Antimicrobial Activity of Protein Fraction from Naja ashei Venom against Staphylococcus epidermidis
Abstract
1. Introduction
2. Results
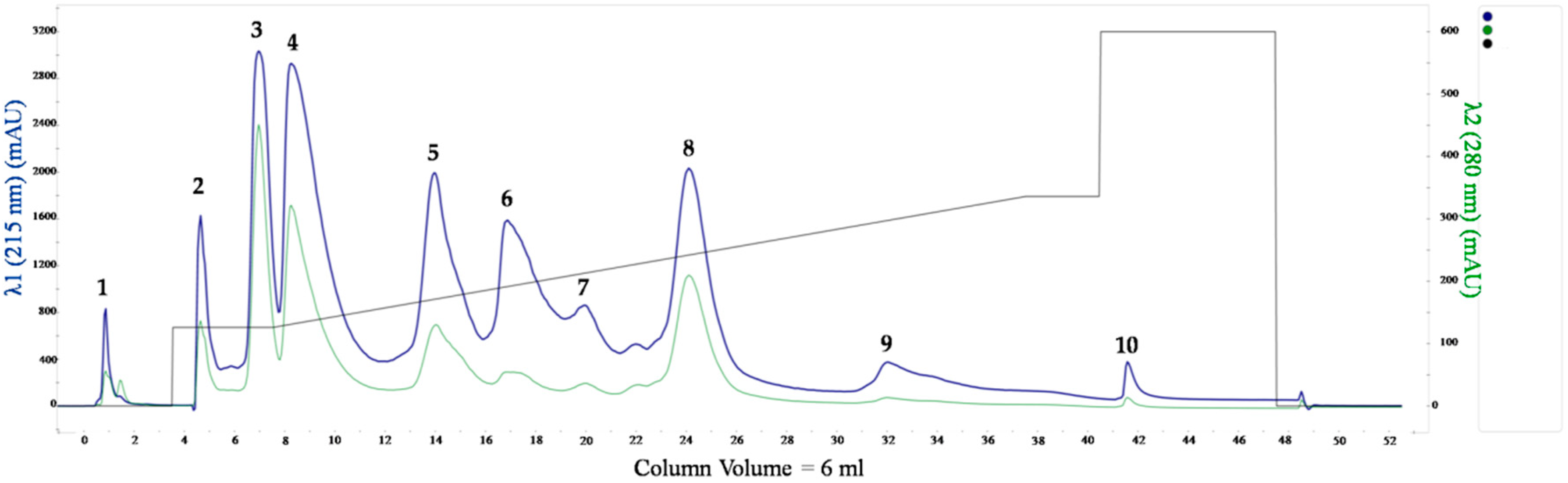
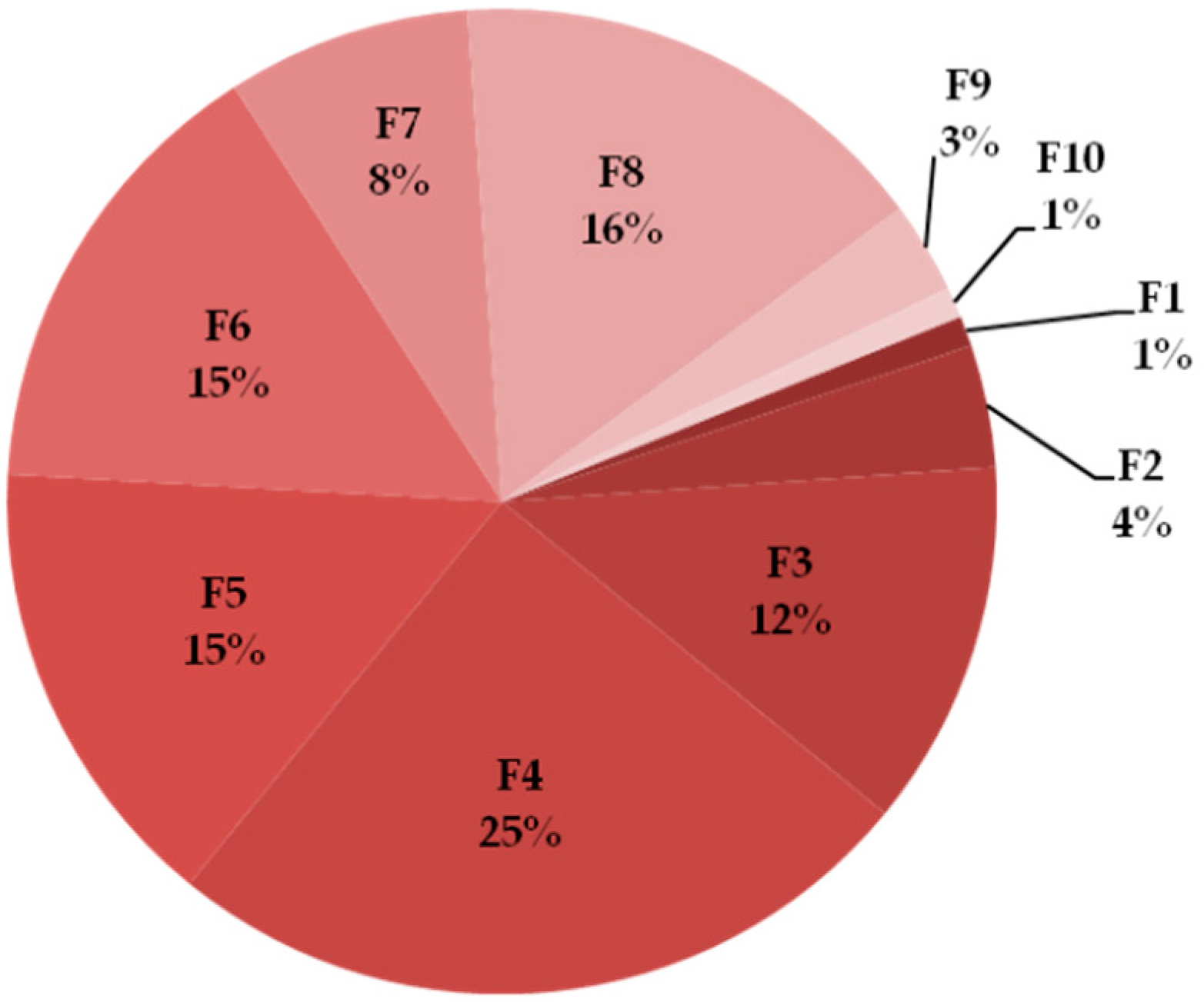
2.1. Naja ashei Venom Fractionation
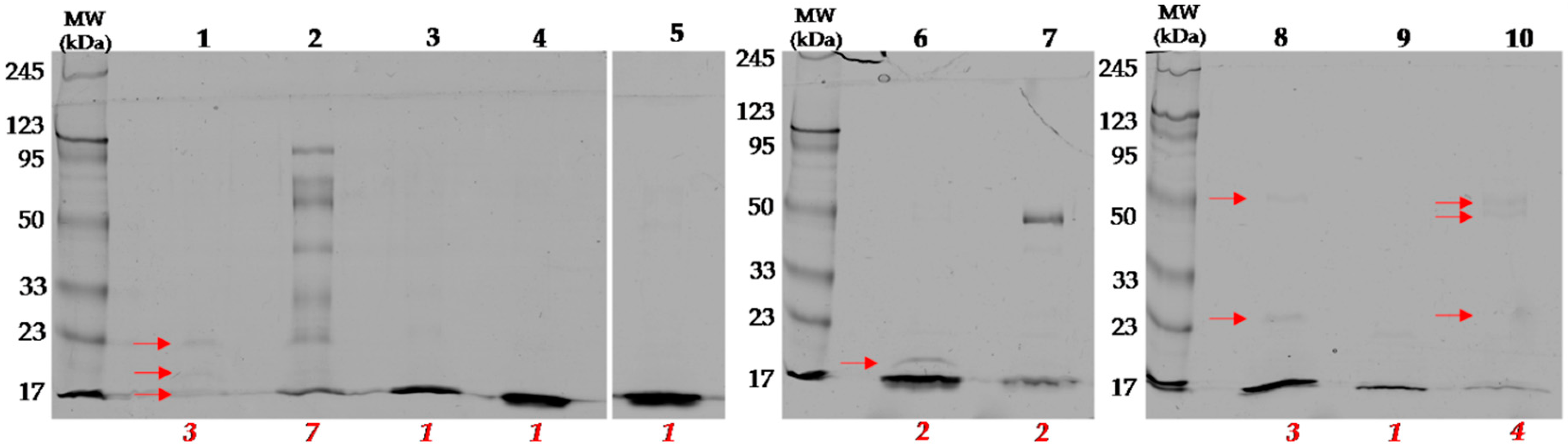
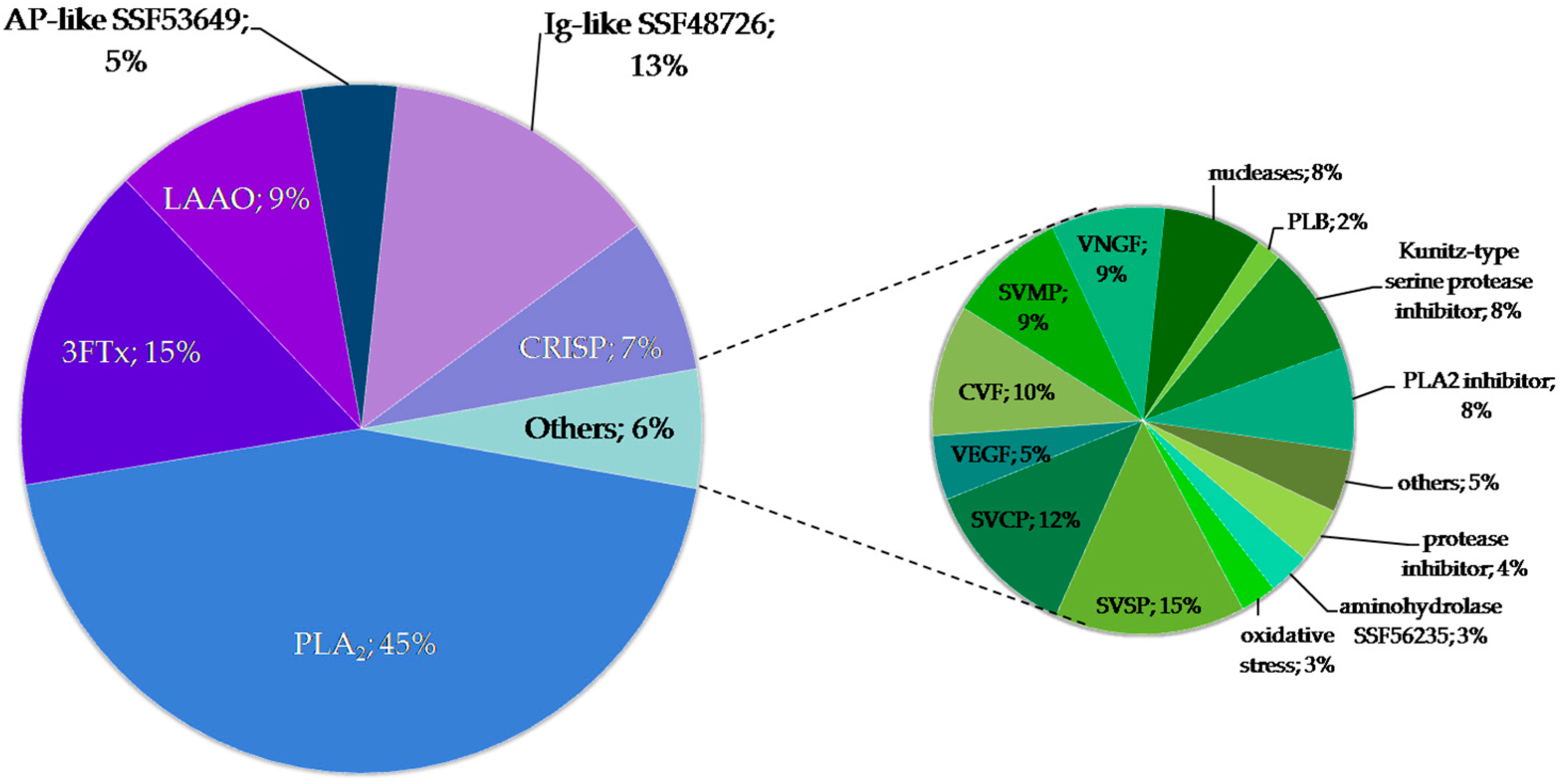
2.2. Identification of Proteins in Obtained Fractions
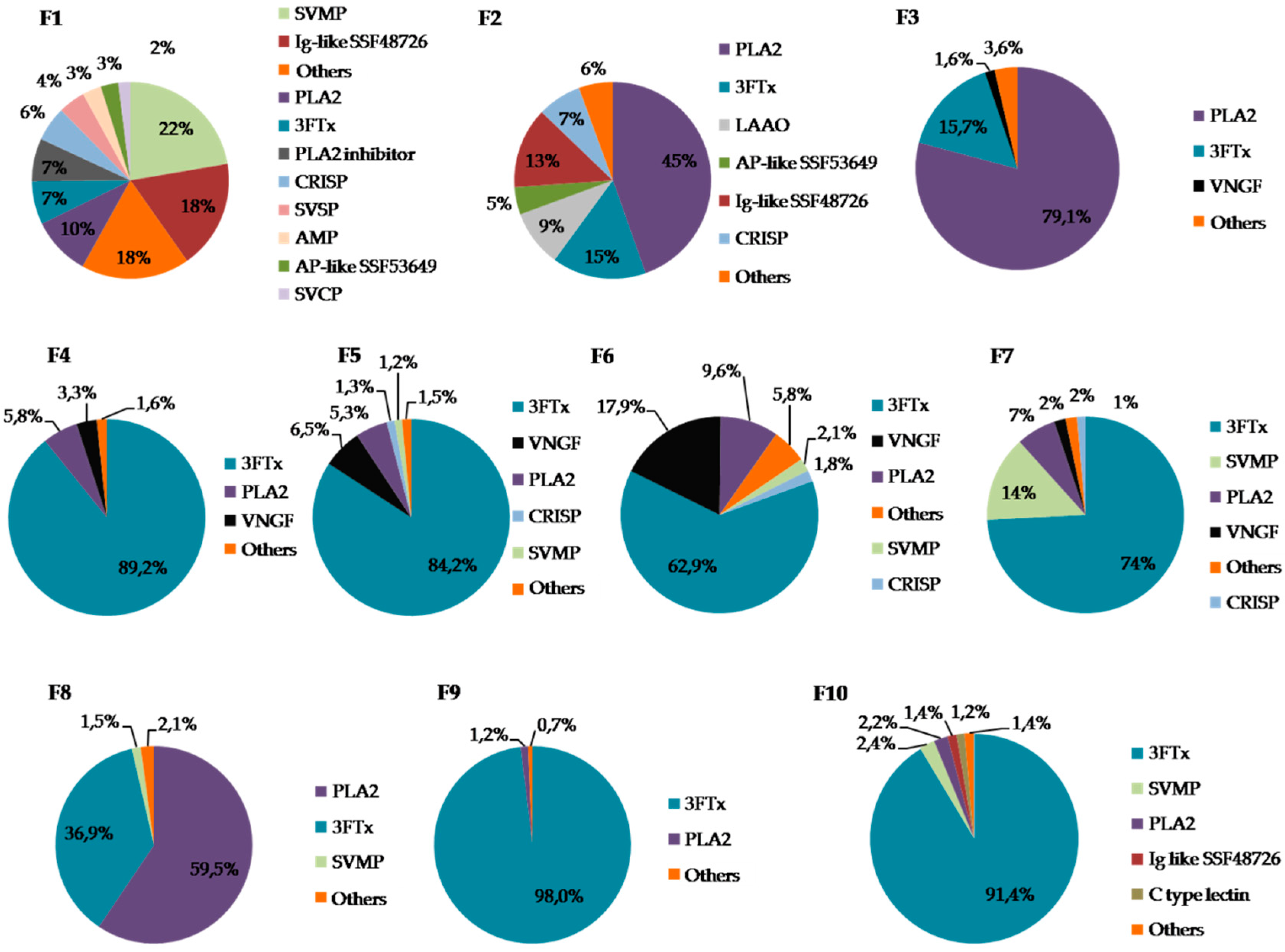
2.2.1. General Characteristics of the Obtained Fractions
2.2.2. Detailed Composition of the F2 Fraction
2.2.3. Proteins with Ig-Like Domain
2.3. Microbiological Tests
2.3.1. Determination of the Minimum Inhibitory Concentration (MIC)
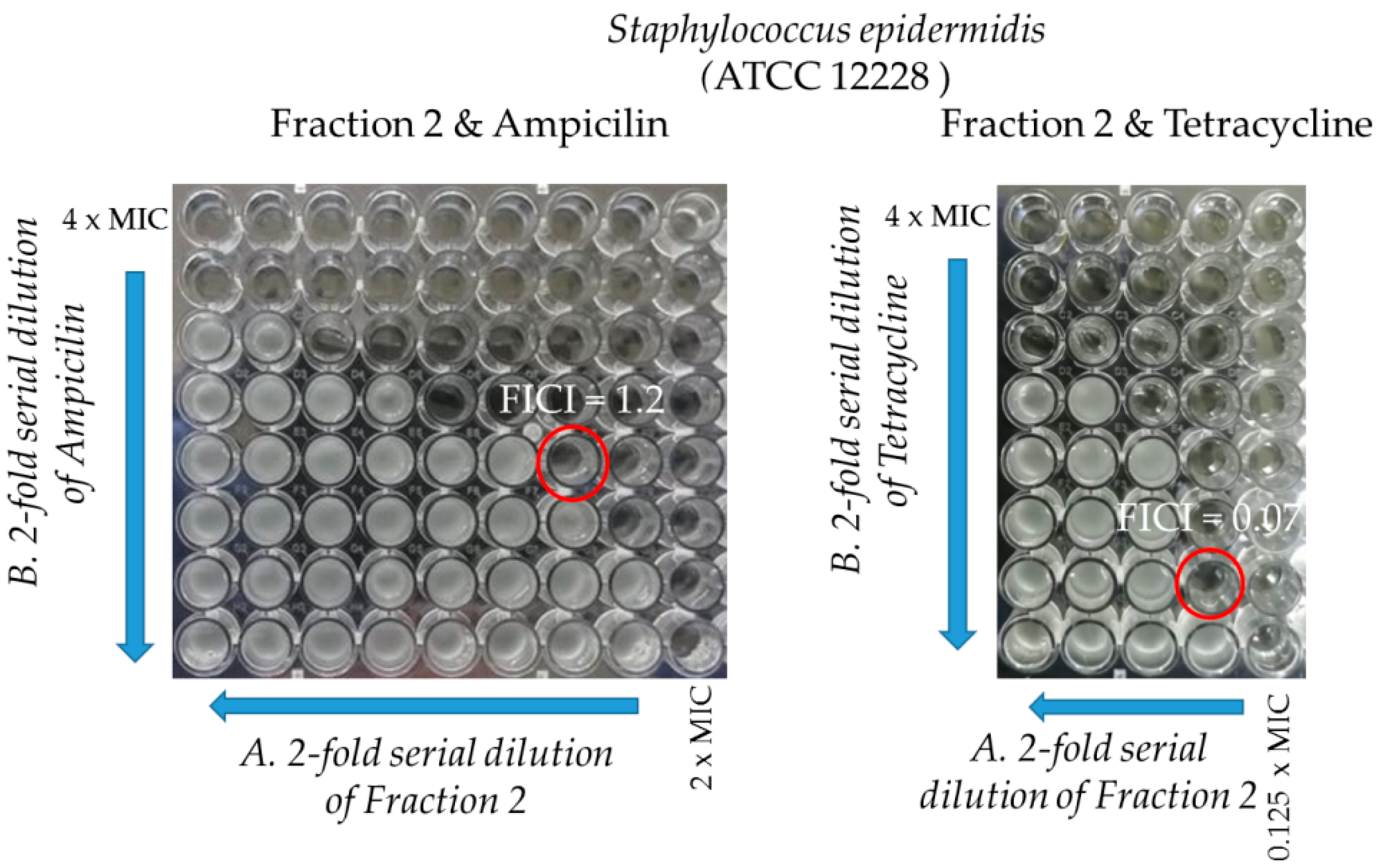
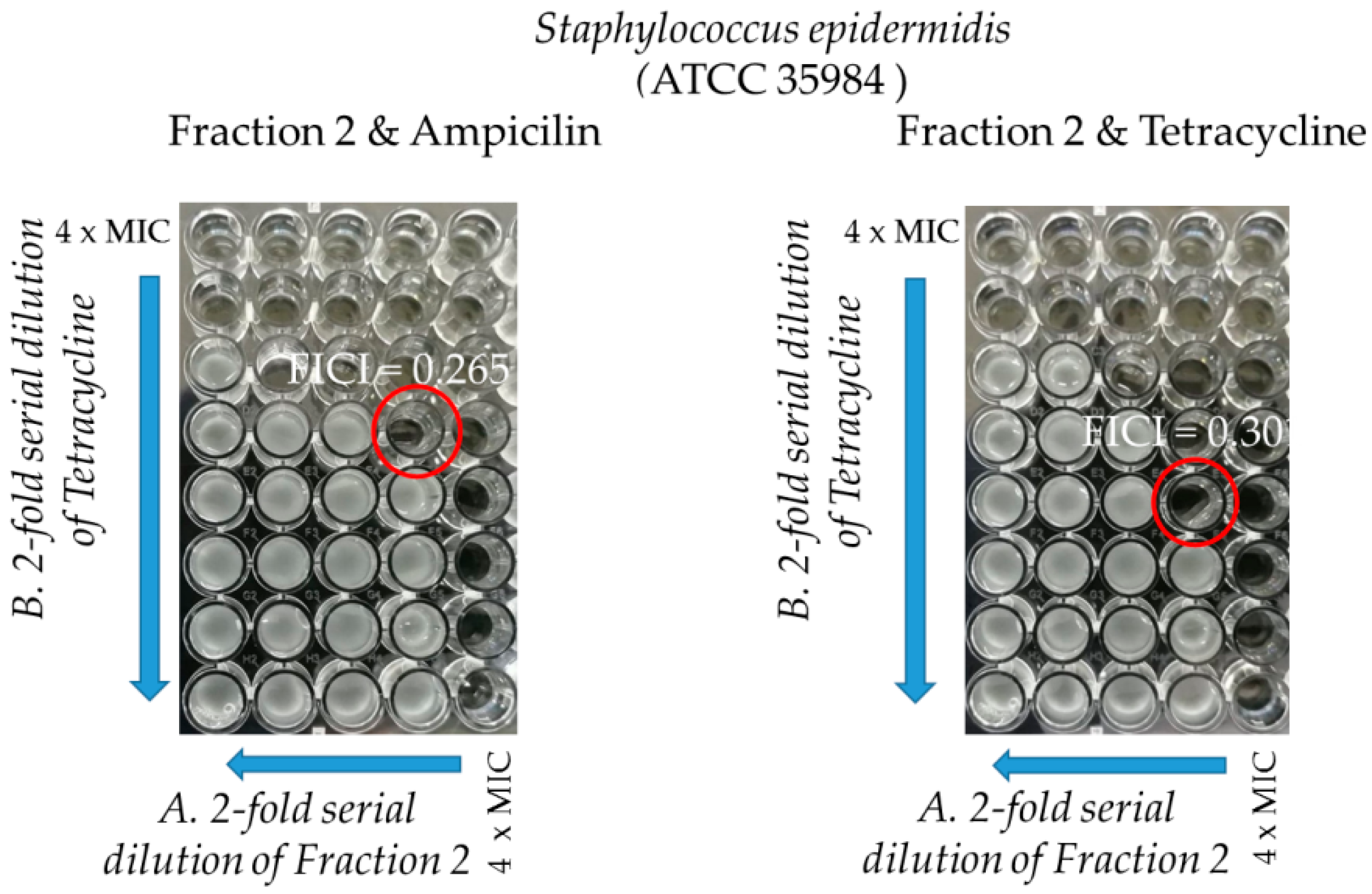
2.3.2. Synergy Testing
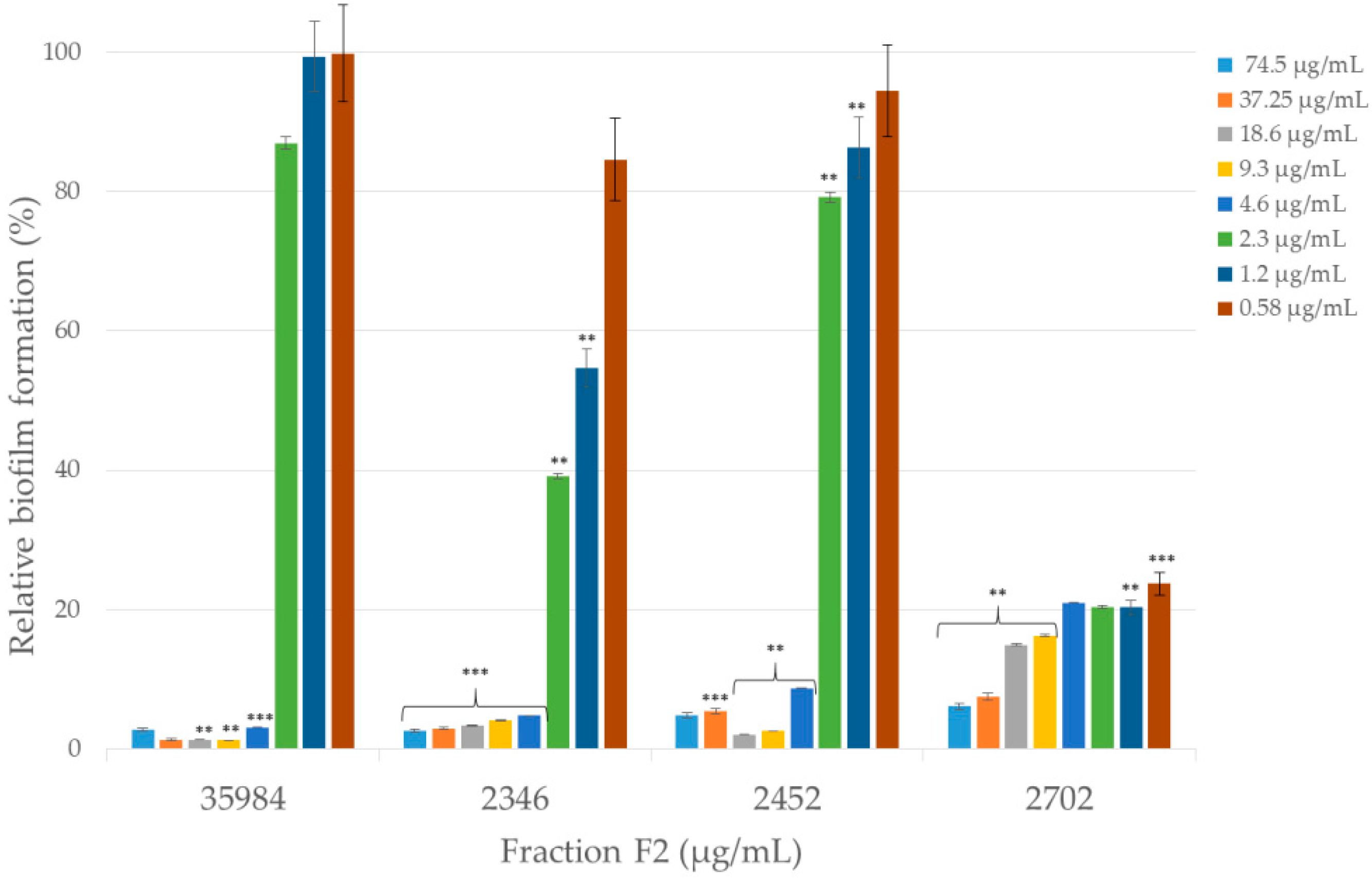
2.3.3. Anti-Biofilm Activity
3. Discussion
4. Materials and Methods
4.1. Venom Fractionation
4.2. Sample Preparation for LC-MS/MS
4.3. Protein Identification by LC-MS/MS
4.4. Antibacterial Activity
4.4.1. Determination of Minimum Inhibitory Concentration (MIC)
4.4.2. Synergy Testing
4.4.3. Anti-Biofilm Activity
4.4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kloos, W.E.; Musselwhite, M.S. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 1975, 30, 381–385. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P.; Glaser, K. Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence 2018, 9, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.L.; Fey, P.D.; Rupp, M.E. Coagulase-negative staphylococcal infections. Infect. Dis. Clin. N. Am. 2009, 23, 73–98. [Google Scholar] [CrossRef] [PubMed]
- De Lalla, F. Antimicrobial chemotherapy in the control of surgical infectious complications. J. Chemother. 1999, 11, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef]
- Xu, K.D.; McFeters, G.A.; Stewart, P.S. Biofilm resistance to antimicrobial agents. Microbiology 2000, 146, 547–549. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Sieradzki, K.; Roberts, R.B.; Serur, D.; Hargrave, J.; Tomasz, A. Recurrent peritonitis in a patient on dialysis and prophylactic vancomycin. Lancet 1998, 351, 880–881. [Google Scholar] [CrossRef]
- Tammelin, A.; Domicel, P.; Hambraeus, A.; Ståhle, E. Dispersal of methicillin-resistant Staphylococcus epidermidis by staff in an operating suite for thoracic and cardiovascular surgery: Relation to skin carriage and clothing. J. Hosp. Infect. 2000, 44, 119–126. [Google Scholar] [CrossRef]
- Raad, I.; Alrahwan, A.; Rolston, K. Staphylococcus epidermidis: Emerging resistance and need for alternative agents. Clin. Infect. Dis. 1998, 26, 1182–1187. [Google Scholar] [CrossRef]
- Villari, P.; Sarnataro, C.; Iacuzio, L. Molecular epidemiology of Staphylococcus epidermidis in a neonatal intensive care unit over a three-year period. J. Clin. Microbiol. 2000, 38, 1740–1746. [Google Scholar] [CrossRef] [PubMed]
- Al Ahmadi, A.J.; Fathi, B.; Jamshidi, A.; Zolfagharian, H.; Mirakabbadi, A.Z. Investigation of the antibacterial effect of venom of the Iranian snake Echiscarinatus. Iran. J. Vet. Scitechnol. 2010, 2, 93–100. [Google Scholar]
- Perumal, S.R.; Stiles, B.G.; Franco, O.L.; Sethi, G.; Lim, L.H.K. Animal venoms as antimicrobial agents. Biochem. Pharmacol. 2017, 134, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Bocian, A.; Hus, K.K. Antibacterial properties of snake venom components. Chem. Pap. 2019. [Google Scholar] [CrossRef]
- Azim, S.; McDowell, D.; Cartagena, A.; Rodriguez, R.; Laughlin, T.F.; Ahmad, Z. Venom peptides cathelicidin and lycotoxin cause strong inhibition of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2016, 87, 246–251. [Google Scholar] [CrossRef]
- Zhao, F.; Lan, X.Q.; Du, Y.; Chen, P.Y.; Zhao, J.; Zhao, F.; Lee, W.H.; Zhang, Y. King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates. Zool. Res. 2018, 39, 87–96. [Google Scholar]
- Koh, D.C.; Armugam, A.; Jeyaseelan, K. Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 2006, 63, 3030–3041. [Google Scholar] [CrossRef]
- Wüster, W.; Broadley, D.G. Get an eyeful of this: A new species of giant spitting cobra from eastern and north-eastern Africa (Squamata: Serpentes: Elapidae: Naja). Zootaxa 2007, 1532, 51–68. [Google Scholar]
- Hus, K.; Buczkowicz, J.; Petrilla, V.; Petrillová, M.; Łyskowski, A.; Legáth, J.; Bocian, A. First Look at the venom of Naja ashei. Molecules 2018, 23, 609. [Google Scholar] [CrossRef]
- Okubo, B.M.; Silva, O.N.; Migliolo, L.; Gomes, D.G.; Porto, W.F.; Batista, C.L.; Ramos, C.S.; Holanda, H.H.; Dias, S.C.; Franco, O.L.; et al. Evaluation of an antimicrobial L-amino acid oxidase and peptide derivaties from Bothropoides mattogrosensis Pitviper venom. PLoS ONE 2012, 7, e33639. [Google Scholar] [CrossRef]
- Bacha, B.A.; Alonazi, M.A.; Elshikh, M.S.; Karray, A. A novel bactericidal homodimeric PLA2 group-I from Walterinnesia aegyptia venom. Int. J. Biolmacromol. 2018, 117, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.W.; Kao, P.H.; Fu, Y.S.; Hu, W.P.; Chang, L.S. Bactericidal effect of Naja nigricollis toxin γ is related to its membrane-damaging activity. Peptides 2011, 32, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.H.; Lin, S.R.; Chang, L.S. Differential binding to phospholipid bilayers modulates membrane-damaging activity of Naja najaatra cardiotoxins. Toxicon 2009, 54, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.L.; Santos, D.O.; Dos Santos, A.L.; Rodrigues, C.R.; De Freitas, C.C.; Cabral, L.M.; Castro, H.C. Comparative analysis of viperidae venoms antibacterial profile: A short communication for proteomics. Evid. Based. Complement. Altern. Med. 2011, 2011, 960267. [Google Scholar] [CrossRef]
- Iğci, N.; Nalbantsoy, A.; Erkan, L.G.; Akça, G.Y.; Yalçın, H.T.; Yalçın, M.; Göçmen, B. Screening of cytotoxic, anti-angiogenic, anti-tumorogenic and antimicrobial activities of Anatolian Vipera ammodytes (Nose-horned viper) venom. Turk. J. Biochem. 2016, 41, 483–491. [Google Scholar] [CrossRef]
- Hegazi, A.G.; EL-Feel, M.A.; Rahman, A.E.H.; Al-Fattah, A.M.A. Antibacterial activity of bee venom collected from Apis mellifera carniolan pure and hybrid races by two collection methods. Int. J. Currmicrobiol. Appl. Sci. 2015, 4, 141–149. [Google Scholar]
- Wang, Y.; Zhang, Z.; Chen, L.; Guang, H.; Li, Z.; Yang, H.; Li, J.; You, D.; Yu, H.; Lai, R. Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PLoS ONE 2011, 6, e22120. [Google Scholar] [CrossRef]
- Dai, C.; Ma, Y.; Zhao, Z.; Zhao, R.; Wang, Q.; Wu, Y.; Cao, Z.; Li, W. Mucroporin, the first cationic host defence peptide from the venom of Lychas mucronatus. Antimicrob. Agents Chemother. 2018, 52, 3967–3972. [Google Scholar] [CrossRef]
- Gauri, S.S.; Bera, K.C.; Bhattacharyya, R.; Mandal, S.M. Identification of an antimicrobial peptide from large freshwater snail (Lymnaea stagnalis): Activity against antibiotics resistant Staphylococcus epidermidis. Int. J. Exp. Res. Rev. 2016, 2, 5–9. [Google Scholar]
- Toyama, M.H.; De Oliveira, D.G.; Beriam, L.O.S.; Novello, J.C.; Rodrigues-Simioni, L.; Marangoni, S. Structural, enzymatic and biological properties of new PLA2 isoform from Crotalus durissus terrificus venom. Toxicon 2003, 41, 1033–1038. [Google Scholar] [CrossRef]
- Vargas, L.J.; Londoño, M.; Quintana, J.C.; Rua, C.; Segura, C.; Lomonte, B.; Núñez, V. An acidic phospholipase A2 with antibacterial activity from Porthidium nasutum snake venom. Comp. Biochem. Physiol. 2012, 161, 341–347. [Google Scholar] [CrossRef]
- Perumal, S.R.; Kandasamy, M.; Gopalakrishnakone, P.; Stiles, B.G.; Rowan, E.G.; Becker, D.; Shanmugam, M.K.; Sethi, G.; Chow, V.T. Wound healing activity and mechanisms of action of an antibacterial protein from the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). PLoS ONE 2014, 9, e80199. [Google Scholar]
- Chen, L.W.; Kao, P.H.; Fu, Y.S.; Lin, S.R.; Chang, L.S. Membrane-damaging activity of Taiwan cobra cardiotoxin 3 is responsible for its bactericidal activity. Toxicon 2011, 58, 46–53. [Google Scholar] [CrossRef]
- Izidoro, L.F.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; Da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. Biomed. Res. Int. 2014, 196754. [Google Scholar] [CrossRef] [PubMed]
- Toyama, M.H.; Toyama, D.D.O.; Passero, L.F.D.; Laurenti, M.D.; Corbett, C.E.; Tomokane, T.Y.; Fonseca, F.V.; Antunes, E.; Joazeiro, P.P.; Beriam, L.O.; et al. Isolation of a new L-amino acid oxidase from Crotalus durissus cascavella venom. Toxicon 2006, 47, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Braga, M.D.M.; Martins, A.M.C.; Amora, D.N.; De Menezes, D.B.; Toyama, M.H.; Toyama, D.O.; Marangoni, S.; Alves, C.D.; Barbosa, P.S.; De Sousa Alves, R.; et al. Purification and biological effects of l-amino acid oxidase isolated from Bothrops insularis venom. Toxicon 2008, 51, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Ande, S.R.; Fussi, H.; Knauer, H.; Murkovic, M.; Ghisla, S.; Fröhlich, K.U.; Macheroux, P. Induction of apoptosis in yeast by L-amino acid oxidase from the Malayan pit viper Calloselasma rhodostoma. Yeast 2008, 25, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Ciscotto, P.; De Avila, M.R.A.; Coelho, E.A.; Oliveira, J.; Diniz, C.G.; Farías, L.M.; De Carvalho, M.A.; Maria, W.S.; Sanchez, E.F.; Borges, A.; et al. Antigenic, microbicidal and antiparasitic properties of an l-amino acid oxidase isolated from Bothrops jararaca snake venom. Toxicon 2009, 53, 330–341. [Google Scholar] [CrossRef]
- Costa Torres, A.F.; Dantas, R.T.; Toyama, M.H.; DizFilho, E.; Zara, F.J.; De Queiroz, R.M.G.; Pinto, N.N.A.; De Oliveira, R.M.; De Oliveira, T.D.; Monteiro, H.S.; et al. Antibacterial and antiparasitic effects of Bothrops marajoensis venom and its fractions: Phospholipase A2 and L-amino acid oxidase. Toxicon 2010, 55, 795–804. [Google Scholar] [CrossRef]
- Lee, M.L.; Tan, N.H.; Fung, S.Y.; Sekaran, S.D. Antibacterial action of a heat-stable form of l-amino acid oxidase isolated from king cobra (Ophiophagus hannah) venom. Comp. Biochem. Physiol. 2011, 153, 237–242. [Google Scholar] [CrossRef]
- Vargas, L.J.; Quintana, J.C.; Pereañez, J.A.; Núñez, V.; Sanz, L.; Calvete, J. Cloning and characterization of an antibacterial L-amino acid oxidase from Crotalus durissus cumanensis venom. Toxicon 2013, 64, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yenugu, S.; Narmadha, G. The human male reproductive tract antimicrobial peptides of the HE2 family exhibit potent synergy with standard antibiotics. J. Pept. Sci. 2010, 16, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Gomez, S.; Japelj, B.; Jerala, R.; Moriyon, I.; Fernandez, A.M.; Leiva, J.; Blondelle, S.E.; Andrä, J.; Brandenburg, K.; Lohner, K.; et al. Structural features governing the activity of lactoferricin-derived peptides that act in synergy with antibiotics against Pseudomonas aeruginosa in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Zharkova, M.S.; Orlov, D.S.; Golubeva, O.Y.; Chakchir, O.B.; Eliseev, I.E.; Grinchuk, T.M.; Shamova, O.V. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics—A novel way to combat antibiotic resistance? Front. Cell. Infect. Microbiol. 2019, 9, 128. [Google Scholar] [CrossRef]
- Canhas, I.N.; Heneine, L.G.D.; Fraga, T.; De Assis, D.C.S.; Borges, M.H.; Chartone-Souza, E.; Nascimento, A.M.A. Antibacterial activity of different types of snake venom from the Viperidae family against Staphylococcus aureus. Acta Sci. Biol. Sci. 2017, 39, 309–319. [Google Scholar] [CrossRef][Green Version]
- Hoerr, V.; Duggan, G.E.; Zbytnuik, L.; Poon, K.K.; Große, C.; Neugebauer, U.; Methling, K.; Löffler, B.; Vogel, H.J. Characterization and prediction of the mechanism of action of antibiotics through NMR metabolomics. BMC Microbiol. 2016, 16, 82. [Google Scholar] [CrossRef]
- Almaaytah, A.; Qaoud, M.T.; Abualhaijaa, A.; Al-Balas, Q.; Alzoubi, K.H. Hybridization and antibiotic synergism as a tool for reducing the cytotoxicity of antimicrobial peptides. Infect. Drug Resist. 2018, 11, 835–847. [Google Scholar] [CrossRef]
- Breitinger, H.-G. Drug synergy–mechanisms and methods of analysis. Toxic. Drug Test. 2012, 143–166. [Google Scholar]
- Jia, J.; Zhu, F.; Ma, X.; Cao, Z.; Li, Y.; Chen, Y.Z. Mechanisms of drug combinations: Interaction and network perspectives. Nat. Rev. Drug Discov. 2009, 8, 111–128. [Google Scholar] [CrossRef]
- Yeh, P.J.; Hegreness, M.J.; Aiden, A.P.; Kishony, R. Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 2009, 7, 460–466. [Google Scholar] [CrossRef]
- Cassone, M.; Otvos, L., Jr. Synergy among antibacterial peptides and between peptides and small-molecule antibiotics. Expert Rev. Anti Infect. Ther. 2010, 8, 703–716. [Google Scholar] [CrossRef]
- Singh, A.P.; Prabha, V.; Rishi, P. Efficacy of cryptdin-2 as an adjunct to antibiotics from various generations against Salmonella. Indian J. Microbiol. 2014, 54, 323–328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feng, Q.; Huang, Y.; Chen, M.; Li, G.; Chen, Y. Functional synergy of alpha-helical antimicrobial peptides and traditional antibiotics against Gram-negative and Gram-positive bacteria in vitro and in vivo. Eur. J. Clinmicrobiol. Infect. Dis. 2015, 34, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Singh, S.; Van Hoek, M.L. Short, synthetic cationic peptides have antibacterial activity against Mycobacterium smegmatis by forming pores in membrane and synergizing with antibiotics. Antibiotics 2015, 4, 358–378. [Google Scholar] [CrossRef] [PubMed]
- Khara, J.S.; Lim, F.K.; Wang, Y.; Ke, X.-Y.; Voo, Z.X.; Yang, Y.Y.; Lakshminarayanan, R.; Ee, P.L.R. Designing α-helical peptides with enhanced synergism and selectivity against Mycobacterium smegmatis: Discerning the role of hydrophobicity and helicity. Acta Biomater. 2015, 28, 99–108. [Google Scholar] [CrossRef]
- Soren, O.; Brinch, K.S.; Patel, D.; Liu, Y.; Liu, A.; Coates, A.; Hu, Y. Antimicrobial peptide novicidin synergizes with rifampin, ceftriaxone, and ceftazidime against antibiotic resistant Enterobacteriaceae in vitro. Antimicrob. Agents Chemother. 2015, 59, 6233–6240. [Google Scholar] [CrossRef]
- Costa, S.S.; Viveiros, M.; Amaral, L.; Couto, I. Multidrug efflux pumps in Staphylococcus aureus: An update. Open Microbiol. J. 2013, 7, 59–71. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H., Jr.; Corriden, R.; Rohde, M.; et al. Synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant gram-negative bacterial pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Yao, Y.; Sturdevant, D.E.; Otto, M. Genome wide analysis of gene expression in Staphylococcus epidermidis biofilms: Insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 2005, 191, 289–298. [Google Scholar] [CrossRef]
- Duguid, I.G.; Evans, E.; Brown, M.R.; Gilbert, P. Effect of biofilm culture upon the susceptibility of Staphylococcus epidermidis to tobramycin. J. Antimicrobiol. Chemother. 1992, 30, 803–810. [Google Scholar] [CrossRef]
- Duguid, I.G.; Evans, E.; Brown, M.R.; Gilbert, P. Growth-rate-independent killing by ciprofloxacin of biofilm-derived Staphylococcus epidermidis: Evidence for cell-cycle dependency. J. Antimicrobiol. Chemother. 1992, 30, 791–802. [Google Scholar] [CrossRef]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; De Leo, F.R.; Otto, M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef]
- Kocianova, S.; Vuong, C.; Yao, Y.; Voyich, J.M.; Fischer, E.R.; DeLeo, F.R.; Otto, M. Key role of poly-γ-dl -glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Investig. 2005, 115, 688–694. [Google Scholar] [CrossRef]
- Kristian, S.A.; Birkenstock, T.A.; Sauder, U.; Mack, D.; Götz, F.; Landmann, R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 2008, 197, 1028–1035. [Google Scholar] [CrossRef]
- Klein, R.C.; Fabres-Klein, M.H.; De Oliveira, L.L.; Feio, R.N.; Malouin, F.; Ribon, A.O.B. A C-Type lectin from Bothrops jararacussu venom disrupts staphylococcal biofilms. PLoS ONE 2015, 10, e0120514. [Google Scholar] [CrossRef]
- Aguilar, A.P.; Onofre, T.S.; Fabres-Klein, M.H.; Klein, R.C.; Feio, R.N.; De Oliveira, M.T.A.; De Oliveira, B.R.A. Carbohydrate-independent biofilm effect of lectin BJcuL on Staphylococcus aureus. Microb. Pathog. 2019, 137, 103745. [Google Scholar] [CrossRef]
- Gopal, R.; Kim, Y.G.; Lee, J.H.; Lee, S.K.; Chae, J.D.; Son, B.K.; Seo, C.H.; Park, Y. Synergistic effects and antibiofilm properties of chimeric peptides against multidrug-resistant Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 2014, 58, 1622–1629. [Google Scholar] [CrossRef]
- De La Fuente, N.C.; Reffuveille, F.; Mansour, S.C.; Reckseidler, Z.S.L.; Hernández, D.; Brackman, G.; Coenye, T.; Hancock, R.E. D-Enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef]
- Bessa, L.J.; Eaton, P.; Dematei, A.; Plácido, A.; Vale, N.; Gomes, P.; Delerue, M.C.; Sa Leite, J.R.; Gameiro, P. Synergistic and antibiofilm properties of ocellatin peptides against multidrug-resistant Pseudomonas aeruginosa. Future Microbiol. 2018, 13, 151–163. [Google Scholar] [CrossRef]
- Gough, J.; Karplus, K.; Hughey, R.; Chothia, C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001, 313, 903–919. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, 351–360. [Google Scholar] [CrossRef]
- Halaby, D.M.; Poupon, A.; Mornon, J.-P. The immunoglobulin fold family: Sequence analysis and 3D structure comparisons. Protein Eng. 1999, 12, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Nishimoto, T.; Kubota, M.; Chaen, H.; Fukuda, S. Cloning, sequencing, and expression of the genes encoding an isocyclomaltooligosaccharide glucanotransferase and an alpha-amylase from a Bacillus circulans strain. Biosci. Biotechnol. Biochem. 2006, 70, 2690–2702. [Google Scholar] [CrossRef] [PubMed]
- Shia, S.; Stamos, J.; Kirchhofer, D.; Fan, B.; Wu, J.; Corpuz, R.T.; Santell, L.; Lazarus, R.A.; Eigenbrot, C. Conformational lability in serine protease active sites: Structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B. J. Mol. Biol. 2005, 346, 1335–1349. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Honjo, K.; Hirota, Y.; Onodera, T. Isolation of the chicken NF-kappa B p65 subunit-encoding cDNA and characterization of its products. Gene 1993, 133, 237–242. [Google Scholar]
- Isom, L.L.; De Jongh, K.S.; Patton, D.E.; Reber, B.F.X.; Offord, J.; Charbonneau, H.; Walsh, K.; Goldin, A.L.; Catterall, W.A. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science 1992, 256, 839–842. [Google Scholar] [CrossRef]
- Rehana, S.; Kini, R.M. Molecular isoforms of cobra venom factor-like proteins in the venom of Austrelaps superbus. Toxicon 2007, 50, 32–52. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Lemmon, A.R.; Margres, M.J.; Aronow, K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 2012, 13, 312. [Google Scholar] [CrossRef]
- Zeng, L.; Sun, Q.Y.; Jin, Y.; Zhang, Y.; Lee, W.H.; Zhang, Y. Molecular cloning and characterization of a complement-depleting factor from king cobra, Ophiophagus hannah. Toxicon 2012, 60, 290–301. [Google Scholar] [CrossRef]
- McGivern, J.J.; Wray, K.P.; Margres, M.J.; Couch, M.E.; Mackessy, S.P.; Rokyta, D.R. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genom. 2014, 15, 1061. [Google Scholar] [CrossRef]
- Aird, S.D.; Da Silva, N.J.; Qiu, L.; Villar-Briones, A.; Saddi, V.A.; Pires De Campos Telles, M.; Grau, M.L.; Mikheyev, A.S. Coral snake Venomics: Analyses, of venom gland transcriptomes and proteomes of six Brazilian taxa. Toxins 2017, 8, 9. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Barsnes, H.; Vaudel, M. SearchGUI: A highly adaptable common interface for proteomics search and de novo engines. J. Proteome Res. 2018, 17, 2552–2555. [Google Scholar] [CrossRef]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.P.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef]
- Zapała, L.; Kosińska, M.; Woźnicka, E.; Byczyński, Ł.; Ciszkowicz, E.; Lecka-Szlachta, K.; Zapała, W.; Chutkowski, M. Comparison of spectral and thermal properties and antibacterial activity of new binary and ternary complexes of Sm(III), Eu(III) and Gd(III) ions with N-phenylanthranilic acid and 1,10-phenanthroline. Thermochim. Acta 2019, 671, 134–148. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Determining Bactericidal Activity of Antimicrobial Agents. Approved Guideline; CLSI document M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999; Volume 19. [Google Scholar]
- Spoorthi, N.J.; Vishwanatha, T.; Reena, V.; Divyashree, B.C.; Aishwarya, S.; Siddhalingeshwara, K.G.; Venugopal, N.; Ramesh, I. Antibiotic synergy test: Checkerboard method on multidrug resistant Pseudomonas aeruginosa. Int. Res. J. Pharm. 2011, 2, 196–198. [Google Scholar]
- Vázquez, R.; García, P. Synergy between two chimeric lysins to kill Streptococcus pneumoniae. Front. Microbiol. 2019, 10, 1251. [Google Scholar] [CrossRef]
- Sopirala, M.M.; Mangino, J.E.; Gebreyes, W.A.; Biller, B.; Bannerman, T.; Balada-Llasat, J.M.; Pancholi, P. Synergy testing by Etest, microdilution checkerboard, and time-kill methods for pan-drug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4678–4683. [Google Scholar] [CrossRef]
- Bielenica, A.; Drzewiecka-Antonik, A.; Rejmak, P.; Stefańska, J.; Koliński, M.; Kmiecik, S.; Lesyng, B.; Włodarczyk, M.; Pietrzyki, P.; Struga, M. Synthesis, structural and antimicrobial studies of type II topoisomerase targeted copper (II) complexes of 1,3-disubstituted thiourea ligands. J. Inorg. Biochem. 2018, 182, 61–70. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
| Fraction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| C-type lectin | − | − | − | − | − | − | − | − | − | + |
| SVSP | + | + | − | − | − | − | − | − | − | + |
| SVCP | + | + | + | − | − | − | − | − | − | − |
| PLA2 inhibitor | + | + | − | − | − | − | − | − | − | + |
| AP-like SSF53649 | + | + | − | − | − | − | − | − | − | − |
| LAAO | − | + | + | − | − | − | − | − | − | + |
| Ig-like SSF48726 | + | + | + | − | − | − | − | − | − | + |
| CRISP | + | + | + | + | + | + | + | + | − | − |
| VNGF | + | + | + | + | + | + | + | + | + | + |
| AMP | + | − | − | − | − | − | − | − | − | − |
| CVF | + | + | + | + | + | + | + | + | − | + |
| Antibiotic | Staphylococcus epidermidis | ||||
|---|---|---|---|---|---|
| ATCC 12228 | ATCC 35984 | 2346 | 2452 | 2702 | |
| MIC µg/mL | |||||
| Chloramphenicol | 7.8 | 15.6 | 125 | 7.8 | 125 |
| Ampicillin | 62.5 | 250 | 500 | 62.5 | 7.8 |
| Kanamycin | 1.9 | −a | −a | 1.9 | 3.9 |
| Streptomycin | −a | −a | 3.9 | 7.8 | 250 |
| Tetracycline | 62.5 | 0.12 | 1.9 | 62.5 | 0.24 |
| Fraction F2 | 37.25 | 9.3 | 4.6 | 9.3 | 37.25 |
| Strains | Ampicillin/FractionF2 | Tetracycline/FractionF2 | ||||
|---|---|---|---|---|---|---|
| MIC (µg/mL) | ||||||
| In Single Use | In Combination | FIC Index | In Single Use | In Combination | FIC Index | |
| ATCC 12228 | 62.5/37.25 | 15.6/37.25 | 1.2 | 62.5/37.25 | 3.9/2.3 | 0.07 |
| ATCC 35984 | 250/9.3 | 62.5/2.3 | 0.26 | 0.12/9.3 | 0.03/2.3 | 0.30 |
| Step | Segment | Initial %B | Final %B | FR (mL/min) | CV |
|---|---|---|---|---|---|
| Equilibration | Isocratic | 0 | 0 | 6 | 3 |
| Sample application | Isocratic | 0 | 0 | 3 | 0.5 |
| Column wash | Isocratic | 0 | 0 | 6 | 3 |
| Elution | Isocratic | 21 | 21 | 6 | 4 |
| Gradient | 21 | 56 | 6 | 30 | |
| Isocratic | 56 | 56 | 6 | 3 | |
| Isocratic | 100 | 100 | 6 | 7 | |
| Column wash | Isocratic | 0 | 0 | 6 | 5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bocian, A.; Ciszkowicz, E.; Hus, K.K.; Buczkowicz, J.; Lecka-Szlachta, K.; Pietrowska, M.; Petrilla, V.; Petrillova, M.; Legáth, Ľ.; Legáth, J. Antimicrobial Activity of Protein Fraction from Naja ashei Venom against Staphylococcus epidermidis. Molecules 2020, 25, 293. https://doi.org/10.3390/molecules25020293
Bocian A, Ciszkowicz E, Hus KK, Buczkowicz J, Lecka-Szlachta K, Pietrowska M, Petrilla V, Petrillova M, Legáth Ľ, Legáth J. Antimicrobial Activity of Protein Fraction from Naja ashei Venom against Staphylococcus epidermidis. Molecules. 2020; 25(2):293. https://doi.org/10.3390/molecules25020293
Chicago/Turabian StyleBocian, Aleksandra, Ewa Ciszkowicz, Konrad K. Hus, Justyna Buczkowicz, Katarzyna Lecka-Szlachta, Monika Pietrowska, Vladimír Petrilla, Monika Petrillova, Ľubomír Legáth, and Jaroslav Legáth. 2020. "Antimicrobial Activity of Protein Fraction from Naja ashei Venom against Staphylococcus epidermidis" Molecules 25, no. 2: 293. https://doi.org/10.3390/molecules25020293
APA StyleBocian, A., Ciszkowicz, E., Hus, K. K., Buczkowicz, J., Lecka-Szlachta, K., Pietrowska, M., Petrilla, V., Petrillova, M., Legáth, Ľ., & Legáth, J. (2020). Antimicrobial Activity of Protein Fraction from Naja ashei Venom against Staphylococcus epidermidis. Molecules, 25(2), 293. https://doi.org/10.3390/molecules25020293








