Plant Xyloglucan Xyloglucosyl Transferases and the Cell Wall Structure: Subtle but Significant
Abstract
- Plant cell walls and structure, and key components
- Roles of xyloglucan xyloglucosyl transferases in cell wall formation and re-modelling
- Catalysis, and remarks on nomenclature and classification
- Enzyme activity assay methods
- Substrate specificity of plant xyloglucan xyloglucosyl transferases
- Homo-transglycosylation reactions
- Hetero-transglycosylation reactions with neutral donor and acceptor substrates
- Hetero-transglycosylation reactions with charged (ionic) acceptor substrates
- Implications of transglycosylation reactions catalysed by xyloglucan xyloglucosyl transferases in the cell wall structure, function and dynamics
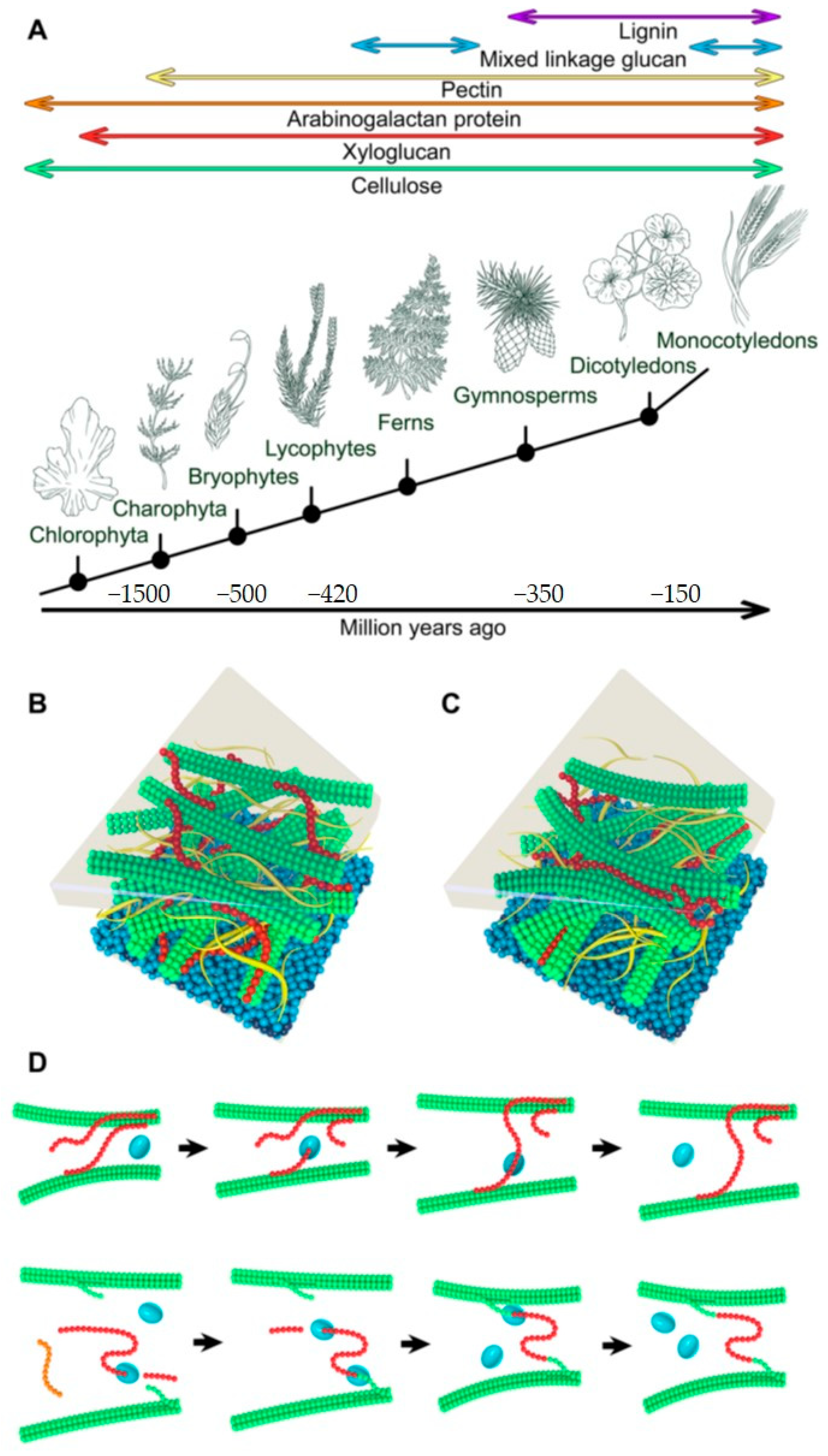
1. Plant Cell Walls and Structure and Key Components
2. Roles of Xyloglucan Xyloglucosyl Transferases in Cell Wall Formation and Re-Modelling
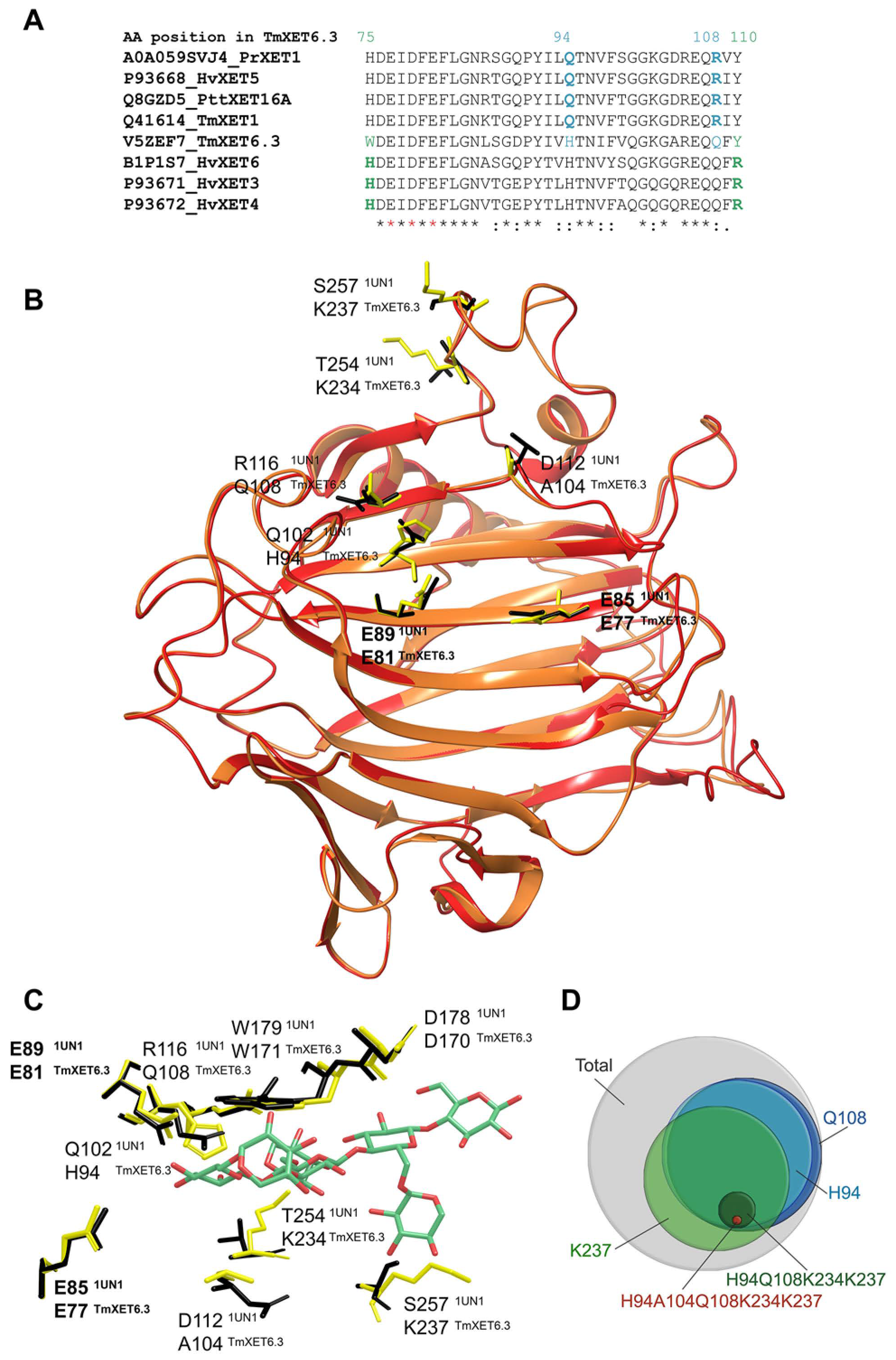
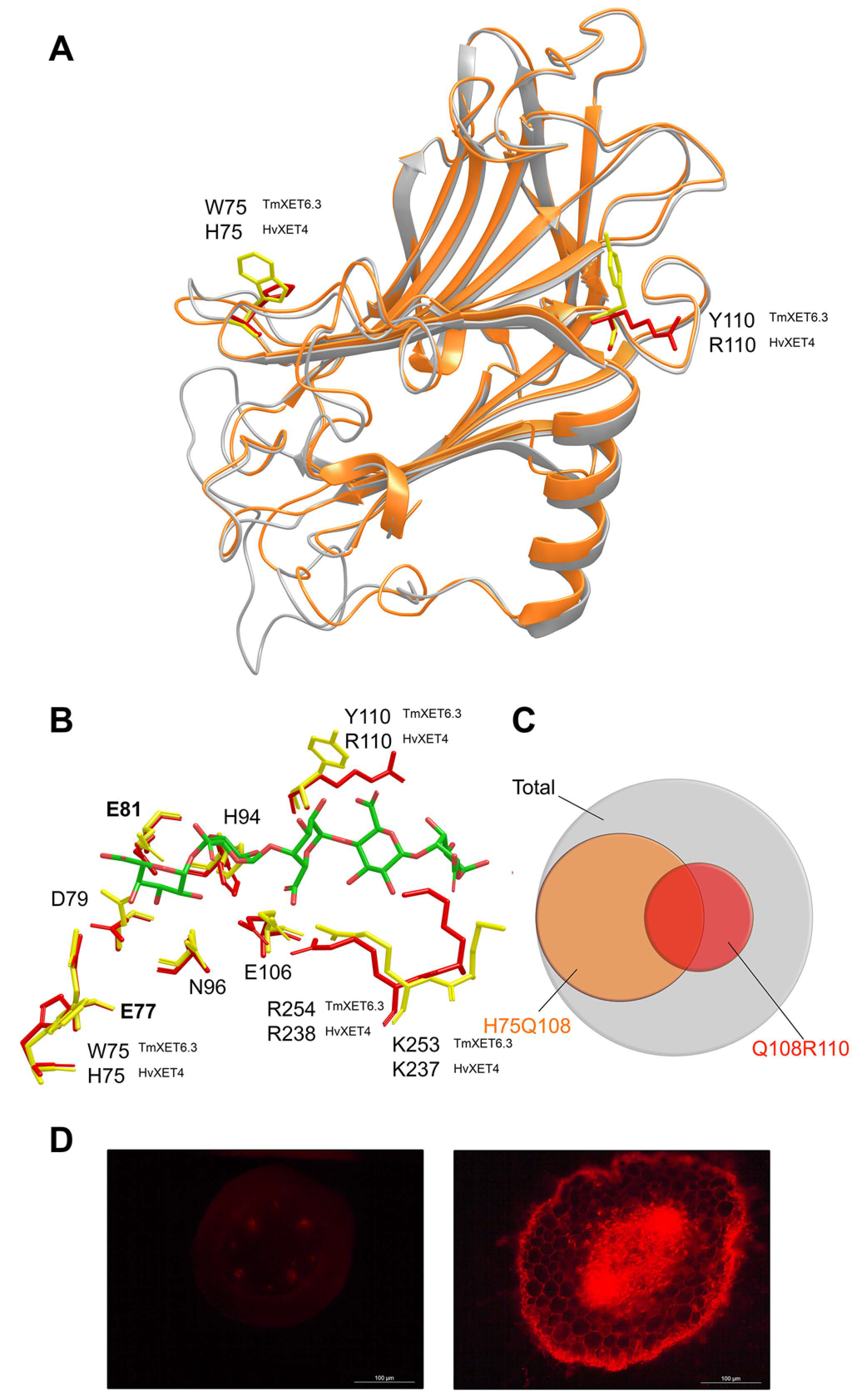
3. Substrate Specificity of Plant Xyloglucan Xyloglucosyl Transferases
4. Implications of Transglycosylation Reactions Catalysed by Xyloglucan Xyloglucosyl Transferases in the Cell Wall Structure, Function and Dynamics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fry, S.C. Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 2004, 161, 641–675. [Google Scholar] [CrossRef]
- Cosgrove, D. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Cabib, E.; Farkaš, V.; Kosík, O.; Blanco, N.; Arroyo, J.; McPhie, P. Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 2008, 283, 29859–29872. [Google Scholar] [CrossRef] [PubMed]
- Farrokhi, N.; Burton, R.A.; Brownfield, L.; Hrmova, M.; Wilson, S.M.; Bacic, A.; Fincher, G.B. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotech. J. 2009, 4, 145–167. [Google Scholar] [CrossRef] [PubMed]
- Blanco, N.; Sanz, A.B.; Rodriguez-Pena, J.M.; Nombela, C.; Farkaš, V.; Hurtado-Guerrero, R.; Arroyo, J. Structural and functional analysis of yeast Crh1 and Crh2 transglycosylases. FEBS J. 2015, 282, 715–731. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall modifying enzymes. J. Exp. Bot. 2015, 67, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Graham, L.E.; Cook, M.E.; Busse, J.S. The origin of plants: Body plan changes contributing to a major evolutionary radiation. PNAS 2000, 97, 4535–4540. [Google Scholar] [CrossRef]
- Niklas, K.J. The cell walls that bind the tree of life. BioScience 2004, 54, 831–841. [Google Scholar] [CrossRef]
- Niklas, K.J.; Kutschera, U. The evolution of the land plant life cycle. New Phytol. 2010, 185, 27–41. [Google Scholar] [CrossRef]
- Sørensen, I.; Pettolino, F.A.; Bacic, A.; Ralph, J.; Lu, F.; O’Neill, M.A.; Fei, Z.; Rose, J.K.C.; Domozych, D.S.; Willats, W.G.T. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011, 68, 201–211. [Google Scholar] [CrossRef]
- Popper, Z.A.; Fry, S.C. Primary cell wall composition of bryophytes and charophytes. Ann. Bot. 2003, 91, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Popper, Z.A.; Fry, S.C. Primary cell wall composition of the pteridophytes and spermatophytes. New Phytol. 2004, 164, 165–174. [Google Scholar] [CrossRef]
- Fincher, G.B. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 2009, 149, 7–37. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Gidley, M.J.; Fincher, G.B. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 2010, 6, 724–732. [Google Scholar] [CrossRef]
- Sørensen, I.; Domozych, D.; Willats, W.G.T. How have plant cell walls evolved? Plant Physiol. 2010, 153, 366–372. [Google Scholar] [CrossRef]
- Fangel, J.U.; Ulvskov, P.; Knox, J.P.; Mikkelsen, M.D.; Harholt, J.; Popper, Z.A.; Willats, W.G.T. Cell wall evolution and diversity. Front. Plant Sci. 2012, 3, 153. [Google Scholar] [CrossRef]
- Kozlova, L.V.; Nazipova, A.R.; Gorshkov, O.V.; Petrova, A.A.; Gorshkova, T.A. Elongating maize root: Zone-specific combinations of polysaccharides from type I and tape II primary cell walls. Sci. Rep. 2020, 10, 10956. [Google Scholar] [CrossRef]
- Iraki, N.M.; Bressan, R.A.; Hasegawa, P.M.; Carpita, N.C. Alteration of the physical and chemical structure of the primary cell wall of growth-limited plant cells adapted to osmotic stress. Plant Physiol. 1989, 91, 39–47. [Google Scholar] [CrossRef]
- Popper, Z.A. Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 2008, 11, 286–292. [Google Scholar] [CrossRef]
- Sarkar, P.; Bosneaga, E.; Auer, M. Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 2009, 60, 3615–3635. [Google Scholar] [CrossRef]
- Popper, Z.A.; Tuohy, M.G. Beyond the green: Understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol. 2010, 153, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Popper, Z.A.; Michel, G.; Hervé, C.; Domozych, D.S.; Willats, W.G.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C.; Nesselrode, B.H.W.A.; Miller, J.G.; Mewburn, B.R. Mixed-linkage (1→3,1→4)-β-D-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 2008, 179, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, I.; Pettolino, F.A.; Wilson, S.M.; Doblin, M.S.; Johansen, B.; Bacic, A.; Willats, W.G. Mixed-linkage (1→ 3),(1→4)-β-D-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. Plant J. 2008, 54, 510–521. [Google Scholar] [CrossRef]
- Xue, X.; Fry, S.C. Evolution of mixed-linkage (1→3, 1→4)-β-D-glucan (MLG) and xyloglucan in Equisetum (horsetails) and other monilophytes. Ann. Bot. 2012, 109, 873–886. [Google Scholar] [CrossRef]
- Burton, R.A.; Fincher, G.B. (1,3;1,4)-β-D-glucans in cell walls of the Poaceae, lower plants, and fungi: A tale of two linkages. Mol. Plant 2009, 2, 873–882. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Popper, Z.A.; Fry, S.C. Widespread occurrence of a covalent linkage between xyloglucan and acidic polysaccharides in suspension-cultured angiosperm cells. Ann. Bot. 2005, 96, 91–99. [Google Scholar] [CrossRef]
- Marcus, S.E.; Verhertbruggen, Y.; Hervé, C.; Ordaz-Ortiz, J.J.; Farkaš, V.; Pedersen, H.L.; Willats, W.G.T.; Knox, J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008, 8, 60. [Google Scholar] [CrossRef]
- McCann, M.C.; Knox, J.P. Plant cell wall biology: Polysaccharides in architectural and developmental contexts. Ann. Plant Rew. Online 2018, 343–366. [Google Scholar] [CrossRef]
- Harholt, J.; Suttangkakul, A.; Scheller, V.H. Biosynthesis of pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signalling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Vincken, J.P.; Schols, H.A.; Oomen, R.J.; Mccann, M.C.; Ulvskov, P.; Voragen, A.G.J.; Visser, R.G.F. If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol. 2003, 132, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant. Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant. Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Weng, J.K.; Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 2010, 187, 273–285. [Google Scholar] [CrossRef]
- Mnich, E.; Bjarnholt, N.; Eudes, A.; Harholt, J.; Holland, C.; Jørgensen, B.; Larsen, F.H.; Liu, M.; Manat, R.; Meyer, A.S.; et al. Phenolic cross-links: Building and de-constructing the plant cell wall. Nat. Prod. Rep. 2020, 37, 919–961. [Google Scholar] [CrossRef]
- Horton, D. Preface. The two most abundant organic substances on Earth, cellulose and starch. Adv. Carbohydr. Chem. Biochem. 2010, 64, xi–xiii. [Google Scholar] [CrossRef]
- Heinze, T. Cellulose: Structure and properties. In Cellulose Chemistry and Properties: Fibers, Nanocelluloses and Advanced Materials. Advances in Polymer Science; Rojas, O., Ed.; Springer: Cham, Switzerland, 2015; Volume 271. [Google Scholar] [CrossRef]
- Kim, S.J.; Chandrasekar, B.; Rea, A.C.; Danhof, L.; Zemelis-Durfee, S.; Thrower, N.; Shepard, Z.S.; Pauly, M.; Brandizzi, F.; Keegstra, K. The synthesis of xyloglucan, an abundant plant cell wall polysaccharide, requires CSLC function. Proc. Natl. Acad. Sci. USA 2020, 117, 20316–20324. [Google Scholar] [CrossRef]
- Ikegaya, H.; Hayashi, T.; Kaku, T.; Iwata, K.; Sonobe, S.; Shimmen, T. Presence of xyloglucan-like polysaccharide in Spirogyra and possible involvement in cell–cell attachment. Phycol. Res. 2008, 56, 216–222. [Google Scholar] [CrossRef]
- Van Sandt, V.S.; Stieperaere, H.; Guisez, Y.; Verbelen, J.P.; Vissenberg, K. XET activity is found near sites of growth and cell elongation in bryophytes and some green algae: New insights into the evolution of primary cell wall elongation. Ann. Bot. 2007, 99, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C.; Aldington, S.; Hetherington, P.R.; Aitken, J. Oligosaccharides as signals and substrates in the plant cell wall. Plant Physiol. 1993, 103, 1–5. [Google Scholar] [CrossRef] [PubMed]
- York, W.S.; Van Halbeek, H.; Darvill, A.G.; Albersheim, P. Structural analysis of xyloglucan oligosaccharides by 1H-n.m.r. spectroscopy and fast-atom-bombardment mass spectrometry. Carbohydr Res. 1990, 200, 9–31. [Google Scholar] [CrossRef]
- York, W.S.; Harvey, L.K.; Guillen, R.; Albersheim, P.; Darvill, A.G. Structural analysis of tamarind seed xyloglucan oligosaccharides using beta-galactosidase digestion and spectroscopic methods. Carbohydr. Res. 1993, 248, 285–301. [Google Scholar] [CrossRef]
- Peña, M.J.; Darvill, A.G.; Eberhard, S.; York, W.S.; O’Neill, M.A. Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology 2008, 18, 891–904. [Google Scholar] [CrossRef]
- Peña, M.J.; Kong, Y.; York, W.S.; O’Neill, M.A. A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 2012, 24, 4511–4524. [Google Scholar] [CrossRef]
- Hsieh, Y.S.Y.; Harris, P.J. Xyloglucans of monocotyledons have diverse structures. Mol. Plant 2009, 2, 943–965. [Google Scholar] [CrossRef]
- Hsieh, Y.S.Y.; Paxton, M.; Ade, C.P.; Harris, P.J. Structural diversity, functions and biosynthesis of xyloglucans in angiosperm cell walls. NZ J. Forest Sci. 2009, 39, 187–196. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Chen, Y.; Wagner, E.; Cosgrove, D.J. Xyloglucan in the primary cell wall: Assessment by FESEM, selective enzyme digestions and nanogold affinity tags. Plant J. 2018, 93, 211–226. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Lerouxel, O.; Drakakaki, G.; Alonso, A.P.; Liepman, A.H.; Keegstra, K.; Raikhel, N.; Wilkerson, C.G. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 8550–8555. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Keegstra, K. Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 2016, 67, 235–259. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T. Xyloglucans in the primary cell wall. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 139–168. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, G.; Wang, X.; Zhang, X.; Friml, J. Evolution of fast root gravitropism in seed plants. Nat. Commun. 2019, 10, 3480. [Google Scholar] [CrossRef] [PubMed]
- Albersheim, P.; Darvill, A.; Roberts, K.; Sederoff, R.; Staehelin, A. Plant Cell Walls, from Chemistry to Biology. Garland Science; Garland Science: New York, NY, USA, 2011; pp. 227–272. ISBN 978-0815319962. [Google Scholar]
- Park, Y.B.; Cosgrove, D.J. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012, 158, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Farkaš, V.; Lahnstein, J.; Fincher, G.B. A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-D-glucans. J. Biol. Chem. 2007, 283, 27344. [Google Scholar] [CrossRef] [PubMed]
- Keegstra, K.; Talmadge, K.W.; Bauer, W.D.; Albersheim, P. The structure of plant cell walls. III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 1973, 51, 188–196. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Pauly, M.; Albersheim, P.; Darvill, A.; York, W.S. Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 1999, 20, 629–639. [Google Scholar] [CrossRef]
- Chanliaud, E.; De Silva, J.; Strongitharm, B.; Jeronimidis, G.; Gidley, M.D. Mechanical effects of plant cell wall enzymes on cellulose/xyloglucan composites. Plant J. 2004, 38, 27–37. [Google Scholar] [CrossRef]
- Chanliaud, E.; Burrows, K.M.; Jeronimidis, G.; Gidley, M.J. Mechanical properties of primary plant cell wall analogues. Planta 2002, 215, 989–996. Available online: https://www.jstor.org/stable/23387052 (accessed on 25 November 2020). [CrossRef] [PubMed]
- Cosgrove, D.J. Re-constructing our models of cellulose and primary cell wall assembly. Curr. Opin. Plant Biol. 2014, 22, 122–131. [Google Scholar] [CrossRef]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Diffuse growth of plant cell walls. Plant Physiol. 2018, 176, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Kuki, H.; Yokoyama, R.; Kuroha, T.; Nishitani, K. Xyloglucan is not essential for the formation and integrity of the cellulose network in the primary cell wall regenerated from Arabidopsis protoplasts. Plants 2020, 9, 629. [Google Scholar] [CrossRef]
- Scheible, W.R.; Pauly, M. Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant. Biol. 2004, 7, 285–295. [Google Scholar] [CrossRef]
- Burton, R.A.; Wilson, S.M.; Hrmova, M.; Harvey, A.J.; Shirley, N.J.; Medhurst, A.; Stone, B.A.; Newbigin, E.J.; Bacic, A.; Fincher, G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 2006, 311, 1940–1942. [Google Scholar] [CrossRef]
- Qu, Y.; Egelund, J.; Gilson, P.R.; Houghton, F.; Gleeson, P.A.; Schultz, C.J.; Bacic, A. Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 2008, 68, 43–59. [Google Scholar] [CrossRef]
- Doblin, M.S.; Pettolino, F.A.; Wilson, S.M.; Campbell, R.; Burton, R.A.; Fincher, G.B.; Newbigin, E.; Bacic, A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5996–6001. [Google Scholar] [CrossRef]
- Wu, A.-M.; Hörnblad, E.; Voxeur, A.; Gerber, L.; Rihouey, C.; Lerouge, P.; Marchant, A. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 2010, 153, 542–554. [Google Scholar] [CrossRef]
- Anders, N.; Wilkinson, M.D.; Lovegrove, A.; Freeman, J.; Tryfona, T.; Pellny, T.K.; Weimar, T.; Mortimer, J.C.; Stott, K.; Baker, J.M.; et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer ontoxylan in grasses. Proc. Natl. Acad. Sci. USA 2012, 17, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.F.; Harholt, J.; Oikawa, A.; Scheller, H.V. Plant glycosyl transferases beyond CAZy: A perspective on DUF families. Front. Plant Sci. 2012, 3, 59. [Google Scholar] [CrossRef] [PubMed]
- Harholt, J.; Sørensen, I.; Fangel, J.U.; Roberts, A.; Willats, W.G.T.; Scheller, H.V.; Larsen, B.; Petersen, B.L.; Banks, J.A.; Ulvskov, P. The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of the cell wall structure. PLoS ONE 2012, 7, e35846. [Google Scholar] [CrossRef] [PubMed]
- Zabotina, O.A.; Avci, U.; Cavalier, D.; Pattathil, S.; Chou, Y.-H.; Eberhard, S.; Danhof, L.; Keegstra, K.; Hahn, M.G. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. 2012, 159, 1367–1384. [Google Scholar] [CrossRef]
- Stonebloom, S.; Ebert, B.; Xiong, G.; Pattathil, S.; Birdseye, D.; Lao, J.; Pauly, M.; Hahn, M.G.; Heazlewood, J.I.; Scheller, H.V. A DUF-246 family glycosyltransferase-like gene affects male fertility and the biosynthesis of pectic arabinogalactans. BMC Plant Biol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Zeng, W.; Lampugnani, E.R.; Picard, K.L.; Song, L.L.; Wu, A.M.; Farion, I.M.; Zhao, J.; Ford, K.; Doblin, M.S.; Bacic, A. Asparagus IRX9, IRX10, and IRX14A are components of an active xylan backbone synthase complex that forms in the Golgi apparatus. Plant Physiol. 2016, 171, 93–109. [Google Scholar] [CrossRef]
- Takenaka, Y.; Kato, K.; Ogawa-Ohnishi, M.; Tsuruhama, K.; Kajiura, H.; Yagyu, K.; Tekeda, A.; Takeda, Y.; Kunieda, T.; Hara-Nishimura, I.; et al. Pectin RG-I rhamnosyl-transferases represent a novel plant-specific glycosyltransferase family. Nat. Plants 2018, 4, 669–676. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Engle, K.A.; Günl, M.; Dieluweit, S.; Schmidt, M.H.; Yang, J.Y.; Moremen, K.W.; Mohnen, D.; Usadel, B. Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol. 2018, 178, 1045–1064. [Google Scholar] [CrossRef]
- Amos, R.A.; Mohnen, D. Critical review of plant cell wall matrix polysaccharide glycosyltransferase activities verified by heterologous protein expression. Front. Plant Sci. 2019, 10, 915. [Google Scholar] [CrossRef]
- Penning, B.W.; McCann, M.C.; Carpita, N.C. Evolution of the cell wall gene families of grasses. Front. Plant Sci. 2019, 10, 1205. [Google Scholar] [CrossRef]
- Yang, J.; Bak, G.; Burgin, T.; Barnes, W.J.; Mayes, H.B.; Peña, M.J.; Urbanowicz, B.R.; Nielsen, E. Biochemical and genetic analysis Identify CSLD3 as a beta-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell 2020, 32, 1749–1767. [Google Scholar] [CrossRef] [PubMed]
- Wachananawat, B.; Kuroha, T.; Takenaka, Y.; Kajiura, H.; Naramoto, S.; Yokoyama, R.; Ishizaki, K.; Nishitani, K.; Ishimizu, T. Diversity of pectin rhamnogalacturonan I rhamnosyltransferases in glycosyltransferase family 106. Front. Plant Sci. 2020, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Dhugga, K.S. Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry 2012, 74, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.W.; Roberts, E.M.; Delmer, D.P. Cellulose synthase (CesA) genes in the green alga Mesotaenium caldariorum. Eukaryot. Cell 2002, 1, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.W.; Roberts, E. Evolution of the Cellulose Synthase (CesA) Gene Family: Insights from Green Algae and Seedless Plants. In Cellulose: Molecular and structural Biology; Brown, R.M., Saxena, I.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 17–34. [Google Scholar] [CrossRef]
- Yin, Y.; Huang, J.; Xu, Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009, 9, 99. [Google Scholar] [CrossRef]
- Endler, A.; Persson, S. Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 2011, 4, 199–211. [Google Scholar] [CrossRef]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O’Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, J.B.; et al. Revised phylogeny of the cellulose synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef]
- Abercrombie, J.M.; O’Meara, B.C.; Moffatt, A.R.; Williams, J.H. Developmental evolution of flowering plant pollen tube cell walls: Callose synthase (CalS) gene expression patterns. EvoDevo 2011, 2, 14. [Google Scholar] [CrossRef]
- Park, S.; Szumlanski, A.L.; Gu, F.; Guo, F.; Nielsen, E. A role for CSLD3 during cell-wall synthesis in a pical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011, 13, 973–980. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Lv, L.M.; Zuo, D.Y.; Wang, X.F.; Cheng, H.L.; Zhang, Y.P.; Wang, Q.L.; Song, G.L.; Ma, Z.Y. Genome-wide identification of the expansin gene family reveals that expansin genes are involved in fibre cell growth in cotton. BMC Plant Biol. 2020, 20, 223. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lyu, T.; Xu, L.; Hu, Z.; Xiong, X.; Liu, T.; Cao, J. Complex molecular evolution and expression of expansin gene families in three basic diploid species of Brassica. Int. J. Mol. Sci. 2020, 21, 3424. [Google Scholar] [CrossRef] [PubMed]
- Herburger, K.; Franková, L.; Picmanova, M.; Loh, J.W.; Valenzuela-Ortega, M.; Meulewaeter, F.; Hudson, A.D.; French, C.E.; Fry, S.C. Hetero-trans-β-Glucanase produces cellulose–xyloglucan covalent bonds in the cell walls of structural plant tissues and is stimulated by expansin. Mol. Plant 2020, 13, 1047–1062. [Google Scholar] [CrossRef] [PubMed]
- Willats, W.G.T.; Orfila, C.; Limberg, G.; Buchholt, H.C.; van Alebeek, G.-J.W.M.; Voragen, A.G.J.; Marcus, S.E.; Christensen, T.M.I.E.; Mikkelsen, J.D.; Murray, B.S.; et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls: Implications for pectin methylesterase action, matrix properties and cell adhesion. J. Biol. Chem. 2001, 276, 19404–19413. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef]
- Franková, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef]
- Thompson, J.E.; Smith, R.C.; Fry, S.C. Xyloglucan undergoes interpolymeric transglycosylation during binding to the plant cell wall in vivo: Evidence from 13C/3H dual labelling and isopycnic centrifugation in caesium trifluoroacetate. Biochem. J. 1997, 327, 699–708. [Google Scholar] [CrossRef]
- Campbell, P.; Braam, J. Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999, 4, 361–366. [Google Scholar] [CrossRef]
- Bourquin, V.; Nishikubo, N.; Abe, H.; Brumer, H.; Denman, S.; Eklund, M.; Christiernin, M.; Teeri, T.T.; Sundberg, B.; Mellerowicz, E.J. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 2002, 14, 3073–3088. [Google Scholar] [CrossRef]
- Nishitani, K. Division of roles among members of the XTH gene family in plants. Plant Biosyst. 2005, 139, 98–101. [Google Scholar] [CrossRef]
- Nishikubo, N.; Takahashi, J.; Roos, A.A.; DerbaMaceluch, M.; Piens, K.; Brumer, H.; Teeri, T.T.; Stalbrand, H.; Mellerowicz, E.J. Xyloglucan endotransglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen. Plant Physiol. 2011, 155, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Farkaš, V.; Sulová, Z.; Stratilová, E.; Hanna, R.; Maclachlan, G. Cleavage of xyloglucan by nasturtium seed xyloglucanase and transglycosylation to xyloglucan subunit oligosaccharides. Arch. Biochem. Biophys. 1992, 298, 365–370. [Google Scholar] [CrossRef]
- Fry, S.; Smith, R.; Renwick, K.; Martin, D.; Hodge, S.; Matthews, K. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, K.; Tominaga, R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 1992, 267, 21058–21064. [Google Scholar] [PubMed]
- Webb, E.C. Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes; Academic Press Inc: Harcourt Brace, Jovanovich Publishers: San Diego, CA, USA, 1992; ISBN 0-12-227164-5. [Google Scholar]
- Placzek, S.; Schomburg, I.; Chang, A.; Jeske, L.; Ulbrich, M.; Tillack, J.; Schomburg, D. BRENDA in 2017: New perspectives and new tools in BRENDA. Nucleic Acids Res. 2017, 45, D380–D388. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Johnston, S.L.; Yauk, Y.K.; Sharma, N.N.; Schröder, R. Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol. Technol. 2009, 51, 149–157. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef]
- Yokoyama, R.; Uwagaki, Y.; Sasaki, H.; Harada, T.; Hiwatashi, Y.; Hasebe, M.; Nishitani, K. Biological implications of the occurrence of 32 members of the XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens. Plant J. 2010, 64, 645–656. [Google Scholar] [CrossRef]
- Muñoz-Bertomeu, J.; Miedes, E.; Lorences, E.P. Expression of xyloglucan endotransglucosylase/hydrolase (XTH) genes and XET activity in ethylene treated apple and tomato fruits. J. Plant Physiol. 2013, 170, 1194–1201. [Google Scholar] [CrossRef]
- Shi, Y.Z.; Zhu, X.F.; Miller, J.G.; Gregson, T.; Zheng, S.J.; Fry, S.C. Distinct catalytic capacities of two aluminium-repressed Arabidopsis thaliana xyloglucan endotransglucosylase/hydrolases, XTH15 and XTH31, heterologously produced in Pichia. Phytochemistry 2015, 112, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.M.; Liu, C.; Wu, F. Genome-wide identification, characterization and expression analysis of xyloglucan endotransglucosylase/hydrolase genes family in barley (Hordeum vulgare). Molecules 2019, 24, 1935. [Google Scholar] [CrossRef] [PubMed]
- Holland, C.; Simmons, T.J.; Meulewaeter, F.; Hudson, A.; Fry, S.C. Three highly acidic Equisetum XTHs differ from hetero-trans-β-glucanase in donor substrate specificity and are predominantly xyloglucan homo-transglucosylases. J. Plant Physiol. 2020, 251, 153210. [Google Scholar] [CrossRef] [PubMed]
- Baran, R.; Sulová, Z.; Stratilová, E.; Farkaš, V. Ping-Pong character of nasturtium-seed xyloglucan endotransglycosylase (XET) Reaction. Gen. Physiol. Biophys. 2000, 19, 427–440. [Google Scholar] [PubMed]
- Saura-Valls, M.; Faure, R.; Ragas, S.; Piens, K.; Brumer, H.; Teeri, T.T.; Cottaz, S.; Driguez, H.; Planas, A. Kinetic analysis using low-molecular mass xyloglucan oligosaccharides defines the catalytic mechanism of a Populus xyloglucan endotransglycosylase. Biochem. J. 2006, 395, 99–106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ait Mohand, F.; Farkaš, V. Screening for hetero-transglycosylating activities in extracts from nasturtium (Tropaeolum majus). Carbohydr. Res. 2006, 34, 577–581. [Google Scholar] [CrossRef]
- Hrmova, M.; Farkaš, V.; Harvey, A.J.; Lahnstein, J.; Wischmann, B.; Kaewthai, N.; Ezcurra, I.; Teeri, T.T.; Fincher, G.B. Substrate specificity and catalytic mechanism of a xyloglucan xyloglucosyl transferase HvXET6 from barley (Hordeum vulgare L.). FEBS J. 2009, 276, 437–456. [Google Scholar] [CrossRef]
- Shinohara, N.; Sunagawa, N.; Tamura, S.; Yokoyama, R.; Ueda, M.; Igarashi, K.; Nishitani, K. The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Nat. Sci. Rep. 2017, 7, 46099–46108. [Google Scholar] [CrossRef]
- Stratilová, B.; Firáková, Z.; Klaudiny, J.; Šesták, S.; Kozmon, S.; Strouhalová, D.; Garajová, S.; Ait-Mohand, F.; Horváthová, Á.; Farkaš, V.; et al. Engineering the acceptor substrate specificity in the xyloglucan endotransglycosylase TmXET6.3 from nasturtium seeds (Tropaeolum majus L.). Plant Mol. Biol. 2019, 100, 181–197. [Google Scholar] [CrossRef]
- Herburger, K.; Franková, L.; Sanhueza, D.; Roig-Sanchez, S.; Meulewaeter, F.; Hudson, A.; Thomson, A.; Laromaine, A.; Budtova, T.; Fry, S.C. Enzymically attaching oligosaccharide-linked ‘cargoes’ to cellulose and other commercial polysaccharides via stable covalent bonds. Int. J. Biol. Macromol. 2020, 164, 4359–4369. [Google Scholar] [CrossRef]
- Stratilová, B.; Šesták, S.; Mravec, J.; Garajová, S.; Pakanová, Z.; Vadinová, K.; Kučerová, D.; Kozmon, S.; Schwerdt, J.G.; Shirley, N.; et al. Another building block in the plant cell wall: Barley xyloglucan xyloglucosyl transferases link covalently xyloglucan and anionic oligosaccharides derived from pectin. Plant J. 2020, 104, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Brumer, H.; Baumann, M.J.; Kallas, A.M.; Henriksson, H.; Denman, S.E.; Teeri, T.T.T.; Jones, T.A. Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 2004, 16, 874–886. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; MacGregor, E.A.; Biely, P.; Stewart, R.J.; Fincher, G.B. Substrate binding and catalytic mechanism of a barley β-D-glucosidase/(1,4)-β-D-glucan exohydrolase. J. Biol. Chem. 1998, 273, 11134–11143. [Google Scholar] [CrossRef]
- Schröder, R.; Wegrzyn, T.F.; Sharma, N.N.; Atkinson, R.G. LeMAN4 endo-beta-mannanase from ripe tomato fruit can act as a mannan transglycosylase or hydrolase. Planta 2006, 224, 1091–1102. [Google Scholar] [CrossRef]
- Schröder, R.; Atkinson, R.G.; Redgwell, R.J. Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann. Bot. 2009, 104, 197–204. [Google Scholar] [CrossRef]
- Franková, L.; Fry, S.C. Phylogenetic variation in glycosidases and glycanases acting on plant cell wall polysaccharides, and the detection of transglycosidase and trans-β-xylanase activities. Plant J. 2011, 67, 662–681. [Google Scholar] [CrossRef]
- Johnston, S.; Prakash, R.; Chen, N.J.; Kumagai, M.H.; Turano, H.M.; Cooney, J.M.; Atkinson, R.G.; Paull, R.E.; Cheetamun, R.; Bacic, A.; et al. An enzyme activity capable of endotransglycosylation of heteroxylan polysaccharides is present in plant primary cell walls. Planta 2013, 237, 173–187. [Google Scholar] [CrossRef]
- Derba-Maceluch, M.; Awano, T.; Takahashi, J.; Lucenius, J.; Ratke, C.; Kontro, I.; Busse-Wicher, M.; Kosík, O.; Tanaka, R.; Winzéll, A.; et al. Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol. 2015, 205, 666–681. [Google Scholar] [CrossRef]
- Fry, S.C.; Mohler, K.E.; Nesselrode, B.H.W.A.; Franková, L. Mixed-linkage β-glucan:xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. Plant J. 2008, 55, 240–252. [Google Scholar] [CrossRef]
- Mohler, K.E.; Simmons, T.J.; Fry, S.C. Mixed-linkage glucan:xyloglucan endotransglucosylase (MXE) re-models hemicelluloses in Equisetum shoots but not in barley shoots or Equisetum callus. New Phytol. 2013, 197, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.J.; Mohler, K.E.; Holland, C.; Goubet, F.; Franková, L.; Houston, D.R.; Hudson, A.D.; Meulewaeter, F.; Fry, S.C. Hetero-trans-β-glucanase, an enzyme unique to Equisetum plants, functionalizes cellulose. Plant J. 2015, 83, 753–769. [Google Scholar] [CrossRef] [PubMed]
- Viborg, A.H.; Terrapon, N.; Lombard, V.; Michel, G.; Gurvan, M.; Czjzek, M.; Henrissat, B.; Brumer, H. A subfamily roadmap for functional glycogenomics of the evolutionarily diverse Glycoside Hydrolase Family 16 (GH16). J. Biol. Chem. 2019, 294, 15973–15986. [Google Scholar] [CrossRef] [PubMed]
- Baumann, M.J.; Eklöf, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Baumann, M.J.; Eklöf, J.M.; Gullfot, F.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Brumer, H.; Czjek, M. Analysis of nasturtium TmNXG1 complexes by crystallography and molecular dynamics provides detailed insight into substrate recognition by family GH16 xyloglucan endo-transglycosylases and endo-hydrolases. Proteins 2009, 75, 820–836. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Farkaš, V.; Fincher, G.B.; Hrmova, M. Barley xyloglucan xyloglucosyl transferases bind xyloglucan-derived oligosaccharides in their acceptor-binding regions in multiple conformational states. Arch. Biochem. Biophys. 2010, 496, 61–68. [Google Scholar] [CrossRef]
- McGregor, N.; Yin, V.; Tung, C.C.; Van Petegem, F.; Brumer, H. Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases. Plant J. 2017, 89, 651–670. [Google Scholar] [CrossRef]
- Kaewthai, N.; Harvey, A.J.; Hrmova, M.; Brumer, H.; Ezcurra, I.; Teeri, T.T.; Fincher, G.B. Heterologous expression of diverse barley XTH genes in the yeast Pichia pastoris. Plant Biotechnol. 2010, 27, 251–258. [Google Scholar] [CrossRef]
- Behar, H.; Graham, S.W.; Brumer, H. Comprehensive cross-genome survey and phylogeny of glycoside hydrolase family 16 members reveals the evolutionary origin of EG 16 and XTH proteins in plant lineages. Plant J. 2018, 95, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C. Novel ’dot-blot’ assays for glycosyltransferases and glycosylhydrolases: Optimization for xyloglucan endotransglycosylase (XET) activity. Plant J. 1997, 11, 1141–1150. [Google Scholar] [CrossRef]
- Farkaš, V.; Ait-Mohand, F.; Stratilová, E. Sensitive detection of transglycosylating activity of xyloglucan endotransglycosylase/hydrolase (XTH) after isoelectric focusing in polyacrylamide gels. Plant Physiol. Biochem. 2005, 43, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Garajová, S.; Flodrová, D.; Ait-Mohand, F.; Farkaš, V.; Stratilová, E. Characterization of two partially purified xyloglucan endotransglycosylases from parsley (Petroselinum crispum) roots. Biologia 2008, 63, 313–319. [Google Scholar] [CrossRef]
- Kosík, O.; Auburn, R.P.; Russell, S.; Stratilová, E.; Garajová, S.; Hrmova, M.; Farkaš, V. Polysaccharide microarrays for high-throughput screening of transglycosylase activities in plant extracts. Glycoconj. J. 2010, 27, 79–87. [Google Scholar] [CrossRef]
- Vissenberg, K.; Martinez-Vilchez, I.M.; Verbelen, J.P.; Miller, J.G.; Fry, S.C. In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 2000, 12, 1229–1237. [Google Scholar] [CrossRef]
- Nishikubo, N.; Awano, T.; Banasiak, A.; Bourquin, V.; Ibatullin, F.; Funada, R.; Brumer, H.; Teeri, T.T.; Hayashi, T.; Sundberg, B.; et al. Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar—A glimpse into the mechanism of the balancing act of trees. Plant Cell Phys. 2007, 48, 843–855. [Google Scholar] [CrossRef]
- Ibatullin, F.M.; Banasiak, A.; Baumann, M.J.; Greffe, L.; Takahashi, J.; Mellerowicz, E.J.; Brumer, H.A. Real-time fluorogenic assay for the visualization of glycoside hydrolase activity in planta. Plant Phys. 2009, 151, 1741–1750. [Google Scholar] [CrossRef]
- Mravec, J.; Kračun, S.K.; Rydahl, M.G.; Westereng, B.; Miart, F.; Clausen, M.H.; Fangel, J.U.; Daugaard, M.; Van Cutsem, P.; De Fine Licht, H.H.; et al. Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 2014, 141, 4841–4850. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Farkaš, V.; Hrmova, M.; Fincher, G.B. Xyloglucan xyloglucosyl transferases from barley (Hordeum vulgare L.) bind oligomeric and polymeric xyloglucan molecules in their acceptor binding sites. Biochim. Biophys. Acta 2010, 1800, 674–684. [Google Scholar] [CrossRef]
- Kosík, O.; Garajová, S.; Matulová, M.; Řehulka, P.; Stratilová, E.; Farkaš, V. Effect of the label of oligosaccharide acceptors on the kinetic parameters of nasturtium seed xyloglucan endotransglycosylase (XET). Carbohydr. Res. 2011, 346, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Sulová, Z.; Lednická, M.; Farkaš, V. A colorimetric assay for xyloglucan endotransglycosylase from germinating seeds. Anal. Biochem. 1995, 229, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Morales-Quintana, L.; Carrasco-Orellana, C.; Beltrán, D.; Moya-León, M.A.; Herrera, R. Molecular insights of a xyloglucan endo-transglycosylase/hydrolase of radiata pine (PrXTH1) expressed in response to inclination: Kinetics and computational study. Plant Physiol. Biochem. 2019, 136, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Strohmeier, M.; Hrmova, M.; Fischer, M.; Harvey, A.J.; Fincher, J.B.; Pleiss, J. Molecular modeling of family GH16 glycoside hydrolases: Potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the Poaceae. Protein Sci. 2004, 13, 3200–3213. [Google Scholar] [CrossRef] [PubMed]
- Stratilová, E.; Ait-Mohand, F.; Řehulka, P.; Garajová, S.; Flodrová, D.; Řehulková, H.; Farkaš, V. Xyloglucan endotransglycosylases (XETs) from germinating nasturtium (Tropaeolum majus) seeds: Isolation and characterization of the major form. Plant Physiol. Biochem. 2010, 48, 207–215. [Google Scholar] [CrossRef]
- Zemková, Z.; Garajová, S.; Flodrová, D.; Řehulka, P.; Zelko, I.; Vadkertiová, R.; Farkaš, V.; Stratilová, E. Incorporation of β-(1,6)-linked glucooligosaccharides (pustulooligosaccharides) into plant cell wall structures. Chem. Pap. 2012, 66, 14–820. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Brummell, D.A.; Bennett, A.B. Two divergent xyloglucan endotransglycosylases exhibit mutually exclusive patterns of expression in nasturtium. Plant Physiol. 1996, 110, 493–499. [Google Scholar] [CrossRef]
- Herburger, K.; Ryan, L.M.; Popper, Z.A.; Holzinger, A. Localisation and substrate specificities of transglycanases in charophyte algae relate to development and morphology. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Schünmann, P.H.D.; Smith, R.C.; Lang, V.; Matthews, R.; Chandler, P.M. Expression of XET-related genes and its relation to elongation in leaves of barley (Hordeum vulgare L.). Plant Cell Environ. 1997, 20, 1439–1450. [Google Scholar] [CrossRef]
- Schröder, R.; Atkinson, R.G.; Langenkamper, G.; Redgwell, R.J. Biochemical and molecular characterisation of xyloglucan endotransglycosylase from ripe kiwifruit. Planta 1998, 204, 242–251. [Google Scholar] [CrossRef]
- Catala, C.; Rose, J.K.C.; York, W.S.; Albersheim, P.; Darvill, A.G.; Bennett, A.B. Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Physiol. 2001, 127, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Yokoyama, R.; Seki, M.; Ito, T.; Shinozaki, K.; Takahashi, T.; Komeda, Y.; Nishitani, K. AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J. 2005, 42, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Maris, A.; Kaewthai, N.; Eklöf, J.M.; Miller, J.G.; Brumer, H.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Differences in enzymatic properties of XTH proteins of Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 261–271. [Google Scholar] [CrossRef]
- Nardi, C.F.; Villarreal, N.M.; Opazo, M.C.; Martinez, G.A.; Moya León, M.A.; Civello, P.M. Expression of FaXTH1 and FaXTH2 genes in strawberry fruit. Cloning of promoter regions and effect of plant growth regulators. Sci. Hortic. 2014, 165, 111–122. [Google Scholar] [CrossRef]
- Han, Y.; Zhu, Q.; Zhang, Z.; Meng, K.; Hou, Y.; Ban, Q.; Suo, J.; Rao, J. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes and diverse roles of isoenzymes during persimmon fruit development and postharvest softening. PLoS ONE 2015, 10, e0123668. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ban, Q.; Hou, Y.; Meng, K.; Suo, J.; Rao, J. Isolation and characterization of two persimmon xyloglucan endotransglycosylase/hydrolase (XTH) genes that have divergent functions in cell wall modification and fruit postharvest softening. Front. Plant Sci. 2016, 7, 624. [Google Scholar] [CrossRef]
- Kushwah, S.; Banasiak, A.; Nishikubo, N.; Derba-Maceluch, M.; Majda, M.; Endo, S.; Kumar, V.; Gomez, L.; Gorzsas, A.; McQueen-Mason, S.; et al. Arabidopsis XTH4 and XTH9 contribute to wood cell expansion and secondary wall formation. Plant Physiol. 2020, 182, 1946–1965. [Google Scholar] [CrossRef]
- Redgwell, R.J.; Fry, S.C. Xyloglucan endotransglycosylase activity increases during kiwifruit (Actinidia deliciosa) ripening. Plant Physiol. 1993, 103, 1399–1406. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Bennett, A.B. Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999, 4, 176–183. [Google Scholar] [CrossRef]
- Oh, M.H.; Romanow, W.G.; Smith, R.C.; Zamski, E.; Sasse, J.; Clouse, S.D. Soybean BRU1 encodes a functional xyloglucan endotransglycosylase that is highly expressed in inner epicotyl tissues during brassinosteroid-promoted elongation. Plant Cell Physiol. 1998, 39, 124–130. [Google Scholar] [CrossRef]
- Campbell, P.; Braam, J. In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 1999, 18, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.E.; Fry, S.C. Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J. 2001, 26, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.J.; Fry, S.C. Bonds broken and formed during the mixed-linkage glucan:xyloglucan endotransglucosylase reaction catalysed by Equisetum hetero-trans-β-glucanase. Biochem. J. 2017, 474, 1055–1070. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.R.; Lou, H.; Aubert, M.K.; Wilkinson, L.G.; Little, A.; Houston, K.; Pinto, S.C.; Shirley, N.J. Exploring the role of cell wall-related genes and polysaccharides during plant development. Plants 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, H.; Yin, C.; Wang, X.; Jiang, Q.; Zhang, R.; Ge, F.; Chen, Y.; Yang, L. Genome-wide identification and characterization of xyloglucan endotransglycosylase/ hydrolase in Ananas comosus during Development. Genes 2019, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xie, F.; He, Q.; Li, J.; Liu, J.; Sun, B.; Luo, Y.; Zhang, Y.; Chen, Q.; Zhang, F.; et al. Expression analysis of XTH in stem swelling of stem mustard and selection of reference genes. Genes 2020, 11, 113. [Google Scholar] [CrossRef]
- Carpita, N.; McCann, M. The cell wall. In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Wilhelm, G., Jones, R.L., Eds.; American Society of Plant Physiologists: Rockville, IL, USA, 2000; pp. 52–108. ISBN 978-0-470-71421-8. [Google Scholar]
- Thompson, D.S. How do cell walls regulate plant growth? J. Exp. Bot. 2005, 56, 2275–2285. [Google Scholar] [CrossRef]
- Bulone, V.; Schwerdt, J.G.; Fincher, G.B. Co-evolution of enzymes involved in plant cell wall metabolism in the grasses. Front. Plant Sci. 2009, 10, 1009. [Google Scholar] [CrossRef]

| Enzyme | Plant Source | Enzyme Purity a | Donor b | Acceptor b | Assay Method c Reference |
|---|---|---|---|---|---|
| HvXET3 | Hordeum vulgare L. | + | XG, HEC | XG-OS, MLG-OS, Cello-OS, Pu-OS, AraXyl-OS, La-OS, Xyl-OS, GlcMan-OS, Ara-OS, [α(1-4)GalAp]5 | R, F [125,144,154] |
| HvXET4 | Hordeum vulgare L. | + | XG, HEC | XG-OS, MLG-OS, Cello-OS, Pu-OS, AraXyl-OS, La-OS, Xyl-OS, GlcMan-OS, Ara-OS, [α(1-4)GalAp]5 | R, F [125,144,154] |
| HvXET5 | Hordeum vulgare L. | + | XG, CMC, HEC, MLG | XG-OS, Cello-OS | R, F [58] |
| HvXET6 | Hordeum vulgare L. | + | XG, CMC, HEC, MLG | XG-OS, MLG-OS, Cello-OS, Pu-OS, AraXyl-OS, La-OS, Xyl-OS, GlcMan-OS, Ara-OS, [α(1-4)GalAp]5 | R, F [121,125,144,154] |
| TmXET6.3 | Tropaeolum majus L. | +/− | XG, HEC | XG-OS, MLG-OS, Cello-OS, Pu-OS, AraXyl-OS, La-OS, Xyl-OS, GlcMan-OS, Ara-OS | F [123,159] |
| PttXET16A | Populus tremulus x tremuloides L. | + | XG | XG-OS | C [127,138] |
| PrXTH1 | Pinus radiata L. | − | XG | XG-OS, Cello-OS | C [157] |
| EfXTH-A | Equisetum fluviatile L. | − | XG, cellulose, MLG | XG-OS | R [117] |
| EfXTH-H | Equisetum fluviatile L. | − | XG, cellulose, MLG | XG-OS | R [117] |
| EfXTH-I | Equisetum fluviatile L. | − | XG, cellulose, MLG | XG-OS | R [117] |
| EfHTG | Equisetum fluviatile L. | +/− | XG, cellulose, MLG | XG-OS | R, F [136] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stratilová, B.; Kozmon, S.; Stratilová, E.; Hrmova, M. Plant Xyloglucan Xyloglucosyl Transferases and the Cell Wall Structure: Subtle but Significant. Molecules 2020, 25, 5619. https://doi.org/10.3390/molecules25235619
Stratilová B, Kozmon S, Stratilová E, Hrmova M. Plant Xyloglucan Xyloglucosyl Transferases and the Cell Wall Structure: Subtle but Significant. Molecules. 2020; 25(23):5619. https://doi.org/10.3390/molecules25235619
Chicago/Turabian StyleStratilová, Barbora, Stanislav Kozmon, Eva Stratilová, and Maria Hrmova. 2020. "Plant Xyloglucan Xyloglucosyl Transferases and the Cell Wall Structure: Subtle but Significant" Molecules 25, no. 23: 5619. https://doi.org/10.3390/molecules25235619
APA StyleStratilová, B., Kozmon, S., Stratilová, E., & Hrmova, M. (2020). Plant Xyloglucan Xyloglucosyl Transferases and the Cell Wall Structure: Subtle but Significant. Molecules, 25(23), 5619. https://doi.org/10.3390/molecules25235619







