The Role of Active-Site Plasticity in Damaged-Nucleotide Recognition by Human Apurinic/Apyrimidinic Endonuclease APE1
Abstract
1. Introduction
2. Results and Discussion
2.1. Relative Cleavage Activity of APE1
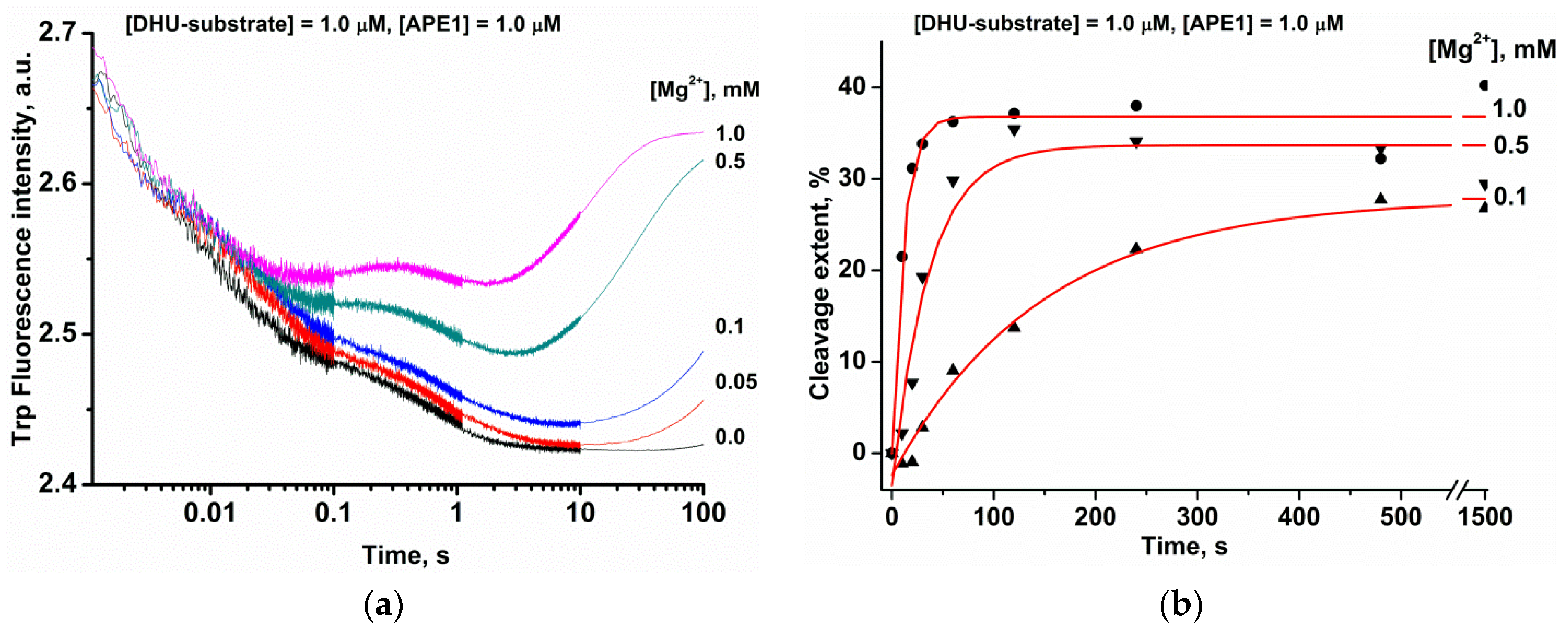
2.2. Effects of Mg2+ Ions on DNA Binding and Catalysis
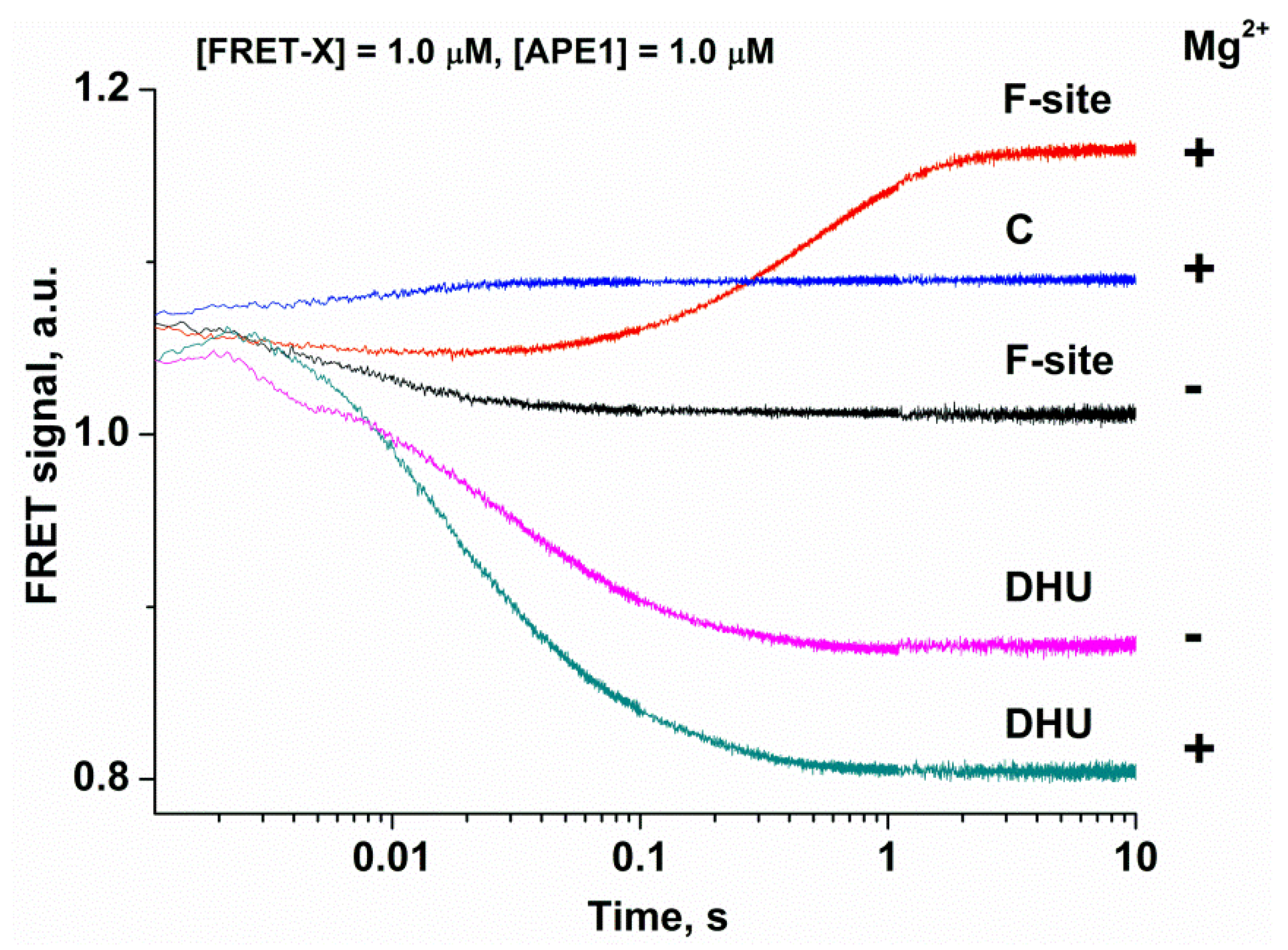
2.3. Analysis of DNA Conformational Changes in the Course of Interaction with APE1
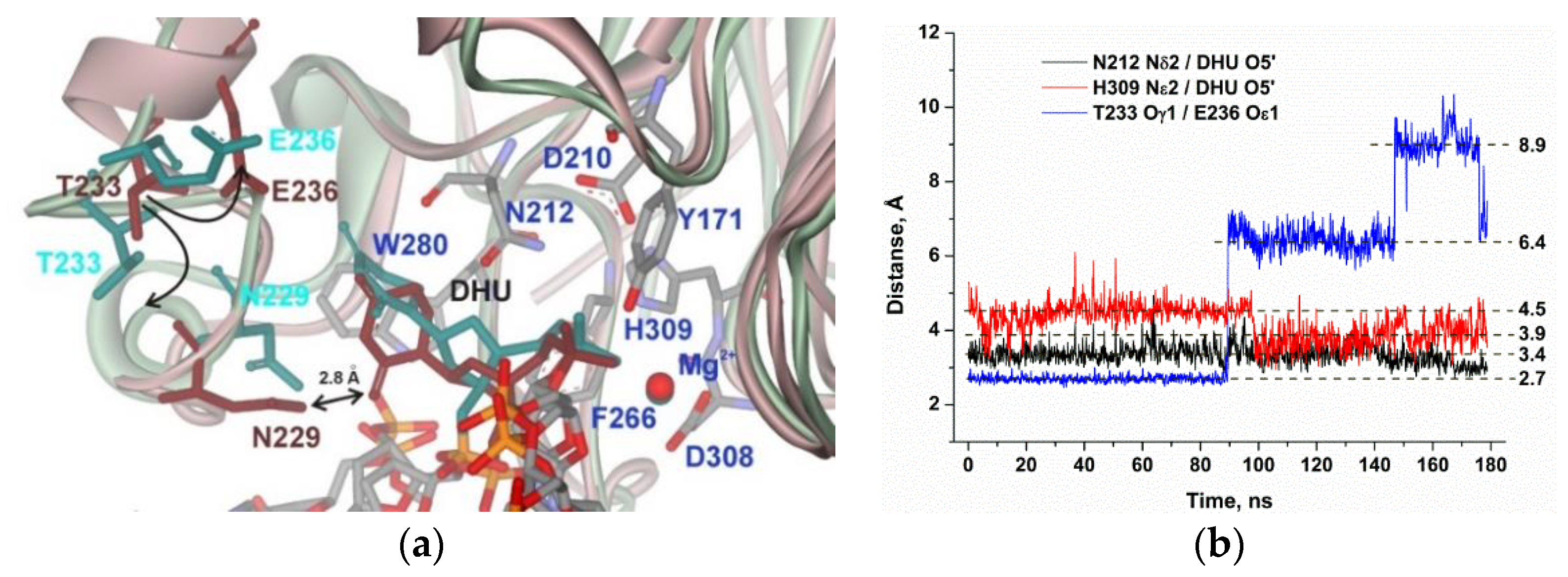
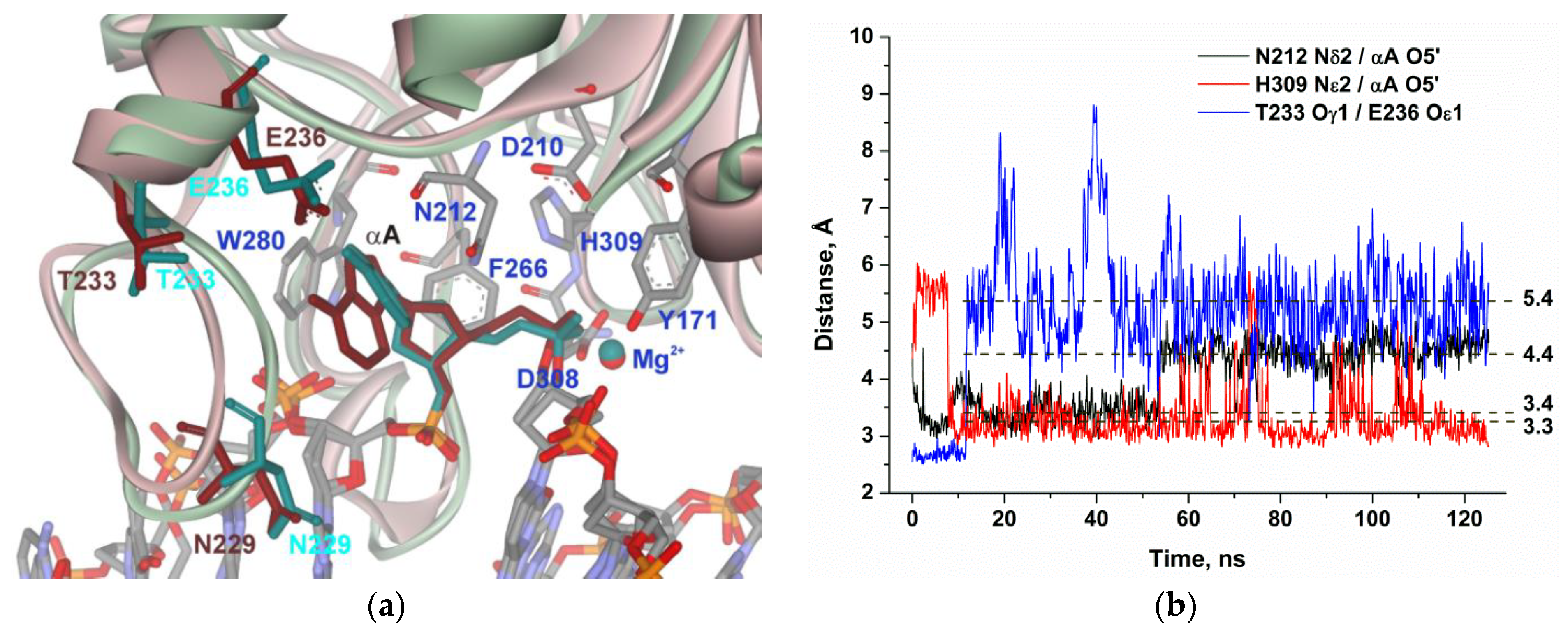
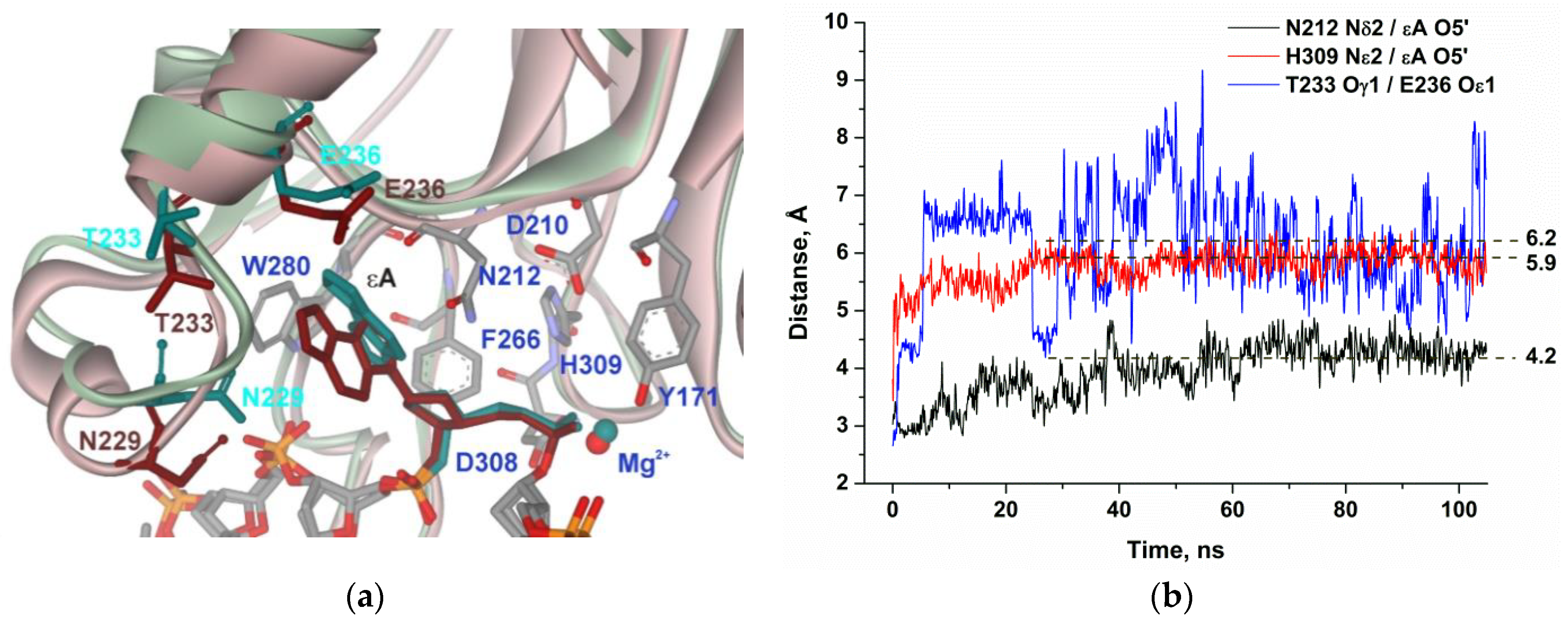
2.4. MD Simulations
3. Materials and Methods
3.1. Protein Expression and Purification
3.2. Oligodeoxynucleotides (ODNs)
3.3. PAGE (Polyacrylamide Gel Electrophoresis) and Enzymatic Analyses
3.4. Stopped-Flow Measurements with Fluorescence Detection
3.5. MD Simulations
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AP-site | apurinic/apyrimidinic site; |
| APE1 | human AP endonuclease; |
| ODN | oligodeoxyribonucleotide. |
References
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D.; Schultz, R.A.; Ellenberger, T. DNA Repair and Mutagenesis; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M., III; Barsky, D. The major human abasic endonuclease: Formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001, 485, 283–307. [Google Scholar] [CrossRef]
- Loeb, L.A.; Preston, B.D. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986, 20, 201–230. [Google Scholar] [CrossRef] [PubMed]
- Schaaper, R.M.; Kunkel, T.A.; Loeb, L.A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc. Natl. Acad. Sci. USA 1983, 80, 487–491. [Google Scholar] [CrossRef] [PubMed]
- David, S.S.; Williams, S.D. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 1998, 98, 1221–1261. [Google Scholar] [CrossRef] [PubMed]
- Mol, C.D.; Parikh, S.S.; Putnam, C.D.; Lo, T.P.; Tainer, J.A. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 101–128. [Google Scholar] [CrossRef]
- Demple, B.; Sung, J.-S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair 2005, 4, 1442–1449. [Google Scholar] [CrossRef]
- Daviet, S.; Couve-Privat, S.; Gros, L.; Shinozuka, K.; Ide, H.; Saparbaev, M.; Ishchenko, A.A. Major oxidative products of cytosine are substrates for the nucleotide incision repair pathway. DNA Repair 2007, 6, 8–18. [Google Scholar] [CrossRef]
- Vrouwe, M.G.; Pines, A.; Overmeer, R.M.; Hanada, K.; Mullenders, L.H. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 2011, 124, 435–446. [Google Scholar] [CrossRef]
- Guliaev, A.B.; Hang, B.; Singer, B. Structural insights by molecular dynamics simulations into specificity of the major human AP endonuclease toward the benzene-derived DNA adduct, pBQ-C. Nucleic Acids Res. 2004, 32, 2844–2852. [Google Scholar] [CrossRef]
- Prorok, P.; Saint-Pierre, C.; Gasparutto, D.; Fedorova, O.S.; Ishchenko, A.A.; Leh, H.; Buckle, M.; Tudek, B.; Saparbaev, M. Highly mutagenic exocyclic DNA adducts are substrates for the human nucleotide incision repair pathway. PLoS ONE 2012, 7, e51776. [Google Scholar] [CrossRef] [PubMed]
- Christov, P.P.; Banerjee, S.; Stone, M.P.; Rizzo, C.J. Selective Incision of the alpha-N-Methyl-Formamidopyrimidine Anomer by Escherichia coli Endonuclease IV. J. Nucleic Acids 2010, 2010, 850234. [Google Scholar] [CrossRef] [PubMed]
- Gros, L.; Ishchenko, A.A.; Ide, H.; Elder, R.H.; Saparbaev, M.K. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004, 32, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Prorok, P.; Alili, D.; Saint-Pierre, C.; Gasparutto, D.; Zharkov, D.O.; Ishchenko, A.A.; Tudek, B.; Saparbaev, M.K. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2013, 110, E3695–E3703. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wilson, D.M., 3rd. Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal. 2014, 20, 678–707. [Google Scholar] [CrossRef] [PubMed]
- Dyrkheeva, N.S.; Khodyreva, S.N.; Lavrik, O.I. Multifunctional human apurinic/apyrimidinic endonuclease 1: The role of additional functions. Mol. Biol. 2007, 41, 450–466. [Google Scholar] [CrossRef]
- Gorman, M.A.; Morera, S.; Rothwell, D.G.; de La Fortelle, E.; Mol, C.D.; Tainer, J.A.; Hickson, I.D.; Freemont, P.S. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997, 16, 6548–6558. [Google Scholar] [CrossRef] [PubMed]
- Beernink, P.T.; Segelke, B.W.; Hadi, M.Z.; Erzberger, J.P.; Wilson, D.M., 3rd; Rupp, B. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: Implications for the catalytic mechanism. J. Mol. Biol. 2001, 307, 1023–1034. [Google Scholar] [CrossRef]
- Manvilla, B.A.; Pozharski, E.; Toth, E.A.; Drohat, A.C. Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg2+ cofactor. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2555–2562. [Google Scholar] [CrossRef]
- Mol, C.D.; Izumi, T.; Mitra, S.; Tainer, J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature 2000, 403, 451–456. [Google Scholar] [CrossRef]
- Mol, C.D.; Hosfield, D.J.; Tainer, J.A. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: The 3′ ends justify the means. Mutat. Res. 2000, 460, 211–229. [Google Scholar] [CrossRef]
- Tsutakawa, S.E.; Shin, D.S.; Mol, C.D.; Izumi, T.; Arvai, A.S.; Mantha, A.K.; Szczesny, B.; Ivanov, I.N.; Hosfield, D.J.; Maiti, B.; et al. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Biol. Chem. 2013, 288, 8445–8455. [Google Scholar] [CrossRef] [PubMed]
- Freudenthal, B.D.; Beard, W.A.; Cuneo, M.J.; Dyrkheeva, N.S.; Wilson, S.H. Capturing snapshots of APE1 processing DNA damage. Nat. Struct. Mol. Biol. 2015, 22, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Matveeva, A.G.; Milov, A.D.; Vorobjev, Y.N.; Dzuba, S.A.; Fedorova, O.S.; Kuznetsov, N.A. Substrate specificity of human apurinic/apyrimidinic endonuclease APE1 in the nucleotide incision repair pathway. Nucleic Acids Res. 2018, 46, 11454–11465. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Kuznetsov, N.A.; Fedorova, O.S. Thermodynamics of Damaged DNA Binding and Catalysis by Human AP Endonuclease 1. Acta Nat. 2016, 8, 103–110. [Google Scholar] [CrossRef]
- Alekseeva, I.V.; Bakman, A.S.; Vorobjev, Y.N.; Fedorova, O.S.; Kuznetsov, N.A. Role of Ionizing Amino Acid Residues in the Process of DNA Binding by Human AP Endonuclease 1 and in Its Catalysis. J. Phys. Chem. B 2019, 123, 9546–9556. [Google Scholar] [CrossRef]
- Alekseeva, I.V.; Davletgildeeva, A.T.; Arkova, O.V.; Kuznetsov, N.A.; Fedorova, O.S. The impact of single-nucleotide polymorphisms of human apurinic/apyrimidinic endonuclease 1 on specific DNA binding and catalysis. Biochimie 2019, 163, 73–83. [Google Scholar] [CrossRef]
- Timofeyeva, N.A.; Koval, V.V.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Kinetic mechanism of human apurinic/apyrimidinic endonuclease action in nucleotide incision repair. Biochemistry 2011, 76, 273–281. [Google Scholar] [CrossRef]
- Timofeyeva, N.A.; Fedorova, O.S. A kinetic mechanism of repair of DNA containing alpha-anomeric deoxyadenosine by human apurinic/apyrimidinic endonuclease 1. Mol. Biosyst. 2016, 12, 3435–3446. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.A.; Fedorova, O.O.S.O.; Kuznetsov, N.N.A.N. Kinetic Features of 3′-5′ Exonuclease Activity of Human AP-Endonuclease APE1. Molecules 2018, 23, 2101. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Novopashina, D.S.; Fedorova, O.S.; Kuznetsov, N.A. Effect of the Substrate Structure and Metal Ions on the Hydrolysis of Undamaged RNA by Human AP Endonuclease APE1. Acta Nat. 2020, 2, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Timofeyeva, N.A.; Koval, V.V.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Lys98 substitution in human AP endonuclease 1 affects the kinetic mechanism of enzyme action in base excision and nucleotide incision repair pathways. PLoS ONE 2011, 6, e24063. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Vorobjev, Y.N.; Kuznetsov, N.A.; Fedorova, O.S. Effects of mono- and divalent metal ions on DNA binding and catalysis of human apurinic/apyrimidinic endonuclease 1. Mol. BioSyst. 2016, 12, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Gelin, A.; Redrejo-Rodríguez, M.; Laval, J.; Fedorova, O.S.; Saparbaev, M.; Ishchenko, A.A. Genetic and biochemical characterization of human AP endonuclease 1 mutants deficient in nucleotide incision repair activity. PLoS ONE 2010. [Google Scholar] [CrossRef] [PubMed]
- Timofeyeva, N.A.; Koval, V.V.; Knorre, D.G.; Zharkov, D.O.; Saparbaev, M.K.; Ishchenko, A.A.; Fedorova, O.S. Conformational dynamics of human AP endonuclease in base excision and nucleotide incision repair pathways. J. Biomol. Struct. Dyn. 2009, 26, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M., 3rd; Takeshita, M.; Grollman, A.P.; Demple, B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995, 270, 16002–16007. [Google Scholar] [CrossRef]
- Strauss, P.R.; Beard, W.A.; Patterson, T.A.; Wilson, S.H. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 1997, 272, 1302–1307. [Google Scholar] [CrossRef]
- Akishev, Z.; Taipakova, S.; Joldybayeva, B.; Zutterling, C.; Smekenov, I.; Ishchenko, A.A.; Zharkov, D.O.; Bissenbaev, A.K.; Saparbaev, M. The major Arabidopsis thaliana apurinic/apyrimidinic endonuclease, ARP is involved in the plant nucleotide incision repair pathway. DNA Repair 2016, 48, 30–42. [Google Scholar] [CrossRef]
- Kanazhevskaya, L.Y.; Koval, V.V.; Zharkov, D.O.; Strauss, P.R.; Fedorova, O.S. Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair. Biochemistry 2010, 49, 6451–6461. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Pre-steady-state fluorescence analysis of damaged DNA transfer from human DNA glycosylases to AP endonuclease APE1. Biochim. Biophys. Acta 2014, 1840, 3042–3051. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Step-by-Step Mechanism of DNA Damage Recognition by Human 8-Oxoguanine DNA Glycosylase. Biochim. Biophys. Acta 2014, 1840, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Vorobjev, Y.N.; Barthes, N.P.F.; Michel, B.Y.; Burger, A.; Fedorova, O.S. New Environment-Sensitive Multichannel DNA Fluorescent Label for Investigation of the Protein-DNA Interactions. PLoS ONE 2014, 9, e100007. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Roy, S.; Thakur, A.R. 20ns molecular dynamics simulation of the antennapedia homeodomain-dna complex: Water interaction and dna structure analysis. J. Biomol. Struct. Dyn. 2010, 27, 443–455. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


| Shorthand | Sequences of DNA Duplexes |
|---|---|
| X-substrate X = F-site, DHU, εA, or αA | 5′-TCTCTCXCCTTCC-3′ 3′-AGAGAGGGGAAGG-5′ |
| FRET-X-substrate Y = F-site, DHU, εA, αA, or C | 5′-FAM-GCTCAXGTACAGAGCTG-3′ 3′-CGAGTGCATGTCTCGAC-BHQ1-5′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulygin, A.A.; Kuznetsova, A.A.; Vorobjev, Y.N.; Fedorova, O.S.; A. Kuznetsov, N. The Role of Active-Site Plasticity in Damaged-Nucleotide Recognition by Human Apurinic/Apyrimidinic Endonuclease APE1. Molecules 2020, 25, 3940. https://doi.org/10.3390/molecules25173940
Bulygin AA, Kuznetsova AA, Vorobjev YN, Fedorova OS, A. Kuznetsov N. The Role of Active-Site Plasticity in Damaged-Nucleotide Recognition by Human Apurinic/Apyrimidinic Endonuclease APE1. Molecules. 2020; 25(17):3940. https://doi.org/10.3390/molecules25173940
Chicago/Turabian StyleBulygin, Anatoly A., Alexandra A. Kuznetsova, Yuri N. Vorobjev, Olga S. Fedorova, and Nikita A. Kuznetsov. 2020. "The Role of Active-Site Plasticity in Damaged-Nucleotide Recognition by Human Apurinic/Apyrimidinic Endonuclease APE1" Molecules 25, no. 17: 3940. https://doi.org/10.3390/molecules25173940
APA StyleBulygin, A. A., Kuznetsova, A. A., Vorobjev, Y. N., Fedorova, O. S., & A. Kuznetsov, N. (2020). The Role of Active-Site Plasticity in Damaged-Nucleotide Recognition by Human Apurinic/Apyrimidinic Endonuclease APE1. Molecules, 25(17), 3940. https://doi.org/10.3390/molecules25173940







