Antisense Oligonucleotides Used to Identify Telomeric G-Quadruplexes in Metaphase Chromosomes and Fixed Cells by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci
Abstract
1. Introduction
2. Results
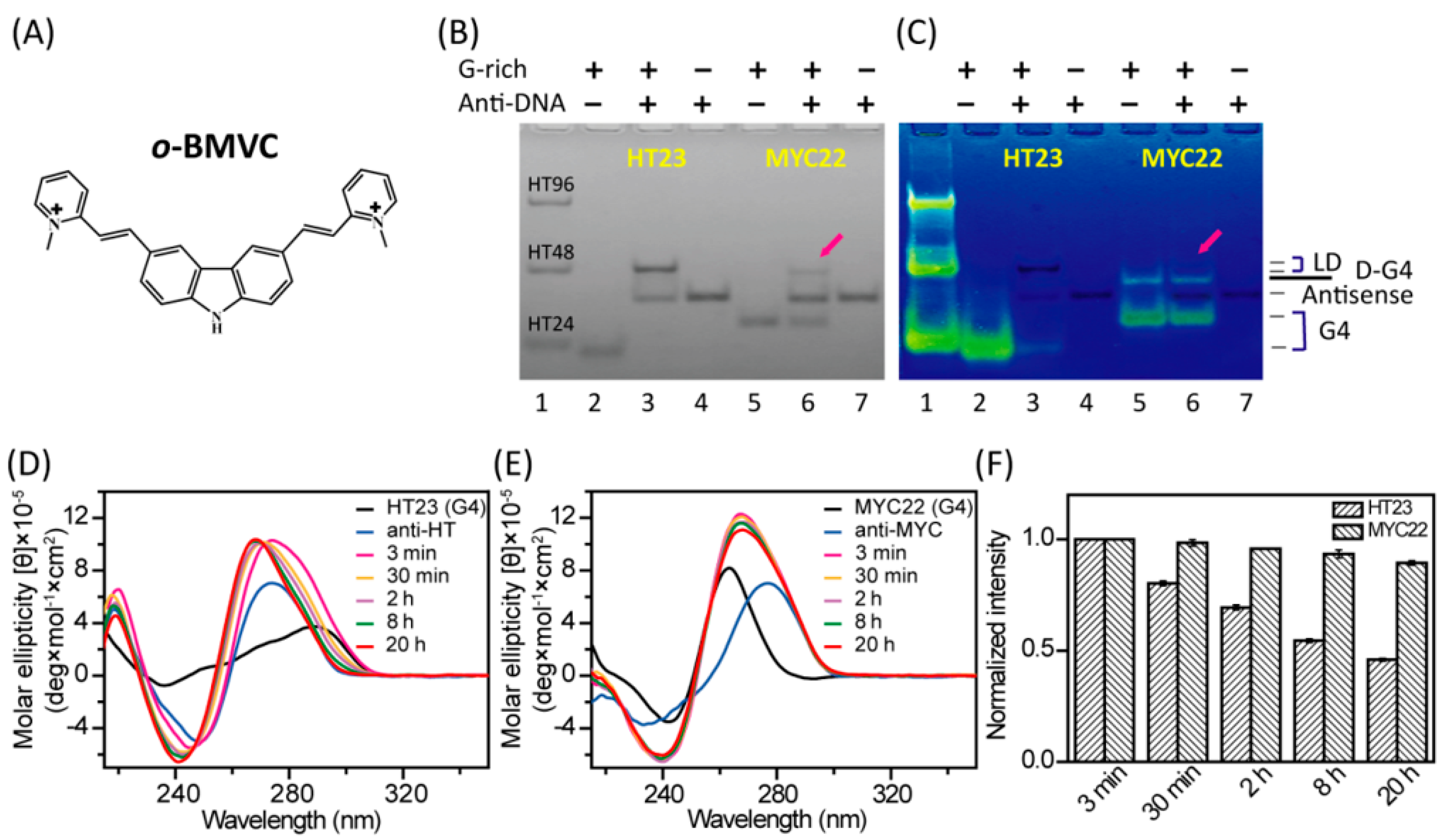
2.1. Unfolding of G4 Structures Induced by their Antisense Sequences In Vitro
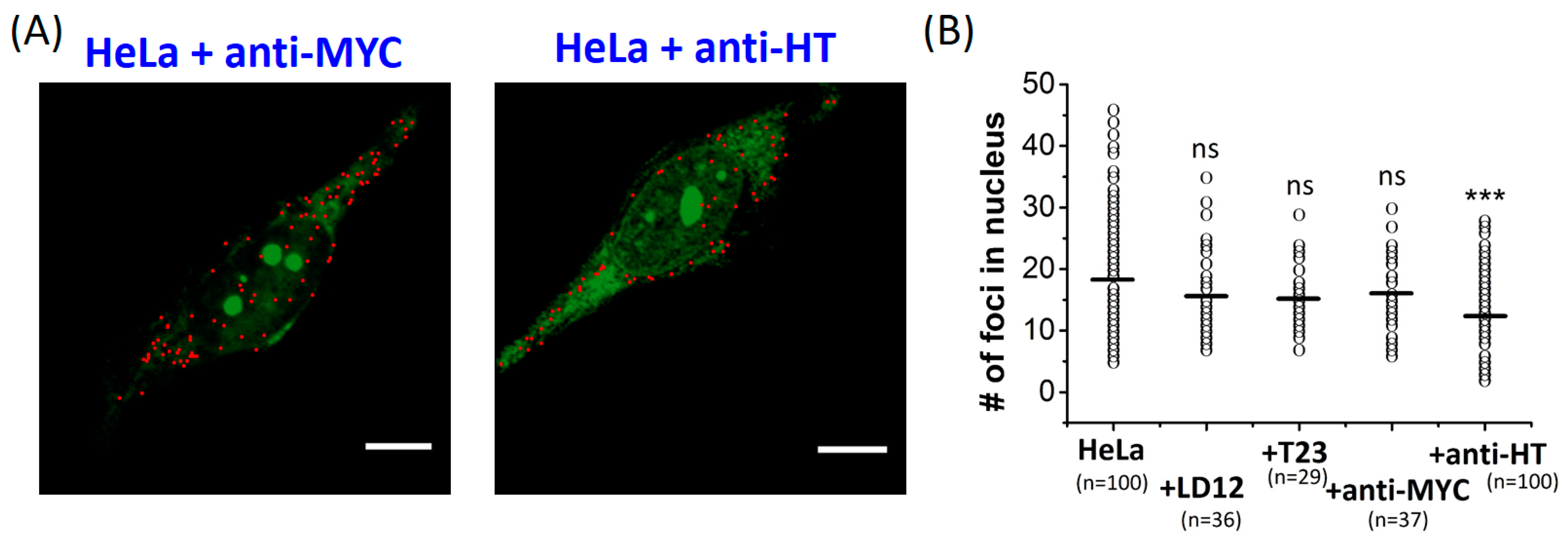
2.2. The Number of G4 Foci in the Nucleus of Fixed HeLa Cells Reduced by Anti-HT Sequences
2.3. The Number of Telomeric G4 Foci in Metaphase Chromosomes Reduced by Anti-HT Sequences
3. Discussion
4. Materials and Methods
4.1. DNA Preparation
4.2. Circular Dichroism (CD)
4.3. Polyacrylamide Gel Electrophoresis (PAGE)
4.4. Cell Cultures
4.5. Fluorescence Lifetime Imaging Microscopy (FLIM)
4.6. Preparation of Metaphase Chromosome Spreads
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Williamson, J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 703–730. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Greider, C.W. Telomeres; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1996. [Google Scholar]
- Wright, W.E.; Tesmer, V.M.; Huffman, K.E.; Levene, S.D.; Shay, J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997, 11, 2801–2809. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. Beginning to understand the end of the chromosome. Cell 2004, 116, 273–279. [Google Scholar] [CrossRef]
- Davis, J.T. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular Chemistry. Angew. Chem. Int. Ed. 2004, 116, 684–716. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS 2010, 277, 1107–1117. [Google Scholar] [CrossRef]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DNA structure. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef]
- Mergny, J.L.; Helene, C. G-quadruplex DNA: A target for drug design. Nat. Med. 1998, 4, 1366–1367. [Google Scholar] [CrossRef]
- Niedle, S.; Parkinson, G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002, 1, 383–393. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Neidle, S. G-quardruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Lipps, H.J.; Rhodes, D. G-quadruplex structure: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Pluckthun, A. In vitro generated antibodies specific for telomeric guanine quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef] [PubMed]
- Granotier, C.; Pennarun, G.; Riou, L.; Hoffschir, F.; Gauthier, L.R.; Cian, A.D.; Gomez, D.; Mandine, E.; Riou, J.F.; Mergny, J.L.; et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005, 33, 4182–4190. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Sun, D.; Guo, K.; Rusche, J.J.; Hurley, L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005, 33, 6070–6080. [Google Scholar] [CrossRef]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef]
- Wei, P.C.; Wang, Z.F.; Lo, W.T.; Su, M.I.; Shew, J.Y.; Chang, T.C.; Lee, W.H. A cis-element with mixed G-quadruplex structure of NPGPx promoter is essential for nucleolin-mediated transactivation on non-targeting siRNA stress. Nucleic Acids Res. 2013, 41, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Huang, F.C.; Kuo, M.H.; Wang, Z.F.; Tseng, T.Y.; Chang, L.C.; Yen, S.J.; Chang, T.C.; Lin, J.J. Inhibition of Cancer Cell Migration and Invasion through Suppressing the Wnt1mediating Signal Pathway by G-quadruplex Structure Stabilizers. J. Biol. Chem. 2014, 289, 14612–14623. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef]
- Weissleder, R.; Pittet, M.J. Imaging in the era of molecular oncology. Nature 2008, 452, 580–589. [Google Scholar] [CrossRef]
- Kobayashi, H.; Ogawa, M.; Alford, R.; Choyke, P.L.; Urano, Y. New Strategies for Fluorescent Probe Design in Medical Diagnostic Imaging. Chem. Rev. 2010, 110, 2620–2640. [Google Scholar] [CrossRef]
- Chen, W.W.; Yi, Y.H.; Chien, C.H.; Hsiung, K.C.; Ma, T.H.; Lin, Y.C.; Lo, S.J.; Chang, T.C. Specific polyunsaturated fatty acids modulate lipid delivery and oocyte development in C. elegans revealed by molecular-selective label-free imaging. Sci. Rep. 2016, 6, 32021. [Google Scholar] [CrossRef]
- Chen, W.W.; Lemieux, G.A.; Camp, C.H., Jr.; Chang, T.C.; Ashrafi, K.; Cicerone, M.T. Spectroscopic coherent Raman imaging of Caenorhabditis elegans reveals lipid particle diversity. Nat. Chem. Biol. 2020. Online ahead of print. [Google Scholar] [CrossRef]
- Chang, C.C.; Kuo, I.C.; Ling, I.F.; Chen, C.T.; Chen, H.C.; Lou, P.J.; Lin, J.J.; Chang, T.C. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004, 76, 4490–4494. [Google Scholar] [CrossRef]
- Chang, C.C.; Chu, J.F.; Kao, F.J.; Chiu, Y.C.; Lou, P.J.; Chen, H.C.; Chang, T.C. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006, 78, 2810–2815. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; Miller, J.; Howat, W.J.; Balasubramanian, S. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS ONE 2014, 9, e102711. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.Y.; Chien, C.H.; Chu, J.F.; Huang, W.C.; Lin, M.Y.; Chang, C.C.; Chang, T.C. Fluorescent probe for visualizing guanine-quadruplex DNA by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 2013, 18, 101309. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Tseng, T.Y.; Chen, Y.T.; Chang, C.C.; Wang, Z.F.; Wang, C.L.; Hsu, T.N.; Li, P.T.; Chen, C.T.; Lin, J.J.; et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015, 43, 10102–10113. [Google Scholar] [CrossRef]
- Tseng, T.Y.; Chen, W.W.; Chu, I.T.; Wang, C.L.; Chang, C.C.; Lin, M.C.; Lou, P.J.; Chang, T.C. The G-quadruplex fluorescent probe 3,6-bis(1-methyl-2-vinyl-pyridinium) carbazole diiodide as a biosensor for human cancers. Sci. Rep. 2018, 8, 16082. [Google Scholar] [CrossRef]
- Tseng, T.Y.; Chu, I.T.; Lin, S.J.; Li, J.; Chang, T.C. Binding of small molecules to G-quadruplex DNA in cells revealed by fluorescence lifetime imaging microscopy of o-BMVC foci. Molecules 2019, 24, 35. [Google Scholar] [CrossRef]
- Shivalingam, A.; Izquierdo, M.A.; Marois, A.L.; Vysniauskas, A.; Suhling, K.; Kuimova, M.K.; Vilar, R. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat. Commun. 2015, 6, 8178. [Google Scholar] [CrossRef]
- Liu, L.Y.; Liu, W.; Wang, K.N.; Zhu, B.C.; Xia, X.Y.; Ji, L.N.; Mao, Z.W. Quantitative detection of G-quadruplex DNA in live cells based on photon counts and complex structure discrimination. Angew. Chem. 2020, 59, 9719–9726. [Google Scholar] [CrossRef]
- Li, W.; Wu, P.; Ohmichi, T.; Sugimoto, N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002, 526, 77–81. [Google Scholar] [CrossRef]
- Zhao, Y.; Kan, Z.Y.; Zeng, Z.X.; Hao, Y.H.; Chen, H.; Tan, Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J. Am. Chem. Soc. 2004, 126, 13255–13264. [Google Scholar] [CrossRef]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Trent, J.O.; Chaires, J.B. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014, 428, 1629–1650. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A threshold selection method from Gray-level histograms. IEEE Trans. Syst. ManCybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Laguerre, A.; Hukezalie, K.; Winckler, P.; Katranji, F.; Chanteloup, G.; Pirrotta, M.; Perrier-Cornet, J.-M.; Wong, J.D.Y.; Monchaud, D. Visualization of RNA-Quadruplexes in Live Cells. J. Am. Chem. Soc. 2015, 137, 8521–8525. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Dai, J.X.; Chen, D.; Jones, R.A.; Hurley, L.H.; Yang, D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006, 34, 5133–5144. [Google Scholar] [CrossRef] [PubMed]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5’-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef]
- Kotar, A.; Wang, B.; Shivalingam, A.; Gonzalez-Garcia, J.; Vilar, R.; Plavec, J. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. Int. Ed. 2016, 55, 12508–12511. [Google Scholar] [CrossRef]
- Li, M.H.; Wang, Z.F.; Kuo, M.H.J.; Hsu, S.T.D.; Chang, T.C. Unfolding kinetics of human telomeric G-quadruplexes studied by NMR spectroscopy. J. Phys. Chem. B 2014, 118, 931–936. [Google Scholar] [CrossRef]
- Moye, A.L.; Porter, K.C.; Cohen, S.B.; Phan, T.; Zyner, K.G.; Sasaki, N.; Lovrecz, G.O.; Beck, J.L.; Bryan, T.M. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015, 6, 7643. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.L.; Liu, H.S.; Xu, Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019, 47, 4940–4947. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Li, Y.; Xiong, W.; Sha, W.; Jing, L.; Tweardy, D.J. G-quartet oligonucleotides: A new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004, 64, 6603–6609. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.Y.; Wang, Z.F.; Chien, C.H.; Chang, T.C. In-cell optical imaging of exogenous G-quadruplex DNA by fluorogenic ligands. Nucleic Acids Res. 2013, 41, 10605–10618. [Google Scholar] [CrossRef] [PubMed]
- Oganesian, L.; Karlseder, J. Telomeric armor: The layers of end protection. J. Cell Sci. 2009, 122, 4013–4025. [Google Scholar] [CrossRef]
- Xu, Y. Chemistry in human telomere biology: Structure, function, and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011, 40, 2719–2740. [Google Scholar] [CrossRef]
- Chaires, J.B.; Gray, R.D.; Dean, W.L.; Monsen, R.; DeLeeuw, L.W.; Stribinskis, V.; Trent, J.O. Human POT1 unfolds G-quadruplexes by conformational selection. Nucleic Acids Res. 2020, 48, 4976–4991. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 0041. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.J.; Shaw, P.G.; Kim, M.-S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of diseases. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Brcic, J.; Plavec, J. ALS and FTD linked GGGGCC-repeat containing DNA oligonucleotide folds into two distinct G-quadruplexes. Biochim. Biophys. Acta 2017, 1861, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Simone, R.; Balendra, R.; Moens, T.G.; Preza, E.; Wilson, K.M.; Heslegrave, A.; Woodling, N.S.; Niccoli, T.; Gilbert-Jaramillo, J.; Abdelkarim, S.; et al. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018, 10, 22–31. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, T.-Y.; Liu, S.-Y.; Wang, C.-L.; Chang, T.-C. Antisense Oligonucleotides Used to Identify Telomeric G-Quadruplexes in Metaphase Chromosomes and Fixed Cells by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci. Molecules 2020, 25, 4083. https://doi.org/10.3390/molecules25184083
Tseng T-Y, Liu S-Y, Wang C-L, Chang T-C. Antisense Oligonucleotides Used to Identify Telomeric G-Quadruplexes in Metaphase Chromosomes and Fixed Cells by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci. Molecules. 2020; 25(18):4083. https://doi.org/10.3390/molecules25184083
Chicago/Turabian StyleTseng, Ting-Yuan, Shin-Ya Liu, Chiung-Lin Wang, and Ta-Chau Chang. 2020. "Antisense Oligonucleotides Used to Identify Telomeric G-Quadruplexes in Metaphase Chromosomes and Fixed Cells by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci" Molecules 25, no. 18: 4083. https://doi.org/10.3390/molecules25184083
APA StyleTseng, T.-Y., Liu, S.-Y., Wang, C.-L., & Chang, T.-C. (2020). Antisense Oligonucleotides Used to Identify Telomeric G-Quadruplexes in Metaphase Chromosomes and Fixed Cells by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci. Molecules, 25(18), 4083. https://doi.org/10.3390/molecules25184083





