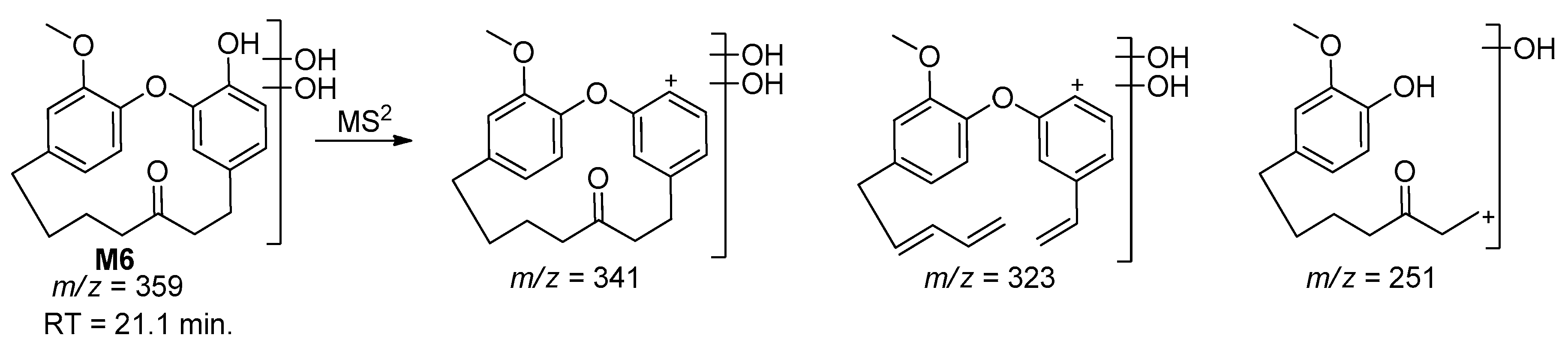
| M6 | 359 | 21.1 | 341, 323, 251 | Dihydroxylation |
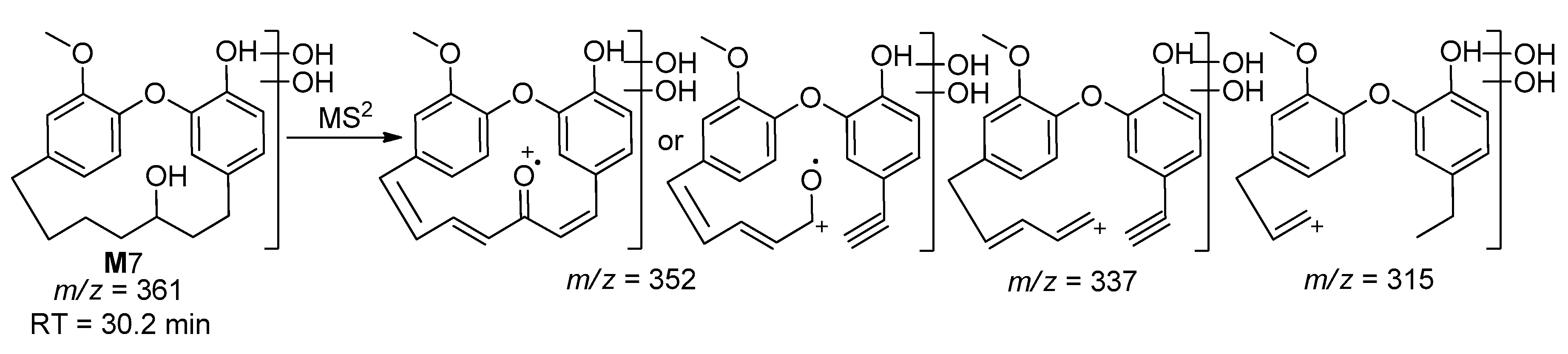
| M7 | 361 | 30.2 | 352, 337, 315 | Dihydroxylation with Reduction |
| Metabolites | Nominal Observed Mass (m/z) | Retention Time (min) | Fragments (MS2 in m/z) | Metabolic Reactions |
|---|---|---|---|---|
| Methoxylamine–Galeon Conjugate | ||||
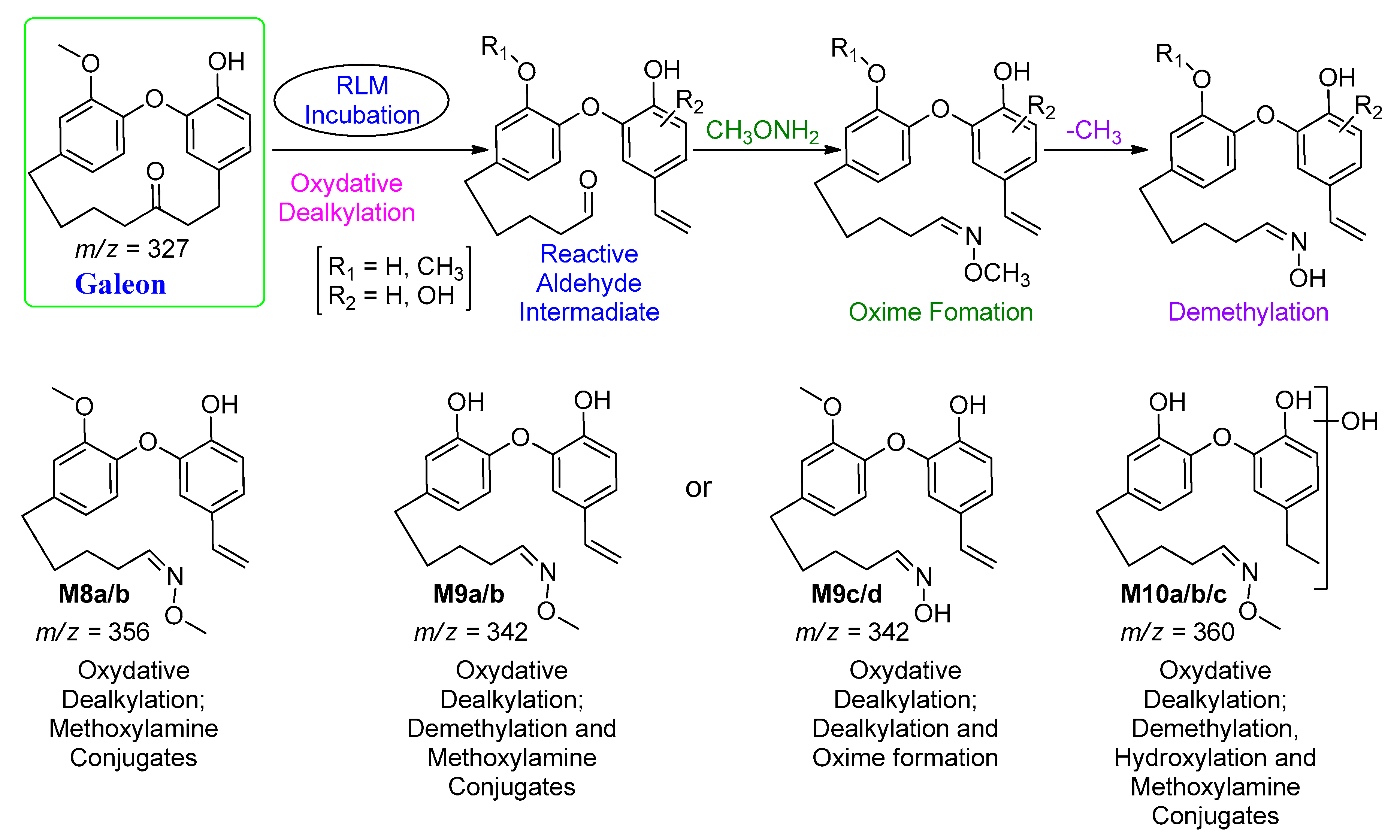
| M8a | 356 | 31.3 | 338, 240 | Oxidative alkylation followed by oxime formation |
| M8b | 31.7 | 294, 192, 129 | ||
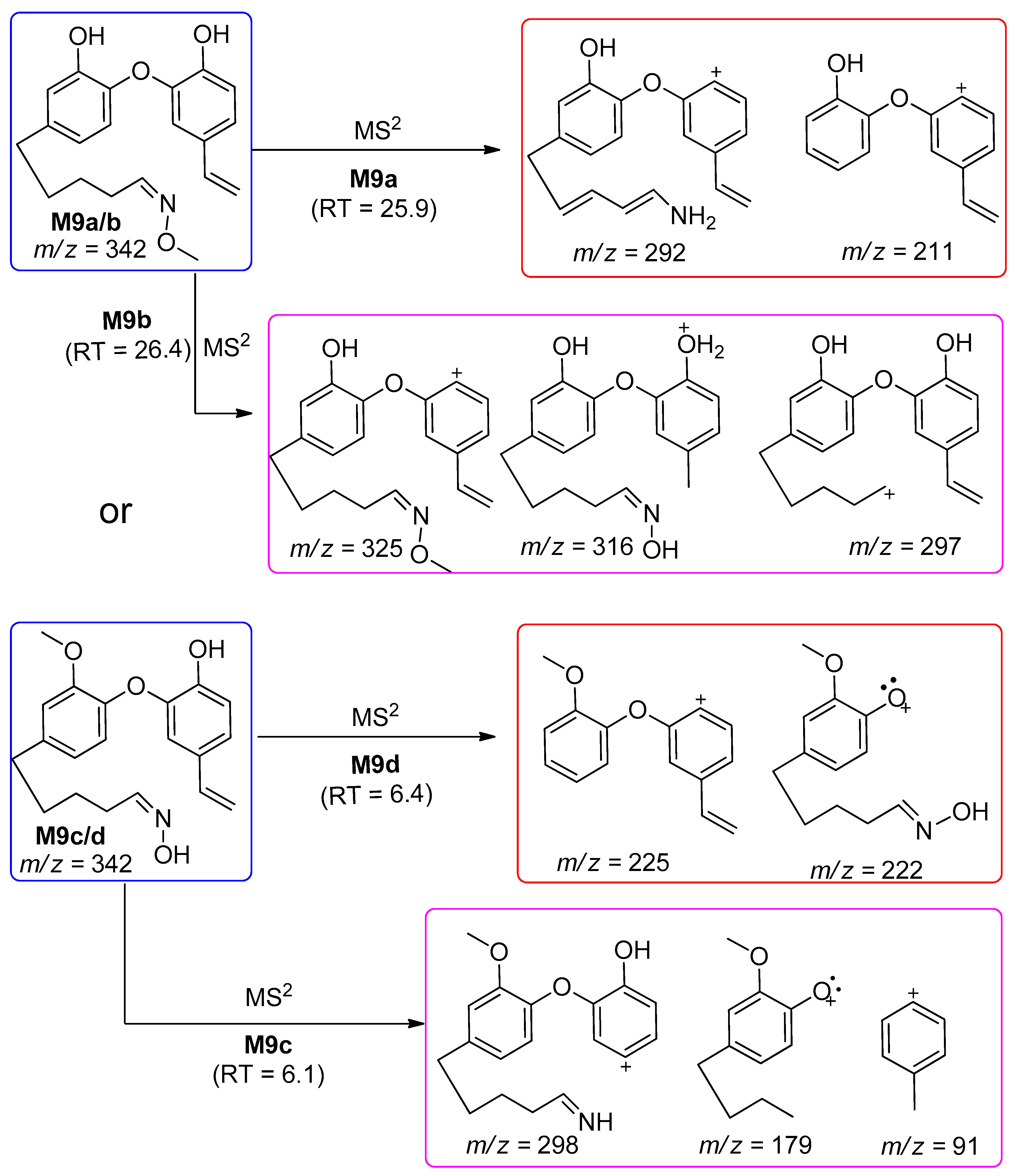
| M9a | 342 | 25.9 | 292, 211 | Oxidative alkylation and demethylation followed by oxime formation |
| M9b | 26.4 | 324, 316, 297 | ||
| M9c | 6.1 | 298, 179, 91 | ||
| M9d | 6.4 | 225, 222 | ||
| M10a | 360 | 20.8 | 343, 325 | Oxidative alkylation, demethylation and hydroxylation followed by oxime formation |
| M10b | 22.8 | 342, 331, 316 | ||
| M10c | 23.8 | 325, 316, 250 | ||
| Glutathione (GHS)–Galeon Conjugate | ||||
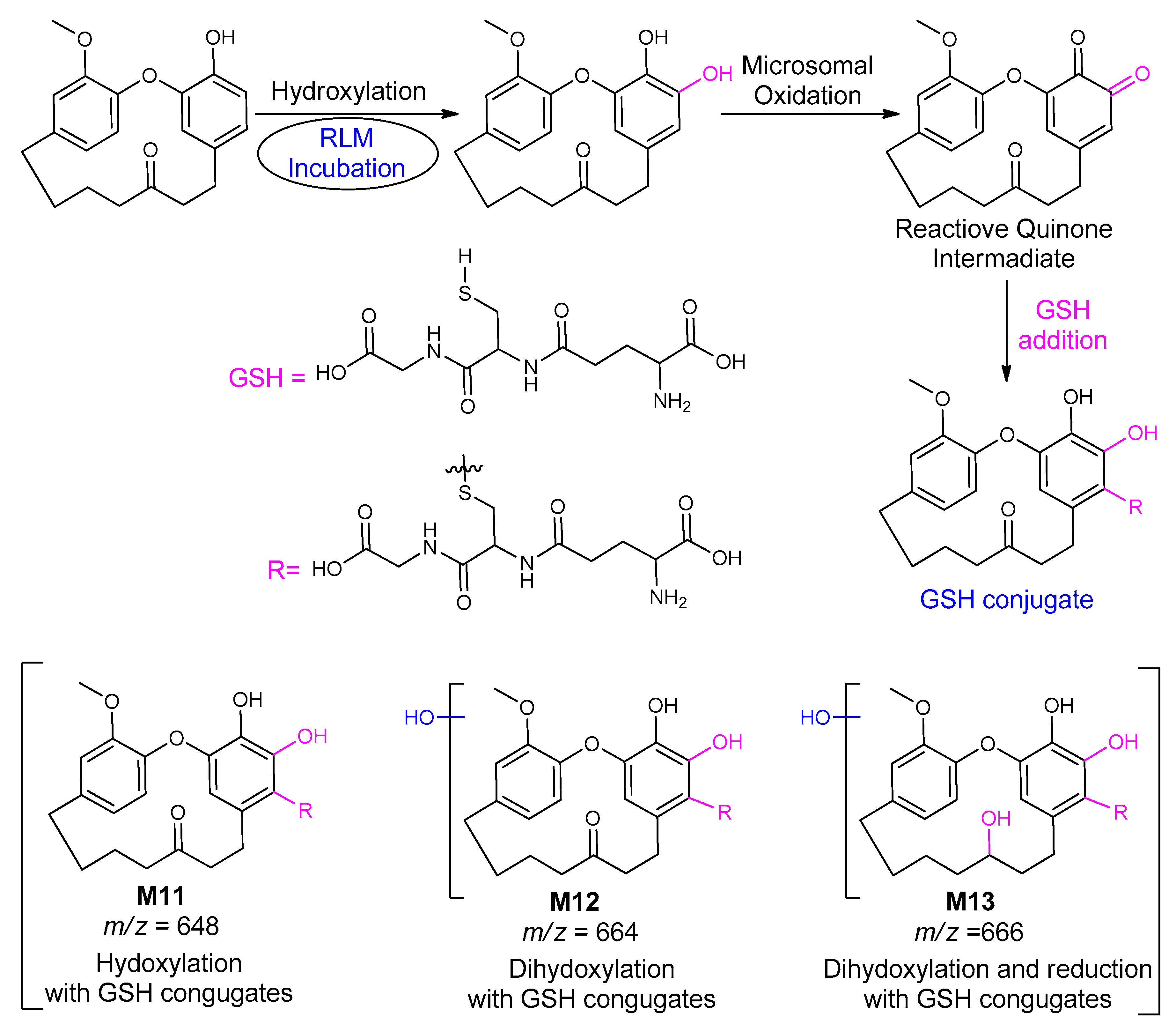
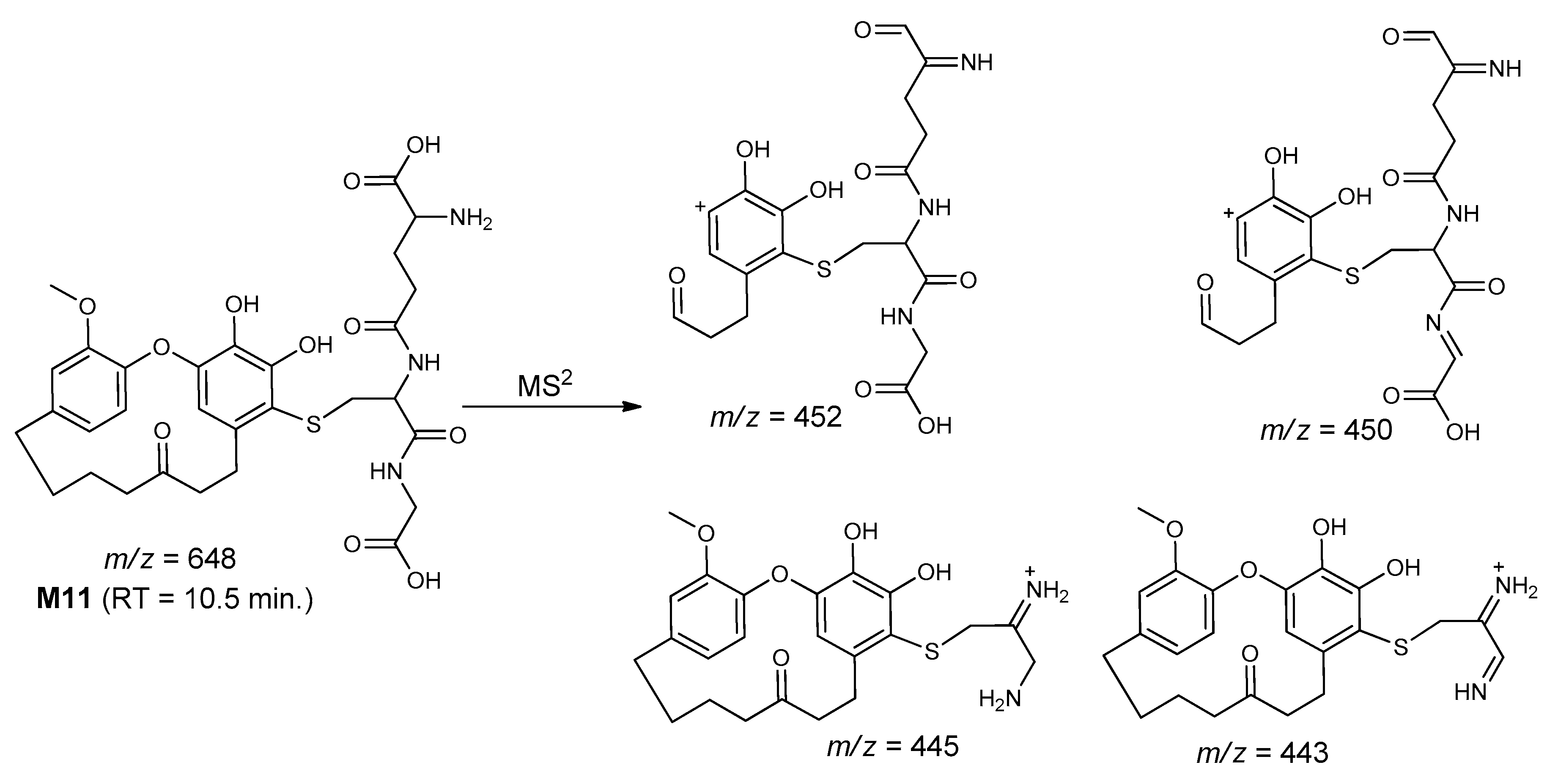
| M11 | 648 | 10.5 | 452, 450, 445, 443 | Mono-hydroxygaleon–GSH conjugate |
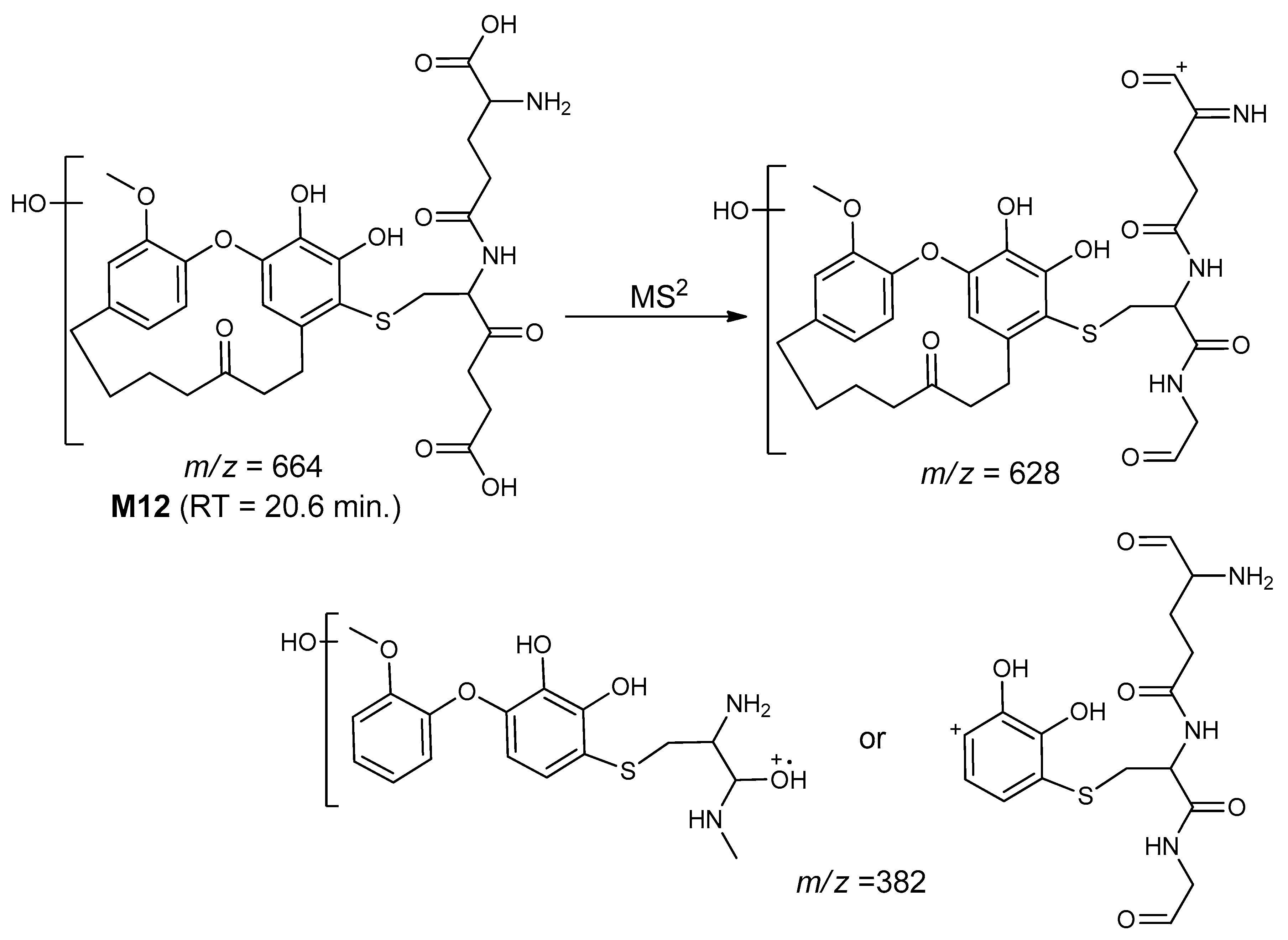
| M12 | 664 | 20.6 | 628, 382 | Di-hydroxygaleon–GSH conjugate |
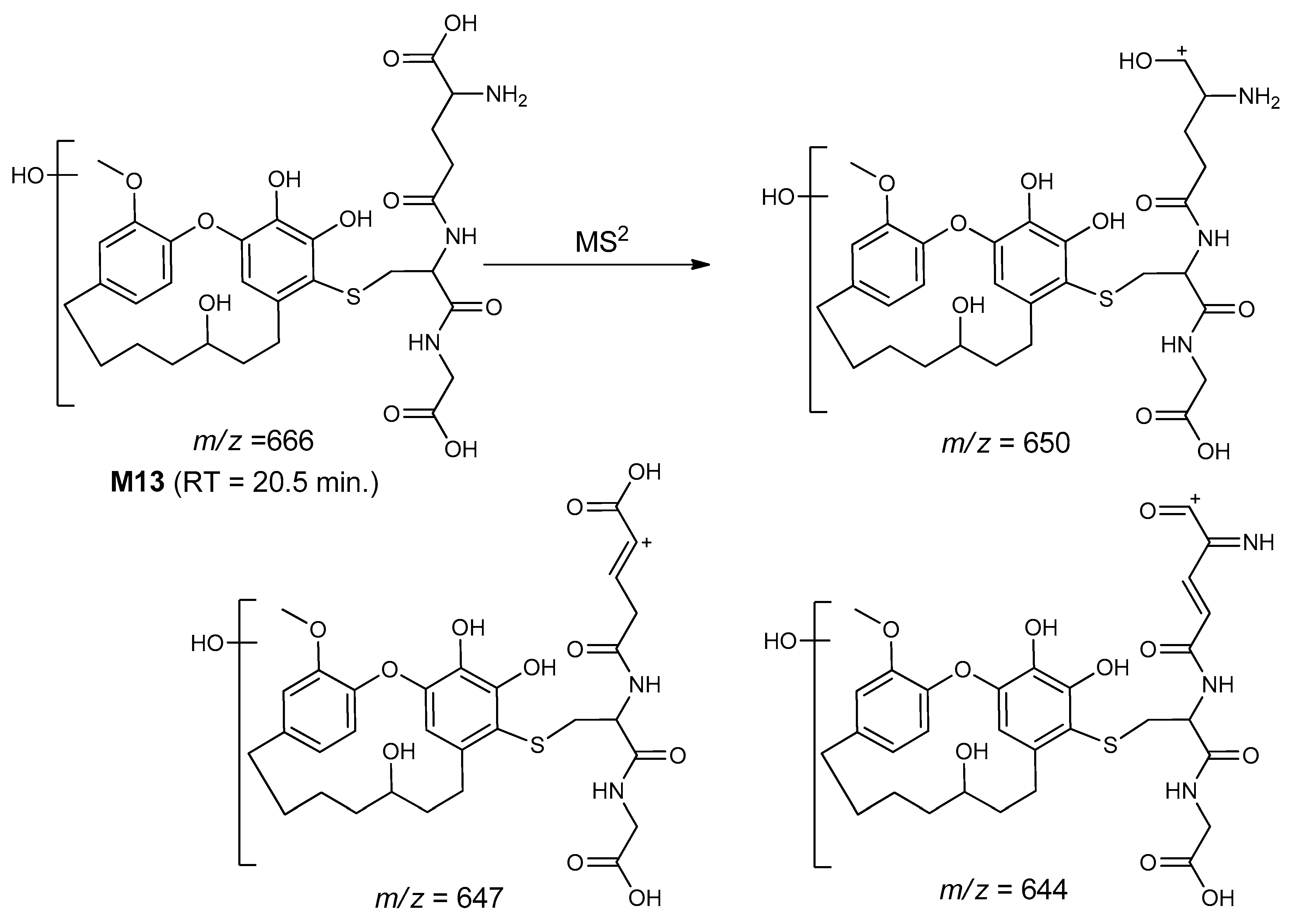
| M13 | 666 | 20.5 | 650, 647, 644 | Reduction of carbonyl and Di-hydroxygaleon–GSH conjugate |
Sample Availability: Galeon is available but metabolites are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, A.F.M.M.; Yin, W.; Kadi, A.A.; Jahng, Y. Galeon: A Biologically Active Molecule with In Silico Metabolite Prediction, In Vitro Metabolic Profiling in Rat Liver Microsomes, and In Silico Binding Mechanisms with CYP450 Isoforms. Molecules 2020, 25, 5903. https://doi.org/10.3390/molecules25245903
Rahman AFMM, Yin W, Kadi AA, Jahng Y. Galeon: A Biologically Active Molecule with In Silico Metabolite Prediction, In Vitro Metabolic Profiling in Rat Liver Microsomes, and In Silico Binding Mechanisms with CYP450 Isoforms. Molecules. 2020; 25(24):5903. https://doi.org/10.3390/molecules25245903
Chicago/Turabian StyleRahman, A. F. M. Motiur, Wencui Yin, Adnan A. Kadi, and Yurngdong Jahng. 2020. "Galeon: A Biologically Active Molecule with In Silico Metabolite Prediction, In Vitro Metabolic Profiling in Rat Liver Microsomes, and In Silico Binding Mechanisms with CYP450 Isoforms" Molecules 25, no. 24: 5903. https://doi.org/10.3390/molecules25245903
APA StyleRahman, A. F. M. M., Yin, W., Kadi, A. A., & Jahng, Y. (2020). Galeon: A Biologically Active Molecule with In Silico Metabolite Prediction, In Vitro Metabolic Profiling in Rat Liver Microsomes, and In Silico Binding Mechanisms with CYP450 Isoforms. Molecules, 25(24), 5903. https://doi.org/10.3390/molecules25245903






