Deletion of the Histone Deacetylase HdaA in Endophytic Fungus Penicillium chrysogenum Fes1701 Induces the Complex Response of Multiple Bioactive Secondary Metabolite Production and Relevant Gene Cluster Expression
Abstract
1. Introduction
2. Results and Discussion
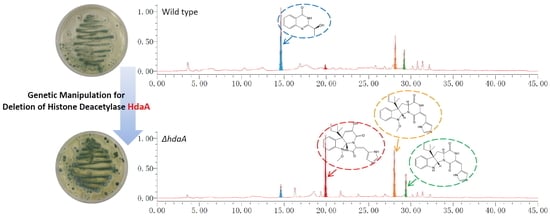
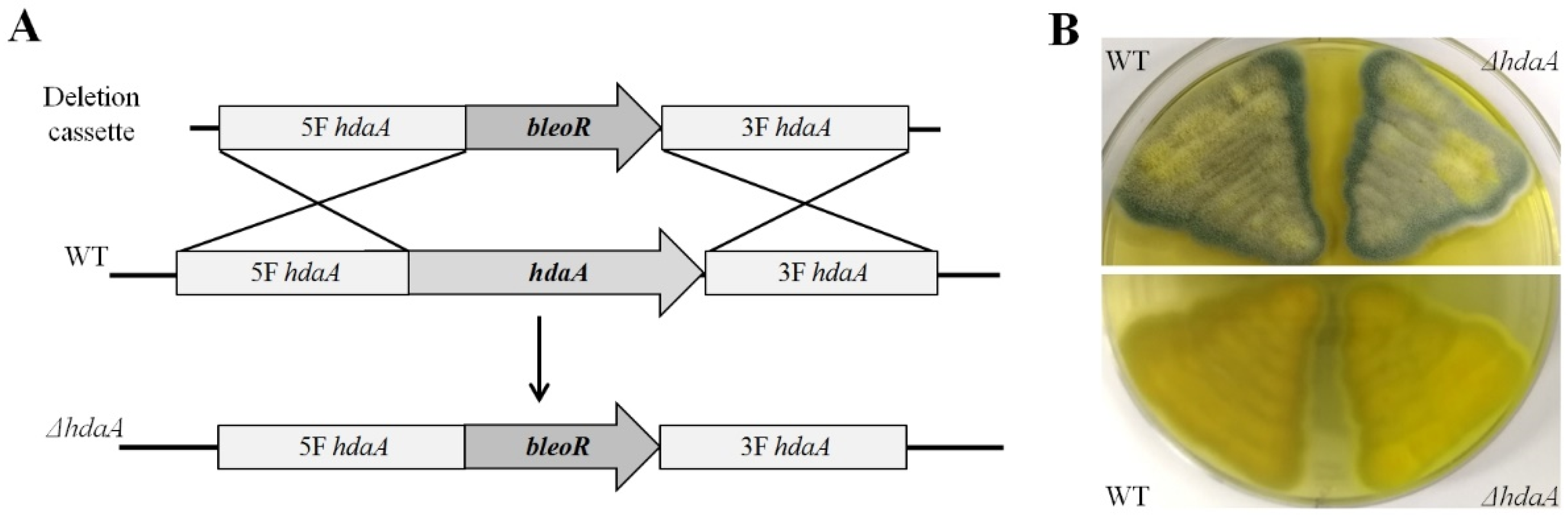
2.1. Identification and Deletion of the hdaA Gene in P. chrysogenum Fes1701
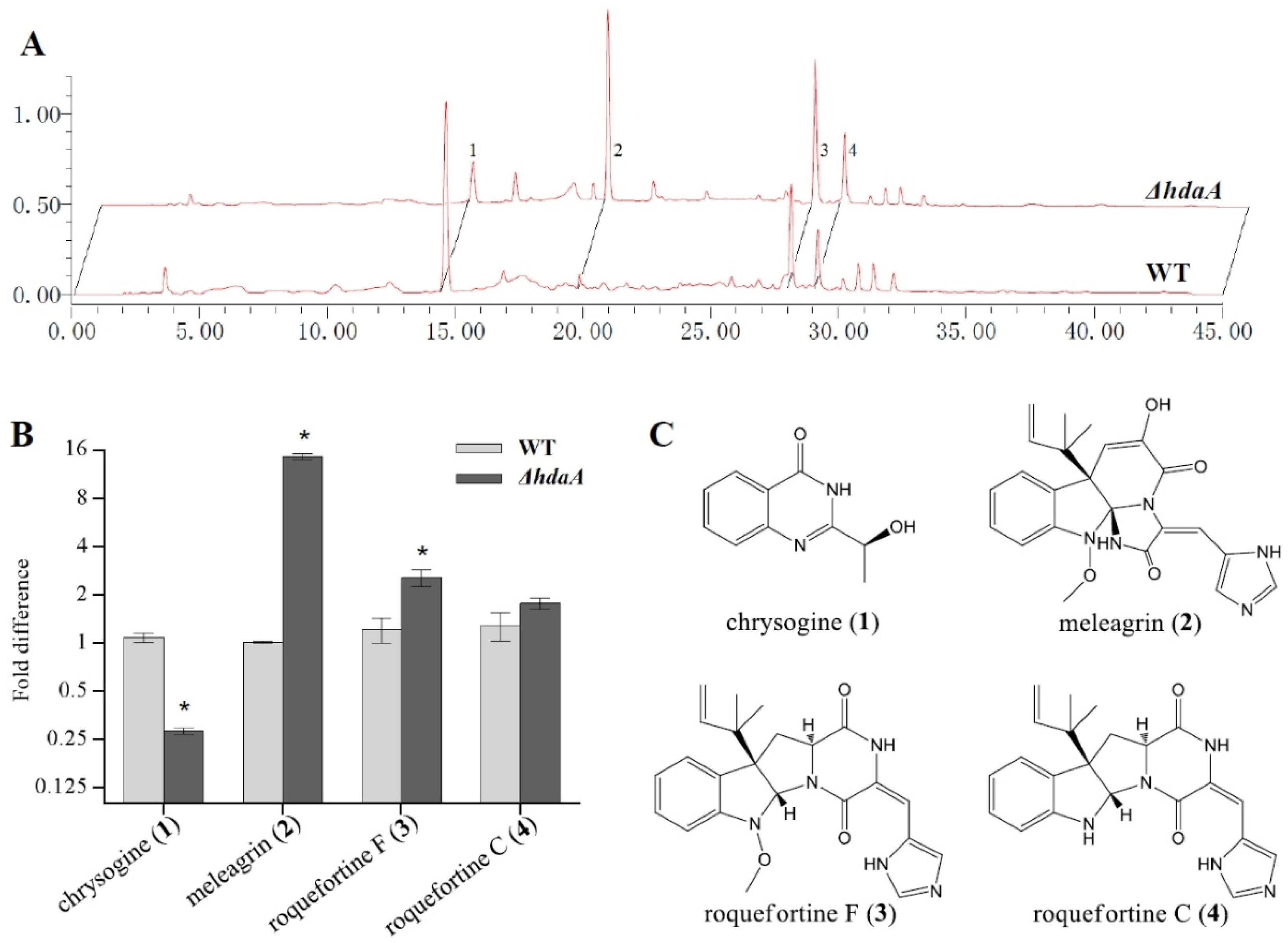
2.2. Effects of hdaA Disruption on SMs Production
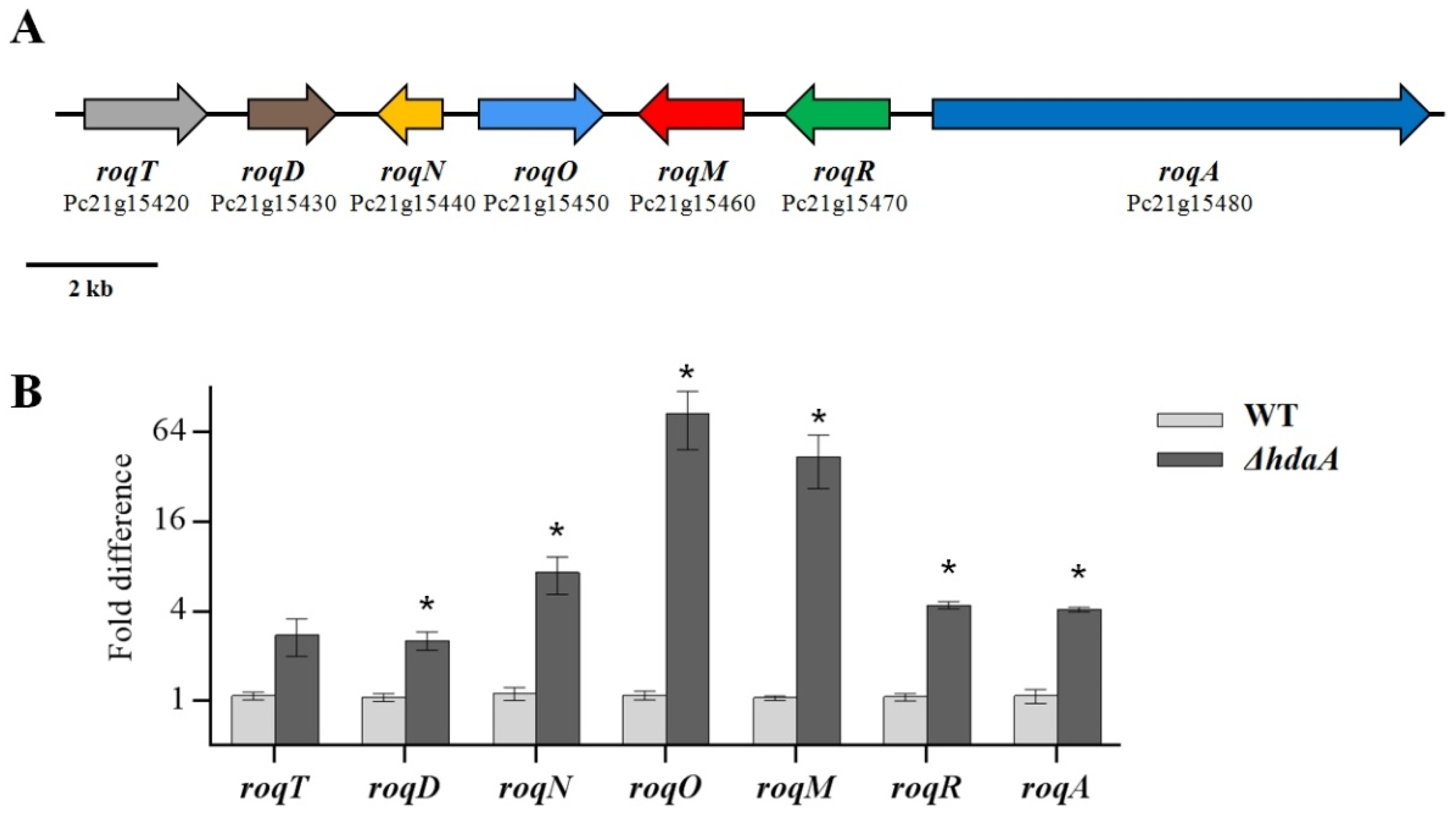
2.3. Effects of hdaA Disruption on the Transcription of SM Biosynthetic Gene Clusters
2.4. Bioactivities of the Metabolites Isolated from the ΔhdaA Strain
3. Materials and Methods
3.1. Strains and Media
3.2. Cloning and Identification of the hdaA Gene in Strain Fes1701
3.3. Creation of the ΔhdaA Strain
3.4. Fermentation Conditions and SM Extraction
3.5. Metabolite Fingerprint Analysis
3.6. Purification and Identification of Natural Products
3.7. RNA Extraction and Real-Time PCR Analysis
3.8. Bioactivity Assay
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat. Prod. Rep. 2007, 24, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Keller, N.P. Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 2014, 41, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.F.; Mao, N.; Xue, X.J.; Qi, Y.X.; Wei, M.Y.; Wang, C.Y.; Shao, C.L. Structures and absolute configurations of diketopiperazine alkaloids chrysopiperazines A–C from the gorgonian-derived Penicillium chrysogenum Fungus. Mar. Drugs 2019, 17, 250. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.L.; Yuan, X.L.; Du, Y.M.; Zhang, Z.F.; Zhang, P. Benzophenone derivatives from an algal-endophytic isolate of Penicillium chrysogenum and their cytotoxicity. Molecules 2018, 23, 3378. [Google Scholar] [CrossRef]
- Qi, B.; Liu, X.; Mo, T.; Zhu, Z.; Li, J.; Wang, J.; Shi, X.; Zeng, K.; Wang, X.; Tu, P.; et al. 3,5-dimethylorsellinic acid derived meroterpenoids from Penicillium chrysogenum MT-12, an endophytic fungus isolated from Huperzia serrata. J. Nat. Prod. 2017, 80, 2699–2707. [Google Scholar] [CrossRef]
- Van den Berg, M.A.; Albang, R.; Albermann, K.; Badger, J.H.; Daran, J.M.; Driessen, A.J.; Garcia-Estrada, C.; Fedorova, N.D.; Harris, D.M.; Heijne, W.H.; et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef]
- Kozlovskii, A.G.; Antipova, T.V.; Zhelifonova, V.P. Biosynthesis of biologically active low-molecular weight compound by fungi of the genus Penicillium (Review). Appl. Biochem. Microbiol. 2015, 51, 236–242. [Google Scholar] [CrossRef]
- Domínguez-Santos, R.; Martín, J.F.; Kosalková, K.; Prieto, C.; Ullán, R.V.; García-Estrada, C. The regulatory factor PcRFX1 controls the expression of the three genes of b-lactam biosynthesis in Penicillium chrysogenum. Fungal Genet. Biol. 2012, 49, 866–881. [Google Scholar] [CrossRef]
- Domínguez-Santos, R.; García-Estrada, C.; Kosalková, K.; Prieto, C.; Santamarta, I.; Martín, J.F. PcFKH1, a novel regulatory factor from the forkhead family, controls the biosynthesis of penicillin in Penicillium chrysogenum. Biochimie 2015, 115, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Kosalková, K.; García-Estrada, C.; Ullán, R.V.; Godio, R.P.; Feltrer, R.; Teijeira, F.; Mauriz, E.; Martín, J.F. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 2009, 91, 214–225. [Google Scholar] [CrossRef]
- Cepeda-García, C.; Domínguez-Santos, R.; García-Rico, R.O.; García-Estrada, C.; Cajiao, A.; Fierro, F.; Martín, J.F. Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2014, 98, 7113–7124. [Google Scholar] [CrossRef] [PubMed]
- Cichewicz, R.H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 2010, 27, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Robyr, D.; Suka, Y.; Xenarios, I.; Kurdistani, S.K.; Wang, A.; Suka, N.; Grunstein, M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 2002, 109, 437–446. [Google Scholar] [CrossRef]
- Bulger, M. Hyperacetylated chromatin domains: Lessons from heterochromatin. J. Biol. Chem. 2005, 280, 21689–21692. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef]
- Lee, I.; Oh, J.H.; Shwab, E.K.; Dagenais, T.R.; Andes, D.; Keller, N.P. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. 2009, 46, 782–790. [Google Scholar] [CrossRef]
- Mao, X.M.; Xu, W.; Li, D.; Yin, W.B.; Chooi, Y.H.; Li, Y.Q.; Tang, Y.; Hu, Y. Epigenetic genome mining of an endophytic fungus leads to the pleiotropic biosynthesis of natural products. Angew. Chem. Int. Ed. 2015, 54, 7592–7596. [Google Scholar] [CrossRef]
- Wu, G.W.; Zhou, H.C.; Zheng, P.; Wang, X.; Li, W.; Zhang, W.; Liu, X.; Liu, H.W.; Keller, N.P.; An, Z.; et al. Polyketide production of pestaloficiols and macrodiolide ficiolides revealed by manipulation of epigenetic regulators in an endophytic fungus. Org. Lett. 2016, 18, 1832–1835. [Google Scholar] [CrossRef]
- Niu, X.; Hao, X.; Hong, Z.; Chen, L.; Yu, X.; Zhu, X. A putative histone deacetylase modulates the biosynthesis of pestalotiollide B and conidiation in Pestalotiopsis microspora. J. Microbiol. Biotechnol. 2015, 25, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Stierle, A.; Strobel, G.A.; Stierle, D.B. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef]
- Strobel, G.A.; Miller, R.V.; Miller, C.; Condron, M.; Teplow, D.B.; Hess, W.M. Cryptocandin, a potent antimycotic from the endophytic fungus Cryptosporiopsis cf. quercina. Microbiology 1999, 145, 1919–1926. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, F.; Zhang, M.; Ou, T.; Wang, R.; Strobel, G.; Xiang, Z.; Zhou, Z.; Xie, J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019, 229, 126328. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Tao, T.; Wang, L.; Zhao, Y.; Huang, H.; Zhang, D.; Liu, M.; Wang, Z.; Han, J. Bioprospecting of novel and bioactive metabolites from endophytic fungi isolated from rubber tree Ficus elastica leaves. J. Microbiol. Biotechnol. 2019, 29, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Tribus, M.; Galehr, J.; Trojer, P.; Brosch, G.; Loidl, P.; Marx, F.; Haas, H.; Graessle, S. HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot Cell 2005, 4, 1736–1745. [Google Scholar] [CrossRef] [PubMed]
- Niederer, D.; Tamm, C.; Zurcher, W. Nitrogen containing metabolites of Fusarium sambucinum. Tetrahedron Lett. 1992, 33, 3997–4000. [Google Scholar] [CrossRef]
- Kawai, K.; Nozawa, K.; Nakajima, S.; Iitaka, Y. Studies on fungal products. VII. The structures of meleagrin and 9-O-p-bromobenzoylmeleagrin. Chem. Pharm. Bull. 1984, 32, 94–98. [Google Scholar] [CrossRef]
- Musuku, A.; Selala, M.I.; de Bruyne, T.; Claeys, M.; Schepens, P.J.C.; Tsatsakis, A.; Shtilman, M.I. Isolation and structure determination of a new roquefortine-related mycotoxin from penicillium verrucosum var. cyclopium isolated from cassava. J. Nat. Prod. 1994, 57, 983–987. [Google Scholar] [CrossRef]
- Du, L.; Li, D.; Zhu, T.; Cai, S.; Wang, F.; Xiao, X.; Gu, Q. New alkaloids and diterpenes from a deep ocean sediment derived fungus Penicillium sp. Tetrahedron 2009, 65, 1033–1039. [Google Scholar] [CrossRef]
- Studt, L.; Schmidt, F.J.; Jahn, L.; Sieber, C.M.; Connolly, L.R.; Niehaus, E.M.; Freitag, M.; Humpf, H.U.; Tudzynski, B. Two histone deacetylases, FfHda1 and FfHda2, are important for Fusarium fujikuroi secondary metabolism and virulence. Appl. Environ. Microb. 2013, 79, 7719–7734. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg, R.D.; Saei, W.; Westphal, K.R.; Klitgaard, C.S.; Nielsen, K.L.; Lysøe, E.; Gardiner, D.M.; Wimmer, R.; Sondergaard, T.E.; Sørensen, J.L. Chrysogine biosynthesis is mediated by a two-module nonribosomal peptide synthetase. J. Nat. Prod. 2017, 80, 2131–2135. [Google Scholar] [CrossRef] [PubMed]
- Viggiano, A.; Salo, O.; Ali, H.; Szymanski, W.; Lankhorst, P.P.; Nygård, Y.; Bovenberg, R.A.L.; Driessen, A.J.M. Pathway for the biosynthesis of the pigment chrysogine by penicillium chrysogenum. Appl. Environ. Microbiol. 2018, 84, e02246-17. [Google Scholar] [CrossRef]
- Guzman-Chavez, F.; Salo, O.; Samol, M.; Ries, M.; Kuipers, J.; Bovenberg, R.A.L.; Vreeken, R.J.; Driessen, A.J.M. Deregulation of secondary metabolism in a histone deacetylase mutant of Penicillium chrysogenum. MicrobiologyOpen 2018, 7, e00598. [Google Scholar] [CrossRef]
- García-Estrada, C.; Ullán, R.V.; Albillos, S.M.; Fernández-Bodega, M.Á.; Durek, P.; von Döhren, H.; Martín, J.F. A single cluster of coregulated genes encodes the biosynthesis of the mycotoxins roquefortine C and meleagrin in Penicillium chrysogenum. Chem. Biol. 2011, 18, 1499–1512. [Google Scholar] [CrossRef] [PubMed]
- García-Rico, R.O.; Fierro, F.; Mauriz, E.; Gómez, A.; Fernández-Bodega, M.A.; Martín, J.F. The heterotrimeric Galpha protein pga1 regulates biosynthesis of penicillin, chrysogenin and roquefortine in Penicillium chrysogenum. Microbiology 2008, 154, 3567–3578. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Feng, T.; Zhao, B.; Li, D.; Cai, S.; Zhu, T.; Wang, F.; Xiao, X.; Gu, Q. Alkaloids from a deep ocean sediment-derived fungus Penicillium sp. and their antitumor activities. J. Antibiot. 2010, 63, 165–170. [Google Scholar] [CrossRef]
- Mady, M.S.; Mohyeldin, M.M.; Ebrahim, H.Y.; Elsayed, H.E.; Houssen, W.E.; Haggag, E.G.; Soliman, R.F.; EI Sayed, K.A. The indole alkaloid meleagrin, from the olive tree endophytic fungus Penicillium chrysogenum, as a novel lead for the control of c-Met-dependent breast cancer proliferation, migration and invasion. Bioorgan. Med. Chem. 2016, 24, 113–122. [Google Scholar] [CrossRef]
- Kopp-Holtwiesche, B.; Rehm, H.J. Antimicrobial action of roquefortine. J. Environ. Pathol. Toxicol. Oncol. 1990, 10, 41–44. [Google Scholar]
- Clark, B.; Capon, R.J.; Lacey, E.; Tennant, S.; Gill, J.H. Roquefortine E, a diketopiperazine from an Australian isolate of Gymnoascus reessii. J. Nat. Prod. 2005, 68, 1661–1664. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.C.; Cui, X.; He, X.; Ding, Z.; Zhu, T.; Tang, Y.; Li, D. Late-Stage terpene cyclization by an integral membrane cyclase in the biosynthesis of isoprenoid epoxycyclohexenone natural products. Org. Lett. 2017, 19, 5376–5379. [Google Scholar] [CrossRef] [PubMed]
- Zilla, M.K.; Qadri, M.; Pathania, A.S.; Strobel, G.A.; Nalli, Y.; Kumar, S.; Guru, S.K.; Bhushan, S.; Singh, S.K.; Vishwakarma, R.A.; et al. Bioactive metabolites from an endophytic Cryptosporiopsis sp. inhabiting Clidemia hirta. Phytochemistry 2013, 95, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.Z.; Li, X.M.; Li, X.; Yang, S.Q.; Meng, L.H.; Wang, B.G. Polyketides from the mangrove-derived endophytic fungus Cladosporium cladosporioides. Mar. Drugs 2019, 17, 296. [Google Scholar] [CrossRef]
- Strobel, G.A. Endophytes as a source of bioactive products. Microbes Infect. 2003, 6, 535–544. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–4 are available from the authors. |


| Compound | Antimicrobial Activity (MIC, μg/mL) | Cytotoxicity (IC50, μM) | ||||
|---|---|---|---|---|---|---|
| E. coli | S. aureus | C. albicans | C. glabrata | K562 | HL-60 | |
| Meleagrin | 128 | 32 | >128 | >128 | 8.9 | 12.7 |
| Roquefortine C | 64 | 16 | >128 | >128 | 27.4 | 28.1 |
| Roquefortine F | 64 | 16 | >128 | >128 | 22.7 | 25.1 |
| Chloramphenicol | 2 | 1 | n.t. | n.t. | n.t. | n.t. |
| Fluconazole | n.t. | n.t. | 1 | 1 | n.t. | n.t. |
| Adriamycin | n.t. | n.t. | n.t. | n.t. | 0.3 | 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Z.; Zhou, H.; Wang, X.; Huang, H.; Wang, H.; Zhang, R.; Wang, Z.; Han, J. Deletion of the Histone Deacetylase HdaA in Endophytic Fungus Penicillium chrysogenum Fes1701 Induces the Complex Response of Multiple Bioactive Secondary Metabolite Production and Relevant Gene Cluster Expression. Molecules 2020, 25, 3657. https://doi.org/10.3390/molecules25163657
Ding Z, Zhou H, Wang X, Huang H, Wang H, Zhang R, Wang Z, Han J. Deletion of the Histone Deacetylase HdaA in Endophytic Fungus Penicillium chrysogenum Fes1701 Induces the Complex Response of Multiple Bioactive Secondary Metabolite Production and Relevant Gene Cluster Expression. Molecules. 2020; 25(16):3657. https://doi.org/10.3390/molecules25163657
Chicago/Turabian StyleDing, Zhuang, Haibo Zhou, Xiao Wang, Huiming Huang, Haotian Wang, Ruiyan Zhang, Zhengping Wang, and Jun Han. 2020. "Deletion of the Histone Deacetylase HdaA in Endophytic Fungus Penicillium chrysogenum Fes1701 Induces the Complex Response of Multiple Bioactive Secondary Metabolite Production and Relevant Gene Cluster Expression" Molecules 25, no. 16: 3657. https://doi.org/10.3390/molecules25163657
APA StyleDing, Z., Zhou, H., Wang, X., Huang, H., Wang, H., Zhang, R., Wang, Z., & Han, J. (2020). Deletion of the Histone Deacetylase HdaA in Endophytic Fungus Penicillium chrysogenum Fes1701 Induces the Complex Response of Multiple Bioactive Secondary Metabolite Production and Relevant Gene Cluster Expression. Molecules, 25(16), 3657. https://doi.org/10.3390/molecules25163657