Mechanism of Type IA Topoisomerases
Abstract
1. Introduction
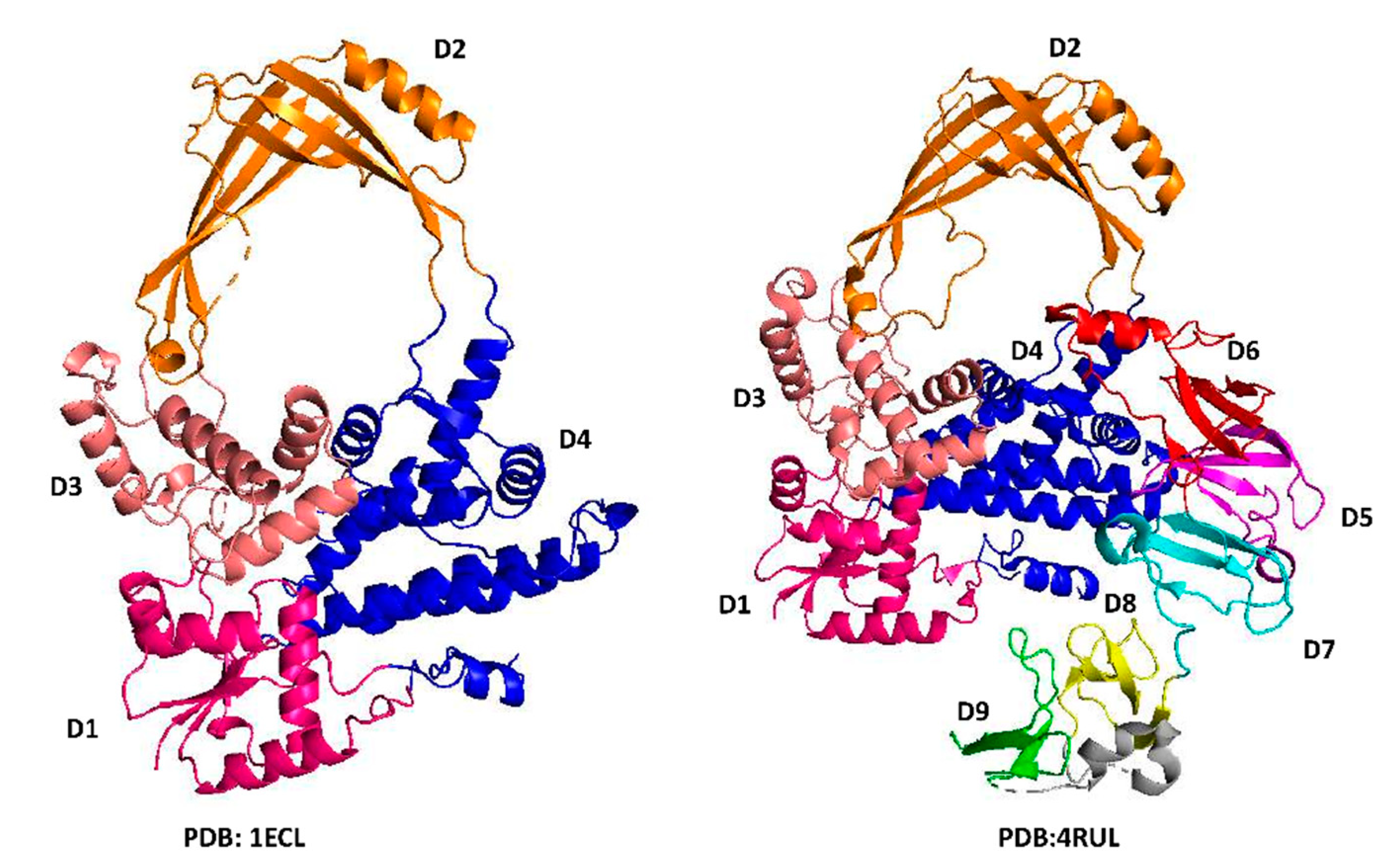
2. General Classification
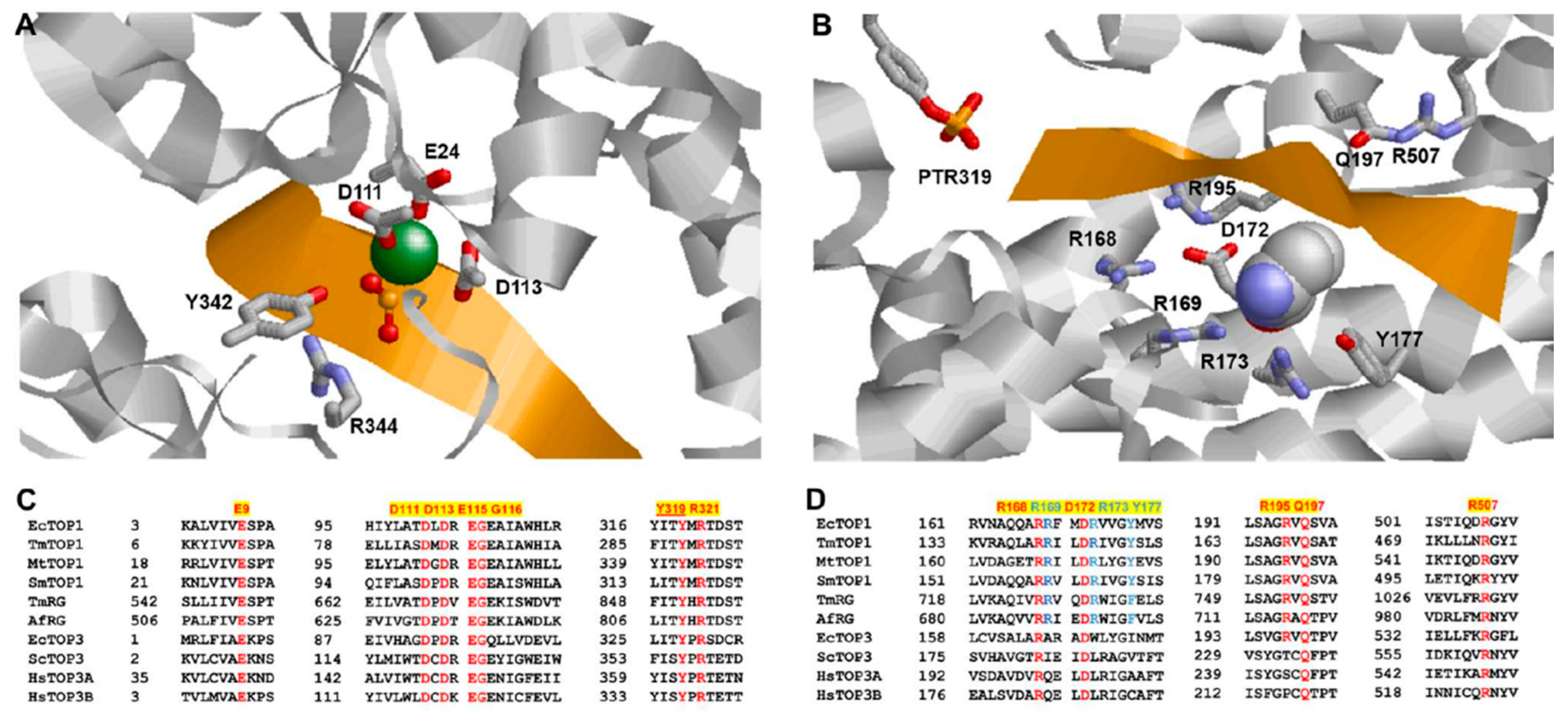
3. N-Terminal Type IA Core Domains—Binding and Positioning of the G-strand for Cleavage at the Active Site
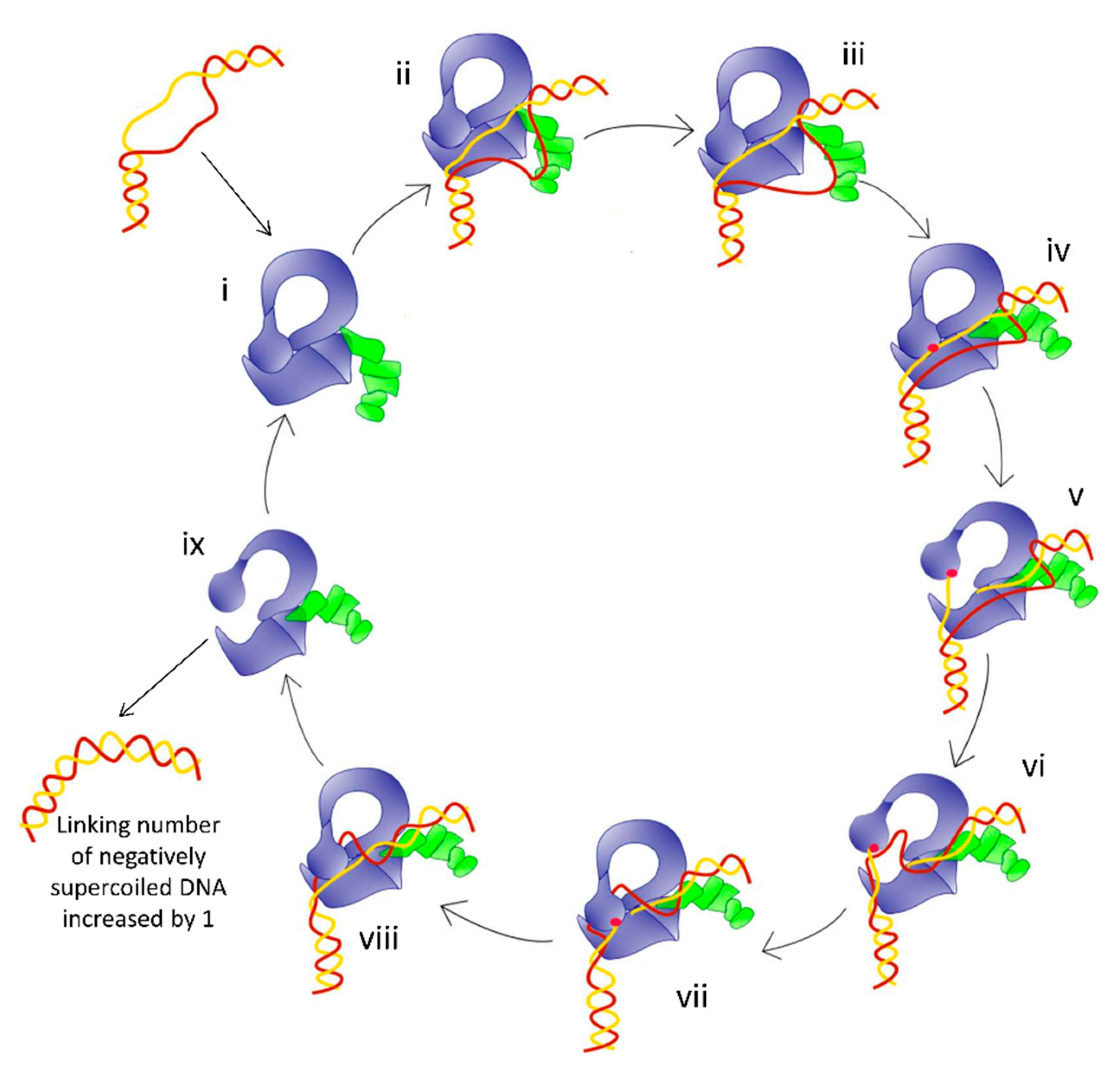
4. Mechanism of the G-strand Cleavage and Religation
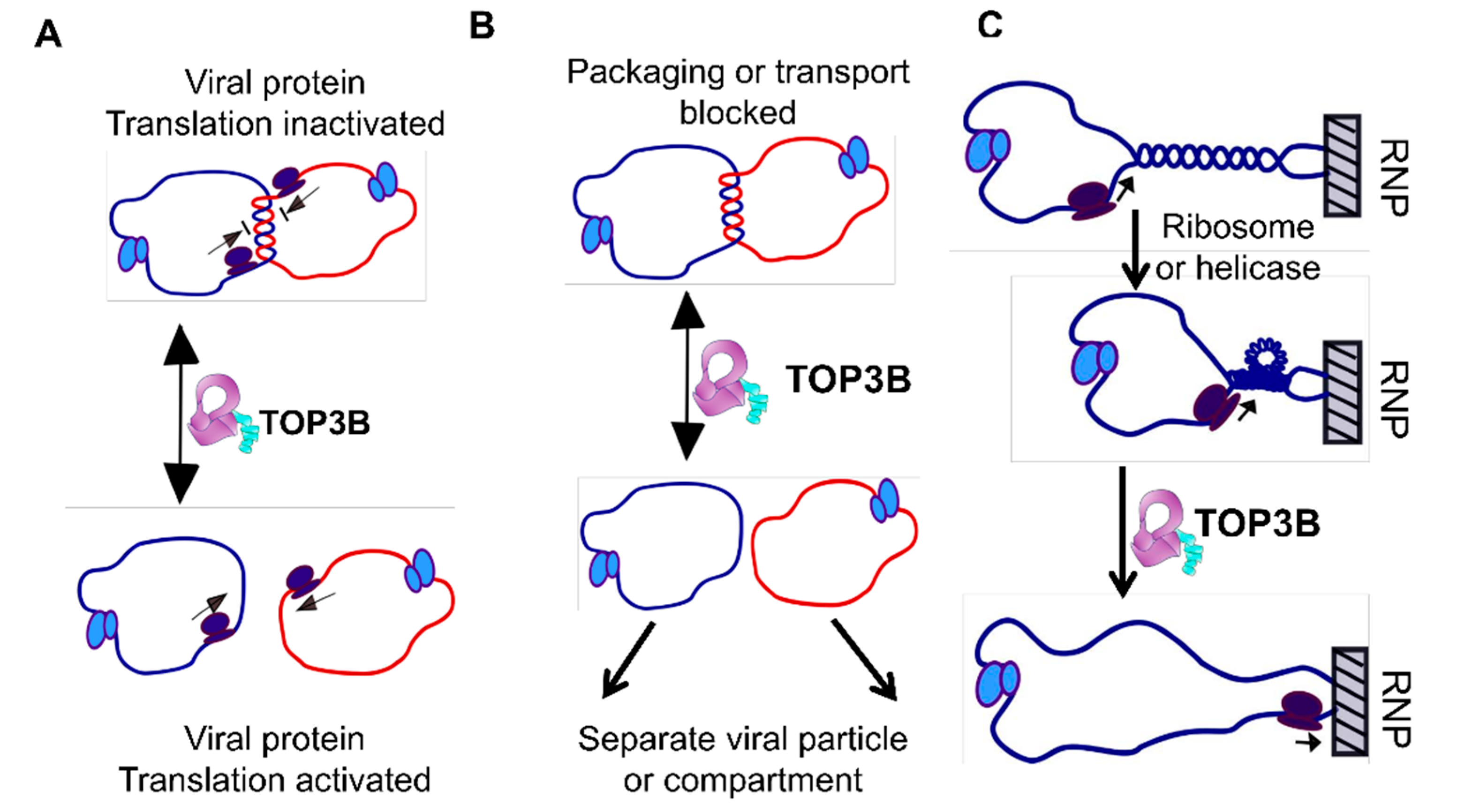
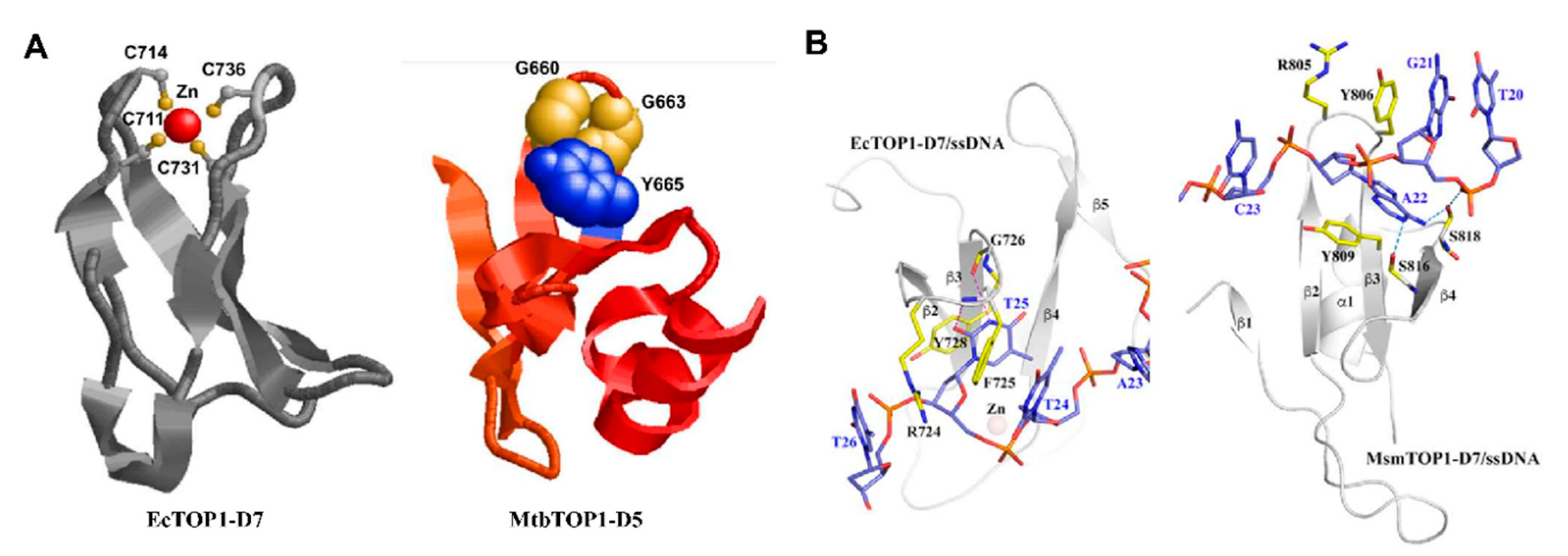
5. C-Terminal Domains—Binding of T-strand
6. Opening and Closing of the G-strand Gate for Strand Passage
7. Key Remaining Questions
Author Contributions
Funding
Conflicts of Interest
References
- Burgers, P.M.J. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 1998, 107, 218–227. [Google Scholar] [CrossRef]
- Nikolov, D.B.; Burley, S.K. RNA polymerase II transcription initiation: A structural view. Proc. Natl. Acad. Sci. USA 1997, 94, 15–22. [Google Scholar] [CrossRef]
- Wang, J.C. DNA Topoisomerases. Annu. Rev. Biochem. 1985, 54, 665–697. [Google Scholar] [CrossRef]
- Champoux, J.J. DNA Topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef]
- Schoeffler, A.J.; Berger, J.M. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008, 41, 41–101. [Google Scholar] [CrossRef]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011, 12, 827–841. [Google Scholar] [CrossRef]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef]
- Chen, S.H.; Chan, N.-L.; Hsieh, T.-S. New Mechanistic and Functional Insights into DNA Topoisomerases. Annu. Rev. Biochem. 2013, 82, 139–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Liu, C.-C.; Alberts, B.M. Type II DNA topoisomerases: Enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 1980, 19, 697–707. [Google Scholar] [CrossRef]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, M.D. DNA supercoiling during transcription. Biophys. Rev. 2016, 8, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-Y.; Shyy, S.; Wang, J.C.; Liu, L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell 1988, 53, 433–440. [Google Scholar] [CrossRef]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef]
- Brochu, J.; Breton, E.V.; Drolet, M. Supercoiling, R-Loops, Replication and the Functions of Bacterial Type 1A Topoisomerases. Genes 2020, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.; Levine, C.; Hassing, H.; Marians, K.J. Topoisomerase III Can Serve as the Cellular Decatenase inEscherichia coli. J. Biol. Chem. 2002, 278, 8653–8660. [Google Scholar] [CrossRef]
- Bizard, A.H.; Yang, X.; Débat, H.; Fogg, J.M.; Zechiedrich, L.; Strick, T.R.; Garnier, F.; Nadal, M. TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase. Nucleic Acids Res. 2017, 46, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, T.J.; Nadalutti, C.A.; Motori, E.; Sommerville, E.W.; Gorman, G.S.; Basu, S.; Hoberg, E.; Turnbull, D.M.; Chinnery, P.F.; Larsson, N.-G.; et al. Topoisomerase 3α Is Required for Decatenation and Segregation of Human mtDNA. Mol. Cell 2018, 69, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Rani, P.; Nagaraja, V. Genome-wide mapping of Topoisomerase I activity sites reveal its role in chromosome segregation. Nucleic Acids Res. 2019, 47, 1416–1427. [Google Scholar] [CrossRef]
- Lee, C.M.; Wang, G.; Pertsinidis, A.; Marians, K.J. Topoisomerase III Acts at the Replication Fork To Remove Precatenanes. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Liu, L.F.; DePew, R.E.; Wang, J.C. Knotted single-stranded DNA rings: A novel topological isomer of circular single-stranded DNA formed by treatment with Escherichia coli ω protein. J. Mol. Biol. 1976, 106, 439–452. [Google Scholar] [CrossRef]
- Brown, P.O.; Cozzarelli, N.R. Catenation and knotting of duplex DNA by type 1 topoisomerases: A mechanistic parallel with type 2 topoisomerases. Proc. Natl. Acad. Sci. USA 1981, 78, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Di Gate, R.J.; Seeman, N.C. An RNA topoisomerase. Proc. Natl. Acad. Sci. USA 1996, 93, 9477–9482. [Google Scholar] [CrossRef] [PubMed]
- Stoll, G.; Pietiläinen, O.P.H.; Linder, B.; Suvisaari, J.; Brosi, C.; Hennah, W.; Leppa, V.; Torniainen, M.; Ripatti, S.; Ala-Mello, S.; et al. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013, 16, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Shen, W.; Guo, R.; Xue, Y.; Peng, W.; Sima, J.; Yang, J.; Sharov, A.; Srikantan, S.; Yang, J.; et al. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013, 16, 1238–1247. [Google Scholar] [CrossRef]
- Ahmad, M.; Shen, W.; Li, W.; Xue, Y.; Zou, S.; Xu, D.; Wang, W. Topoisomerase 3β is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic Acids Res. 2017, 45, 2704–2713. [Google Scholar] [CrossRef]
- Kaufman, C.S.; Genovese, A.; Butler, M.G. Deletion of TOP3B Is Associated with Cognitive Impairment and Facial Dysmorphism. Cytogenet. Genome Res. 2016, 150, 106–111. [Google Scholar] [CrossRef]
- Daghsni, M.; Lahbib, S.; Fradj, M.; Sayeb, M.; Kelmemi, W.; Kraoua, L.; Kchaou, M.; Maazoul, F.; Echebbi, S.; Ben Ali, N.; et al. TOP3B: A Novel Candidate Gene in Juvenile Myoclonic Epilepsy? Cytogenet. Genome Res. 2018, 154, 1–5. [Google Scholar] [CrossRef]
- Ahmad, M.; Xue, Y.; Lee, S.K.; Martindale, J.L.; Shen, W.; Li, W.; Zou, S.; Ciaramella, M.; Débat, H.; Nadal, M.; et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016, 44, 6335–6349. [Google Scholar] [CrossRef]
- Ahmad, M.; Xu, D.; Wang, W. Type IA topoisomerases can be “magicians” for both DNA and RNA in all domains of life. RNA Biol. 2017, 14, 854–864. [Google Scholar] [CrossRef]
- Rani, P.; Kalladi, S.; Bansia, H.; Rao, S.; Jha, R.K.; Jain, P.; Bhaduri, T.; Nagaraja, V. A Type IA DNA/RNA topoisomerase with RNA hydrolysis activity participates in ribosomal RNA processing. J. Mol. Biol. 2020. [Google Scholar] [CrossRef]
- Yang, Y.; McBride, K.M.; Hensley, S.; Lu, Y.; Chedin, F.; Bedford, M.T. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol. Cell 2014, 53, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, Z.; Narayanan, N.; Yang, Y. Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 2018, 46, 3061–3074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wallis, M.; Petrovic, V.; Challis, J.; Kalitsis, P.; Hudson, D.F. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019, 9, 190222. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Xue, Y.; Shen, W.; Zhang, Y.; Joo, Y.; Ahmad, M.; Chinen, M.; Ding, Y.; Ku, W.L.; De, S.; et al. Topoisomerase 3β interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat. Commun. 2018, 9, 4946. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K.R.; Hirano, M.; Fagg, W.S.; McAnarney, E.T.; Shan, C.; Xie, X.; Hage, A.; Pietzsch, C.A.; Bukreyev, A.; Rajsbaum, R.; et al. Topoisomerase III-ss is required for efficient replication of positive-sense RNA viruses. Antivir. Res. 2020, 182, 104874. [Google Scholar] [CrossRef]
- Siaw, G.E.-L.; Liu, I.-F.; Lin, P.-Y.; Been, M.D.; Hsieh, T.-S. DNA and RNA topoisomerase activities of Top3β are promoted by mediator protein Tudor domain-containing protein 3. Proc. Natl. Acad. Sci. USA 2016, 113, E5544–E5551. [Google Scholar] [CrossRef]
- Diosa-Toro, M.; Prasanth, K.R.; Bradrick, S.S.; Garcia-Blanco, M.A. Role of RNA-binding proteins during the late stages of Flavivirus replication cycle. Virol. J. 2020, 17, 60. [Google Scholar] [CrossRef]
- Bizard, A.H.; Hickson, I.D. The many lives of type IA topoisomerases. J. Biol. Chem. 2020, 295, 7138–7153. [Google Scholar] [CrossRef]
- Bergerat, A.; Gadelle, D.; Forterre, P. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 1994, 269, 27663–27669. [Google Scholar]
- Gadelle, D.; Krupovic, M.; Raymann, K.; Mayer, C.; Forterre, P. DNA topoisomerase VIII: A novel subfamily of type IIB topoisomerases encoded by free or integrated plasmids in Archaea and Bacteria. Nucleic Acids Res. 2014, 42, 8578–8591. [Google Scholar] [CrossRef]
- Forterre, P.; Gadelle, D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009, 37, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, K.; Wang, J.C. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 1985, 185, 625–637. [Google Scholar] [CrossRef]
- Dekker, N.H.; Rybenkov, V.V.; Duguet, M.; Crisona, N.J.; Cozzarelli, N.R.; Bensimon, D.; Croquette, V. The mechanism of type IA topoisomerases. Proc. Natl. Acad. Sci. USA 2002, 99, 12126–12131. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.D.; Wang, J.C.; Mondragón, A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nat. Cell Biol. 1994, 367, 138–146. [Google Scholar] [CrossRef]
- Taneja, B.; Schnurr, B.; Slesarev, A.; Marko, J.F.; Mondragón, A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 14670–14675. [Google Scholar] [CrossRef]
- Stewart, L.; Redinbo, M.R.; Qiu, X.; Hol, W.G.J.; Champoux, J.J. A Model for the Mechanism of Human Topoisomerase I. Science 1998, 279, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.M.; Rajan, R.; Mondragón, A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009, 37, 693–701. [Google Scholar] [CrossRef]
- Mondragón, A.; DiGate, R. The structure of Escherichia coli DNA topoisomerase III. Structure 1999, 7, 1373–1383. [Google Scholar] [CrossRef][Green Version]
- Hansen, G.; Harrenga, A.; Wieland, B.; Schomburg, I.; Reinemer, P. Crystal Structure of Full Length Topoisomerase I from Thermotoga maritima. J. Mol. Biol. 2006, 358, 1328–1340. [Google Scholar] [CrossRef]
- Tan, K.; Cao, N.; Cheng, B.; Joachimiak, A.; Tse-Dinh, Y.-C. Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold. J. Mol. Biol. 2016, 428, 182–193. [Google Scholar] [CrossRef]
- Jones, J.A.; Hevener, K.E. Crystal structure of the 65-kilodalton amino-terminal fragment of DNA topoisomerase I from the gram-positive model organism Streptococcus mutans. Biochem. Biophys. Res. Commun. 2019, 516, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Cao, N.; Tan, K.; Zuo, X.; Annamalai, T.; Tse-Dinh, Y.-C. Mechanistic insights from structure of Mycobacterium smegmatis topoisomerase I with ssDNA bound to both N- and C-terminal domains. Nucleic Acids Res. 2020, 48, 4448–4462. [Google Scholar] [CrossRef] [PubMed]
- Bocquet, N.; Bizard, A.H.; Abdulrahman, W.; Larsen, N.B.; Faty, M.; Cavadini, S.; Bunker, R.D.; Kowalczykowski, S.C.; Cejka, P.; Hickson, I.D.; et al. Structural and mechanistic insight into Holliday-junction dissolution by Topoisomerase IIIα and RMI1. Nat. Struct. Mol. Biol. 2014, 21, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Goto-Ito, S.; Yamagata, A.; Takahashi, T.S.; Sato, Y.; Fukai, S. Structural basis of the interaction between Topoisomerase IIIβ and the TDRD3 auxiliary factor. Sci. Rep. 2017, 7, 42123. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.C.; Stock, D. Crystal structure of reverse gyrase: Insights into the positive supercoiling of DNA. EMBO J. 2002, 21, 418–426. [Google Scholar] [CrossRef]
- Rudolph, M.G.; del Duany, Y.T.; Jungblut, S.P.; Ganguly, A.; Klostermeier, D. Crystal structures of Thermotoga maritima reverse gyrase: Inferences for the mechanism of positive DNA supercoiling. Nucleic Acids Res. 2012, 41, 1058–1070. [Google Scholar] [CrossRef]
- Changela, A.; DiGate, R.J.; Mondragón, A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature 2001, 411, 1077–1081. [Google Scholar] [CrossRef]
- Perry, K.; Mondragón, A. Structure of a Complex between E. coli DNA Topoisomerase I and Single-Stranded DNA. Structure 2003, 11, 1349–1358. [Google Scholar] [CrossRef]
- Cheng, B.; Annamalai, T.; Sorokin, E.; Abrenica, M.; Aedo, S.; Tse-Dinh, Y.-C. Asp-to-Asn Substitution at the First Position of the DxD TOPRIM Motif of Recombinant Bacterial Topoisomerase I Is Extremely Lethal to E. coli. J. Mol. Biol. 2009, 385, 558–567. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, B.; Tse-Dinh, Y.-C. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 2011, 108, 6939–6944. [Google Scholar] [CrossRef]
- Tan, K.; Zhou, Q.; Cheng, B.; Zhang, Z.; Joachimiak, A.; Tse-Dinh, Y.-C. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res. 2015, 43, 11031–11046. [Google Scholar] [CrossRef]
- Cao, N.; Tan, K.; Annamalai, T.; Joachimiak, A.; Tse-Dinh, Y.-C. Investigating mycobacterial topoisomerase I mechanism from the analysis of metal and DNA substrate interactions at the active site. Nucleic Acids Res. 2018, 46, 7296–7308. [Google Scholar] [CrossRef]
- Cheng, B.; Feng, J.; Gadgil, S.; Tse-Dinh, Y.-C. Flexibility at Gly-194 Is Required for DNA Cleavage and Relaxation Activity ofEscherichia coliDNA Topoisomerase I. J. Biol. Chem. 2004, 279, 8648–8654. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Feng, J.; Mulay, V.; Gadgil, S.; Tse-Dinh, Y.-C. Site-directed Mutagenesis of Residues Involved in G Strand DNA Binding byEscherichia coliDNA Topoisomerase I. J. Biol. Chem. 2004, 279, 39207–39213. [Google Scholar] [CrossRef]
- Bachar, A.; Itzhaki, E.; Gleizer, S.; Shamshoom, M.; Milo, R.; Antonovsky, N. Point mutations in topoisomerase I alter the mutation spectrum in E. coli and impact the emergence of drug resistance genotypes. Nucleic Acids Res. 2020, 48, 761–769. [Google Scholar] [CrossRef]
- Tse, Y.C.; Kirkegaard, K.; Wang, J.C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J. Biol. Chem. 1980, 255, 5560–5565. [Google Scholar] [PubMed]
- Kovalsky, O.I.; Kozyavkin, S.A.; Slesarev, A.I.; Koyyavkin, S. Archaebacterial reverse gyrase cleavage-site specificity is similar to that of eubacterial DNA topoisomerases I. Nucleic Acids Res. 1990, 18, 2801–2805. [Google Scholar] [CrossRef]
- Annamalai, T.; Dani, N.; Cheng, B.; Tse-Dinh, Y.-C. Analysis of DNA relaxation and cleavage activities of recombinant Mycobacterium tuberculosis DNA topoisomerase I from a new expression and purification protocol. BMC Biochem. 2009, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Narula, G.; Tse-Dinh, Y.-C. Residues of E. coli topoisomerase I conserved for interaction with a specific cytosine base to facilitate DNA cleavage. Nucleic Acids Res. 2012, 40, 9233–9243. [Google Scholar] [CrossRef]
- Liu, D.; Shao, Y.; Chen, G.; Tse-Dinh, Y.-C.; Piccirilli, J.A.; Weizmann, Y. Synthesizing topological structures containing RNA. Nat. Commun. 2017, 8, 14936. [Google Scholar] [CrossRef]
- Viard, T.; De La Tour, C.B. Type IA topoisomerases: A simple puzzle? Biochimie 2007, 89, 456–467. [Google Scholar] [CrossRef]
- Terekhova, K.; Marko, J.F.; Mondragón, A. Studies of bacterial topoisomerases I and III at the single-molecule level. Biochem. Soc. Trans. 2013, 41, 571–575. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, C.-X.; Roche, C.J.; Papanicolaou, N.; Dipietrantonio, A.; Tse-Dinh, Y.-C. Site-directed Mutagenesis of Conserved Aspartates, Glutamates and Arginines in the Active Site Region ofEscherichia coliDNA Topoisomerase I. J. Biol. Chem. 1998, 273, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Wang, J.C. Identification of Active Site Residues inEscherichia coliDNA Topoisomerase I. J. Biol. Chem. 1998, 273, 6050–6056. [Google Scholar] [CrossRef]
- Roston, D.; Demapan, D.; Cui, Q. Extensive free-energy simulations identify water as the base in nucleotide addition by DNA polymerase. Proc. Natl. Acad. Sci. USA 2019, 116, 25048–25056. [Google Scholar] [CrossRef] [PubMed]
- Narula, G.; Annamalai, T.; Aedo, S.; Cheng, B.; Sorokin, E.; Wong, A.; Tse-Dinh, Y.-C. The Strictly Conserved Arg-321 Residue in the Active Site of Escherichia coli Topoisomerase I Plays a Critical Role in DNA Rejoining. J. Biol. Chem. 2011, 286, 18673–18680. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Sun, Y.; Huang, S.-Y.; Jo, U.; Zhang, H.; Tse-Dinh, Y.-C.; Pommier, Y. Topoisomerase 3B (TOP3B) DNA and RNA Cleavage Complexes and Pathway to Repair TOP3B-Linked RNA and DNA Breaks. Mol. Cell 2020. [Google Scholar] [CrossRef]
- Aravind, L.; Leipe, D.D.; Koonin, E.V. Toprim—A conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998, 26, 4205–4213. [Google Scholar] [CrossRef]
- Bhat, A.G.; Leelaram, M.N.; Hegde, S.M.; Nagaraja, V. Deciphering the Distinct Role for the Metal Coordination Motif in the Catalytic Activity of Mycobacterium smegmatis Topoisomerase I. J. Mol. Biol. 2009, 393, 788–802. [Google Scholar] [CrossRef]
- Cheng, B.; Sorokin, E.P.; Tse-Dinh, Y.-C. Mutation adjacent to the active site tyrosine can enhance DNA cleavage and cell killing by the TOPRIM Gly to Ser mutant of bacterial topoisomerase I. Nucleic Acids Res. 2008, 36, 1017–1025. [Google Scholar] [CrossRef]
- Cheng, B.; Shukla, S.; Vasunilashorn, S.; Mukhopadhyay, S.; Tse-Dinh, Y.-C. Bacterial Cell Killing Mediated by Topoisomerase I DNA Cleavage Activity. J. Biol. Chem. 2005, 280, 38489–38495. [Google Scholar] [CrossRef]
- Narula, G.; Becker, J.; Cheng, B.; Dani, N.; Abrenica, M.V.; Tse-Dinh, Y.-C. The DNA relaxation activity and covalent complex accumulation of Mycobacterium tuberculosis topoisomerase I can be assayed in Escherichia coli: Application for identification of potential FRET-dye labeling sites. BMC Biochem. 2010, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Perry, K.; Mondragón, A. Biochemical Characterization of an Invariant Histidine Involved inEscherichia coliDNA Topoisomerase I Catalysis. J. Biol. Chem. 2002, 277, 13237–13245. [Google Scholar] [CrossRef] [PubMed]
- Strzałka, A.; Szafran, M.J.; Strick, T.; Jakimowicz, D. C-terminal lysine repeats in Streptomyces topoisomerase I stabilize the enzyme-DNA complex and confer high enzyme processivity. Nucleic Acids Res. 2017, 45, 11908–11924. [Google Scholar] [CrossRef]
- Szafran, M.J.; Strick, T.; Strzałka, A.; Zakrzewska-Czerwińska, J.; Jakimowicz, D. A highly processive topoisomerase I: Studies at the single-molecule level. Nucleic Acids Res. 2014, 42, 7935–7946. [Google Scholar] [CrossRef]
- Ahmed, W.; Bhat, A.G.; Leelaram, M.N.; Menon, S.; Nagaraja, V. Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage. Nucleic Acids Res. 2013, 41, 7462–7471. [Google Scholar] [CrossRef] [PubMed]
- Beran-Steed, R.K.; Tse-Dinh, Y.-C. The carboxyl terminal domain ofEscherichia coli DNA topoisomerase I confers higher affinity to DNA. Proteins 1989, 6, 249–258. [Google Scholar] [CrossRef]
- Grishin, N.V. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J. Mol. Biol. 2000, 299, 1165–1177. [Google Scholar] [CrossRef]
- Yu, L.; Zhu, C.-X.; Tse-Dinh, Y.-C.; Fesik, S.W. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry 1995, 34, 7622–7628. [Google Scholar] [CrossRef]
- Zumstein, L.; Wang, J.C. Probing the structural domains and function in vivo of Escherichia coli DNA topoisomerase I by mutagenesis. J. Mol. Biol. 1986, 191, 333–340. [Google Scholar] [CrossRef]
- Viard, T.; Cossard, R.; Duguet, M.; De La Tour, C.B. Thermotoga maritima-Escherichia coli chimeric topoisomerases. Answers about involvement of the carboxyl-terminal domain in DNA topoisomerase I-mediated catalysis. J. Biol. Chem. 2004, 279, 30073–30080. [Google Scholar] [CrossRef] [PubMed]
- Ahumada, A.; Tse-Dinh, Y.-C. The Zn(II) Binding Motifs of E. coli DNA Topoisomerase I Is Part of a High-Affinity DNA Binding Domain. Biochem. Biophys. Res. Commun. 1998, 251, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.X.; Samuel, M.; Pound, A.; Ahumada, A.; Tse-Dinh, Y.C. Expression and DNA-binding properties of the 14K carboxyl terminal fragment of Escherichia coli DNA topoisomerase I. Biochem. Mol. Boil. Int. 1995, 35, 375–385. [Google Scholar]
- Dorn, A.; Röhrig, S.; Papp, K.; Schröpfer, S.; Hartung, F.; Knoll, A.; Puchta, H. The topoisomerase 3α zinc-finger domain T1 of Arabidopsis thaliana is required for targeting the enzyme activity to Holliday junction-like DNA repair intermediates. PLoS Genet. 2018, 14, e1007674. [Google Scholar] [CrossRef]
- Ahumada, A.; Tse-Dinh, Y.-C. The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I. BMC Biochem. 2002, 3, 13. [Google Scholar] [CrossRef]
- Wilson, T.M.; Chen, A.D.; Hsieh, T. Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J. Biol. Chem. 2000, 275, 1533–1540. [Google Scholar] [CrossRef]
- Godin, K.S.; Varani, G. How Arginine-Rich Domains Coordinate mRNA Maturation Events. RNA Biol. 2007, 4, 69–75. [Google Scholar] [CrossRef][Green Version]
- Zhou, Q.; Zhou, Y.N.; Jin, D.J.; Tse-Dinh, Y.-C. Deacetylation of topoisomerase I is an important physiological function of E. coli CobB. Nucleic Acids Res. 2017, 45, 5349–5358. [Google Scholar] [CrossRef]
- Zhou, Q.; Hernandez, M.E.G.; Fernandez-Lima, F.; Tse-Dinh, Y.-C. Biochemical Basis of E. coli Topoisomerase I Relaxation Activity Reduction by Nonenzymatic Lysine Acetylation. Int. J. Mol. Sci. 2018, 19, 1439. [Google Scholar] [CrossRef]
- Tiwari, P.B.; Chapagain, P.P.; Banda, S.; Darici, Y.; Uren, A.; Tse-Dinh, Y.-C. Characterization of molecular interactions between Escherichia coli RNA polymerase and topoisomerase I by molecular simulations. FEBS Lett. 2016, 590, 2844–2851. [Google Scholar] [CrossRef]
- Banda, S.; Cao, N.; Tse-Dinh, Y.-C. Distinct Mechanism Evolved for Mycobacterial RNA Polymerase and Topoisomerase I Protein–Protein Interaction. J. Mol. Biol. 2017, 429, 2931–2942. [Google Scholar] [CrossRef]
- Cheng, B.; Zhu, C.-X.; Ji, C.; Ahumada, A.; Tse-Dinh, Y.-C. Direct Interaction between Escherichia coli RNA Polymerase and the Zinc Ribbon Domains of DNA Topoisomerase I. J. Biol. Chem. 2003, 278, 30705–30710. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Sala, C.; Hegde, S.R.; Jha, R.K.; Cole, S.T.; Nagaraja, V. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet. 2017, 13, e1006754. [Google Scholar] [CrossRef] [PubMed]
- Drolet, M. Growth inhibition mediated by excess negative supercoiling: The interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006, 59, 723–730. [Google Scholar] [CrossRef]
- Massé, E.; Drolet, M. Escherichia coli DNA Topoisomerase I Inhibits R-loop Formation by Relaxing Transcription-induced Negative Supercoiling. J. Biol. Chem. 1999, 274, 16659–16664. [Google Scholar] [CrossRef]
- Kondekar, S.M.; Gunjal, G.V.; Radicella, J.P.; Rao, D.N. Molecular dissection of Helicobacter pylori Topoisomerase I reveals an additional active site in the carboxyl terminus of the enzyme. DNA Repair 2020, 91–92, 102853. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.-X.; Roche, C.J.; Tse-Dinh, Y.-C. Effect of Mg(II) Binding on the Structure and Activity ofEscherichia coliDNA Topoisomerase I. J. Biol. Chem. 1997, 272, 16206–16210. [Google Scholar] [CrossRef]
- Leelaram, M.N.; Bhat, A.G.; Godbole, A.A.; Bhat, R.S.; Manjunath, R.; Nagaraja, V. Type IA topoisomerase inhibition by clamp closure. FASEB J. 2013, 27, 3030–3038. [Google Scholar] [CrossRef]
- Fouque, K.J.D.; Garabedian, A.; Leng, F.; Tse-Dinh, Y.-C.; Fernandez-Lima, F. Microheterogeneity of Topoisomerase IA/IB and Their DNA-Bound States. ACS Omega 2019, 4, 3619–3626. [Google Scholar] [CrossRef]
- Mills, M.; Tse-Dinh, Y.-C.; Neuman, K.C. Direct observation of topoisomerase IA gate dynamics. Nat. Struct. Mol. Biol. 2018, 25, 1111–1118. [Google Scholar] [CrossRef]
- Li, Z.; Mondragon, A.; Hiasa, H.; Marians, K.J.; DiGate, R.J. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol. Microbiol. 2000, 35, 888–895. [Google Scholar] [CrossRef]
- Gunn, K.H.; Marko, J.F.; Mondragón, A. An orthogonal single-molecule experiment reveals multiple-attempt dynamics of type IA topoisomerases. Nat. Struct. Mol. Biol. 2017, 24, 484–490. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dasgupta, T.; Ferdous, S.; Tse-Dinh, Y.-C. Mechanism of Type IA Topoisomerases. Molecules 2020, 25, 4769. https://doi.org/10.3390/molecules25204769
Dasgupta T, Ferdous S, Tse-Dinh Y-C. Mechanism of Type IA Topoisomerases. Molecules. 2020; 25(20):4769. https://doi.org/10.3390/molecules25204769
Chicago/Turabian StyleDasgupta, Tumpa, Shomita Ferdous, and Yuk-Ching Tse-Dinh. 2020. "Mechanism of Type IA Topoisomerases" Molecules 25, no. 20: 4769. https://doi.org/10.3390/molecules25204769
APA StyleDasgupta, T., Ferdous, S., & Tse-Dinh, Y.-C. (2020). Mechanism of Type IA Topoisomerases. Molecules, 25(20), 4769. https://doi.org/10.3390/molecules25204769




